Overexpression of the Golden SNP-Carrying Orange Gene Enhances Carotenoid Accumulation and Heat Stress Tolerance in Sweetpotato Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plasmid Construction and Agrobacterium-Mediated Transformation
2.3. Generation of Transgenic Sweetpotato Plants
2.4. RNA Extraction and qRT-PCR Analysis of Carotenogenesis-Related Genes
2.5. Analysis of Carotenoid Contents
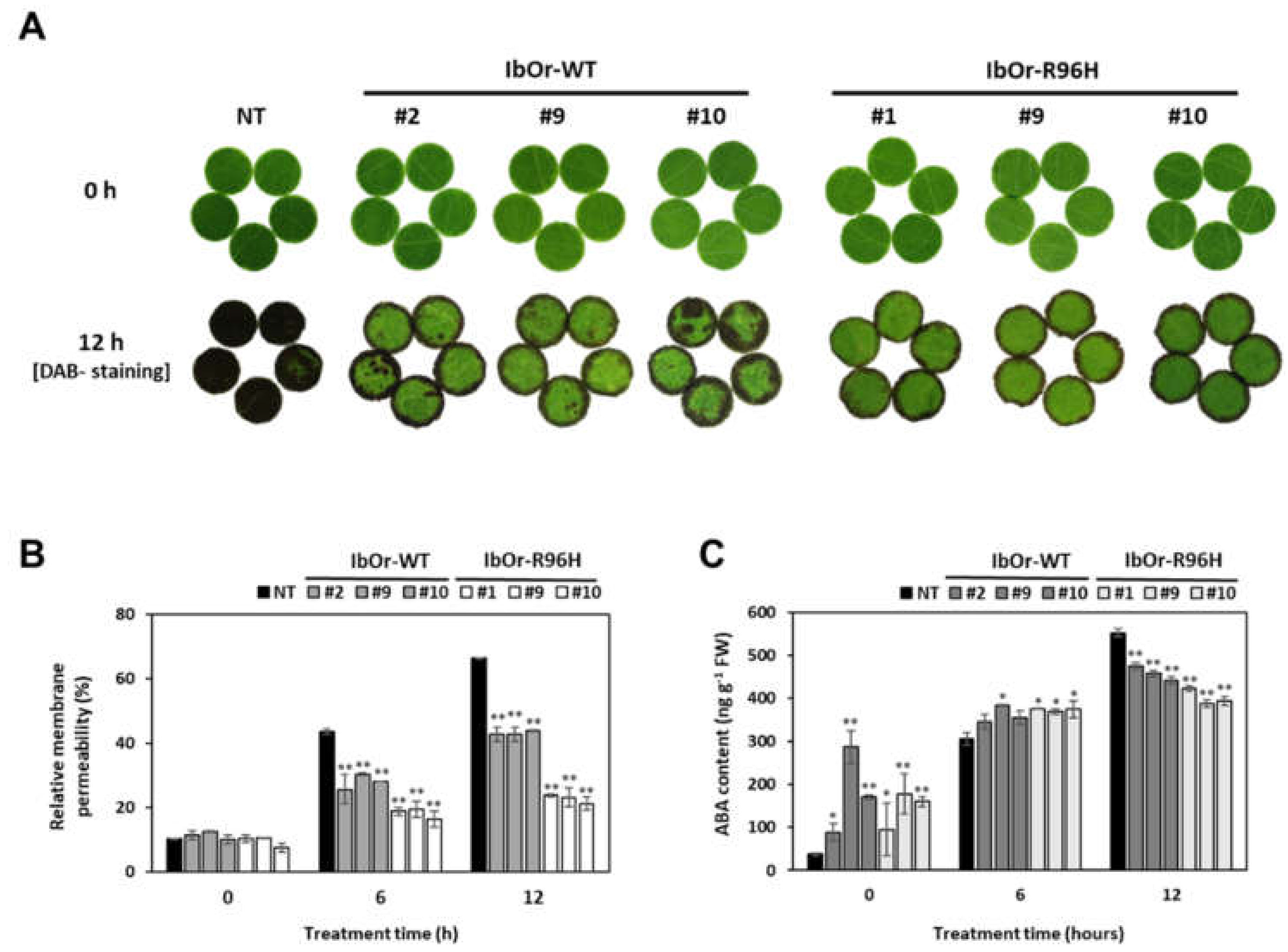
2.6. Analysis of Heat Tolerance
2.7. Determination of ABA Content
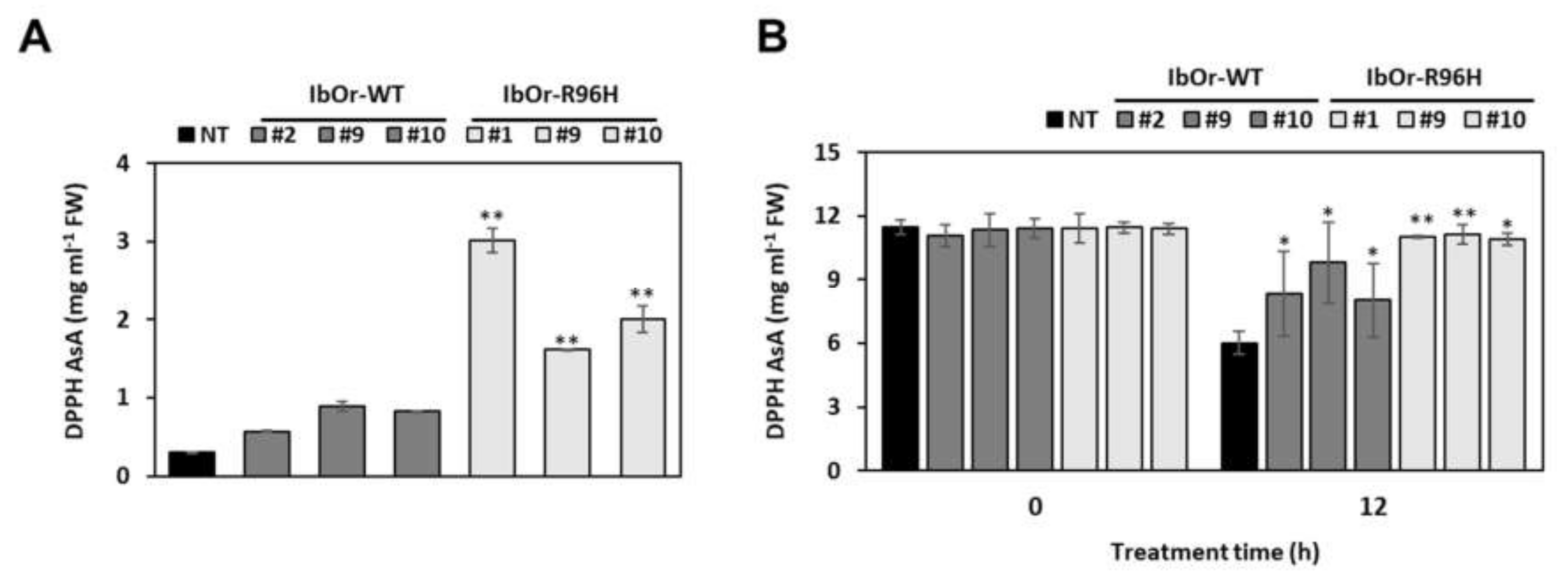
2.8. Antioxidant Activity Analysis
2.9. Statistical Analysis
3. Results
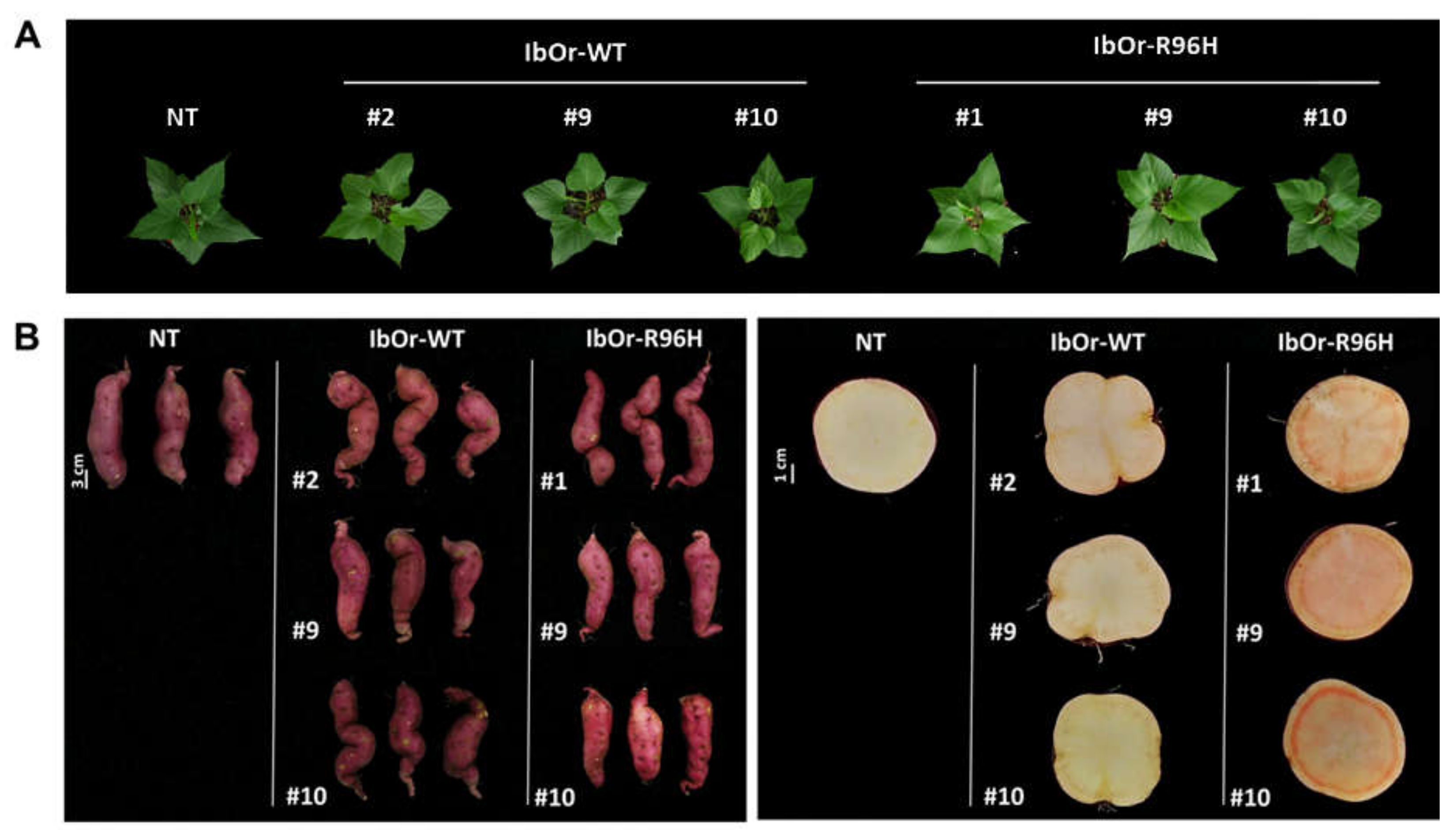
3.1. IbOr-R96H Transgenic Sweetpotato Plants Exhibited a Light-Orange Color
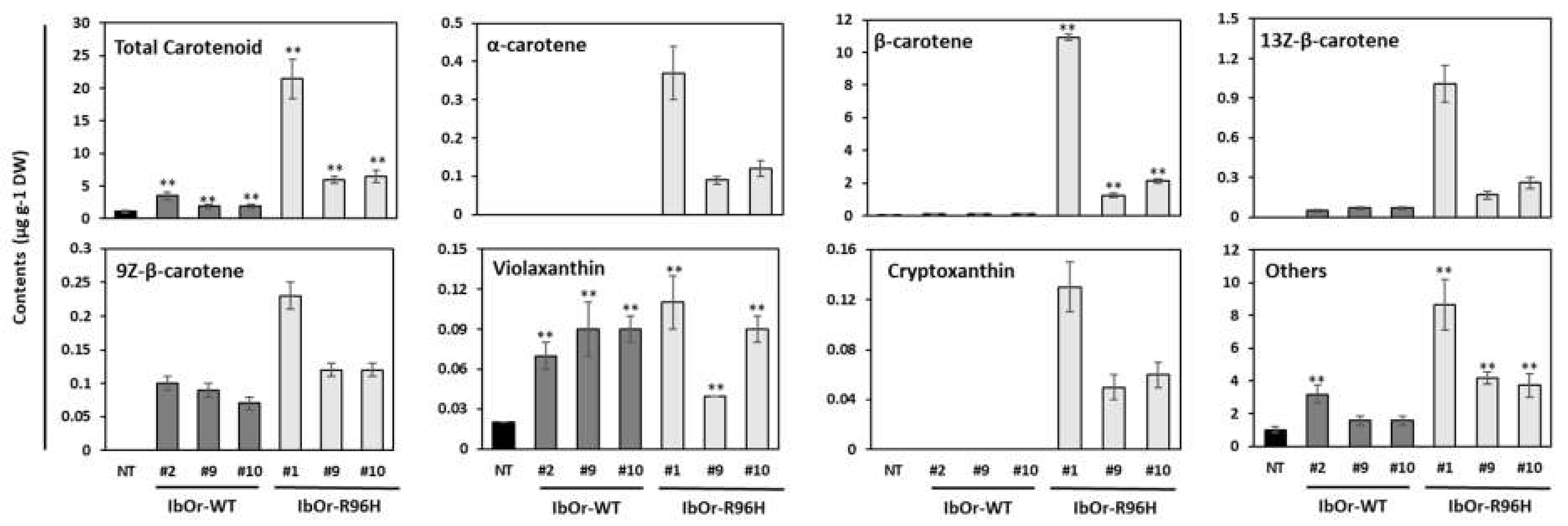
3.2. IbOr-R96H Overexpression Induces Overaccumulation of Carotenoids in Storage Roots
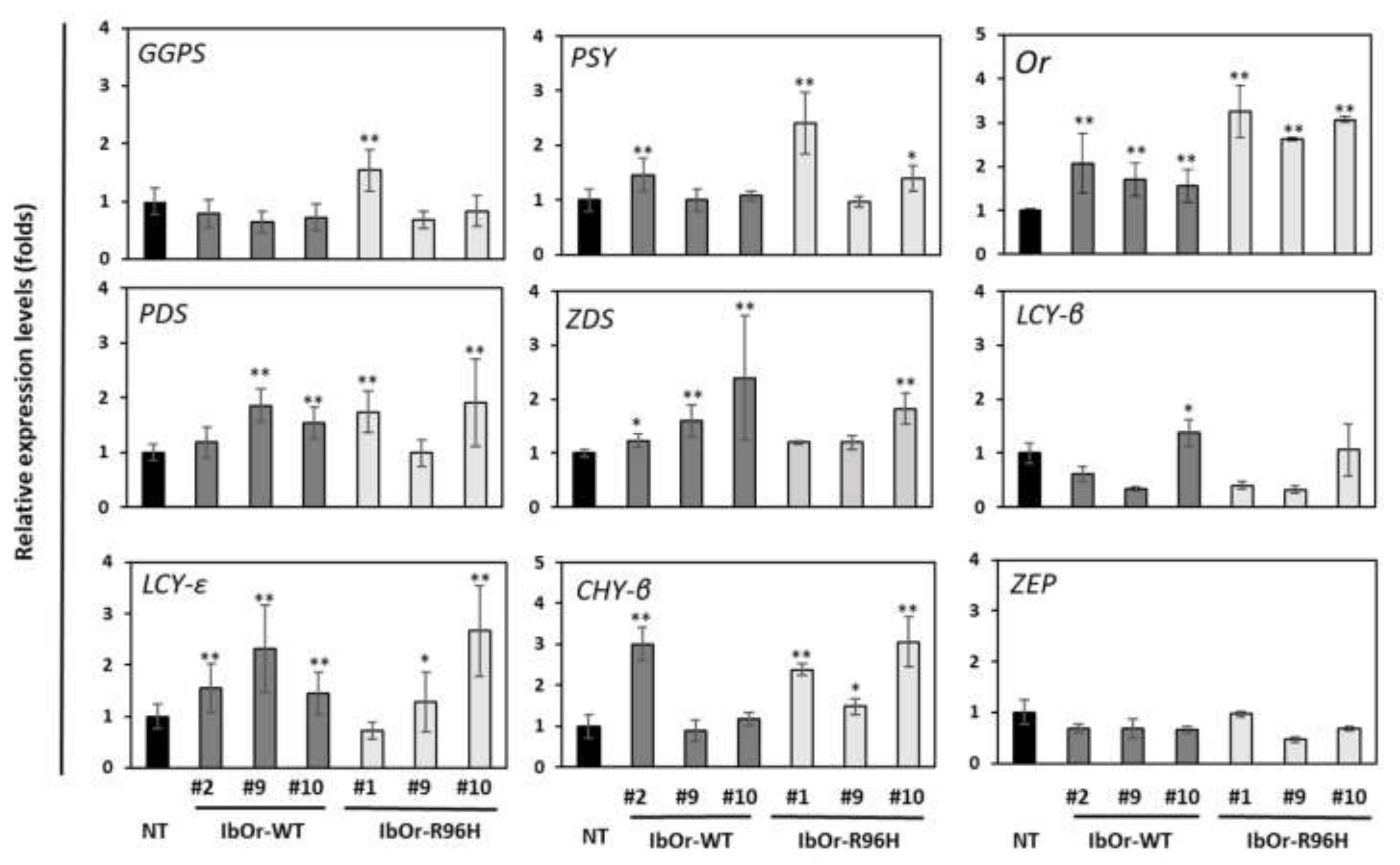
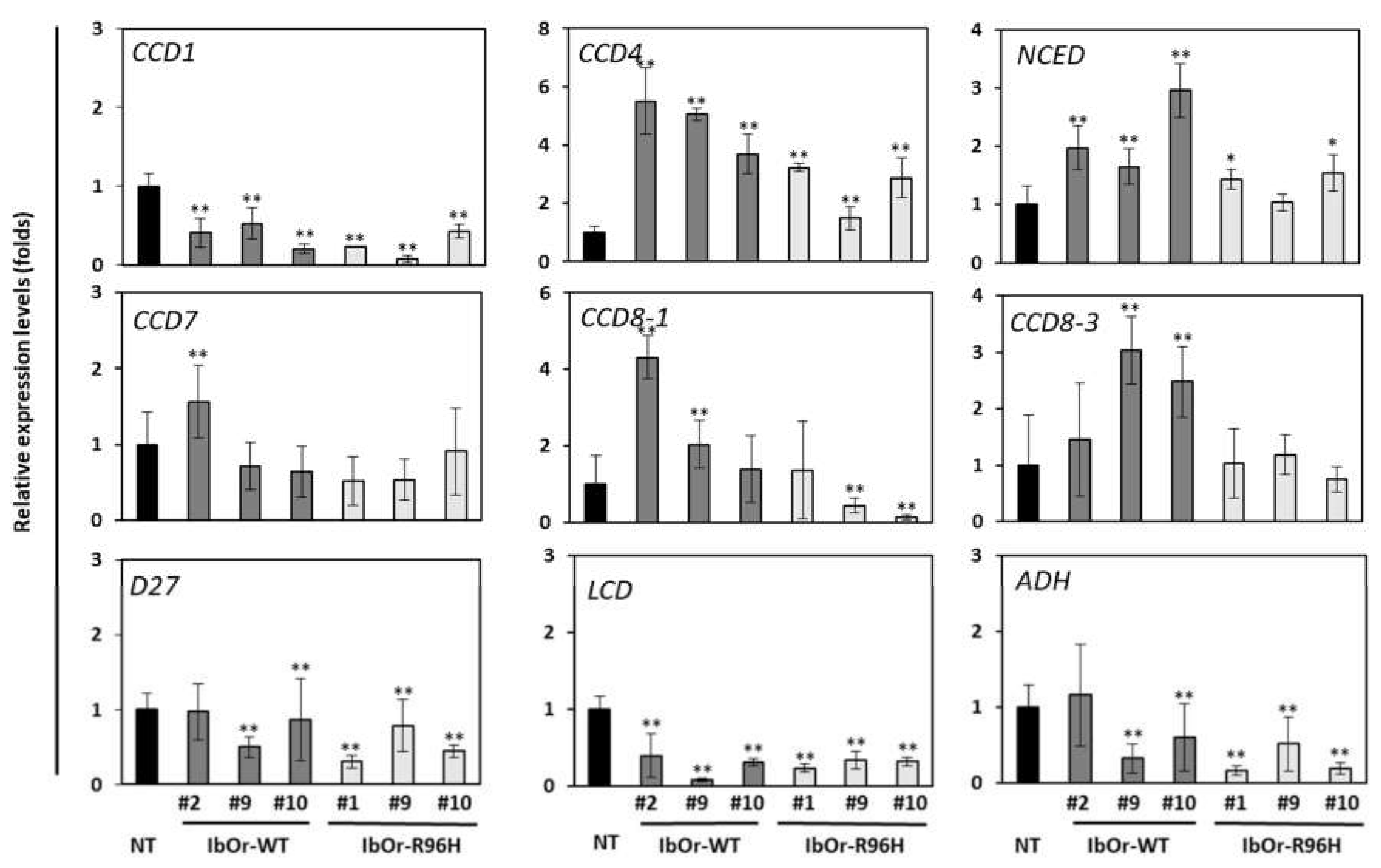
3.3. IbOr-R96H Overexpression Significantly Altered the Expression of Carotenoid Metabolism-Related Genes
3.4. IbOr-R96H Transgenic Plants Exhibit Increased Antioxidant Activity and Heat Stress Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yabuzaki, J. Carotenoids database: Structures, chemical fingerprints and distribution among organisms. Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed]
- Watkins, J.L.; Pogson, B.J. Prospects for carotenoid biofortification targeting retention and catabolism. Trends Plant Sci. 2020, 25, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Giuliano, G.; Al-Babili, S. Carotenoid biofortification in crop plants: Citius, altius, fortius. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158664. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Uragami, C.; Cogdell, R.J. Carotenoids and photosynthesis. Subcell. Biochem. 2016, 79, 111–139. [Google Scholar]
- Black, R.E.; Allen, L.H.; A Bhutta, Z.; E Caulfield, L.; De Onis, M.; Ezzati, M.; Mathers, C.; Rivera, J. Maternal and child undernutrition: Global and regional exposures and health consequences. Lancet 2008, 371, 243–260. [Google Scholar] [CrossRef]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef]
- Giuliano, G. Provitamin A biofortification of crop plants: A gold rush with many miners. Curr. Opin. Biotechnol. 2017, 44, 169–180. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Bovell-Benjamin, A.C. Sweetpotato: A review of its past, present, and future role in human nutrition. Adv. Food Nutr. Res. 2007, 52, 1–59. [Google Scholar]
- Kwak, S.S. Biotechnology of sweetpotato: Ensuring the food and nutrition security in the face of climate change. Plant Cell Rep. 2019, 38, 1361–1363. [Google Scholar] [CrossRef] [PubMed]
- Paine, J.A.; Shipton, A.C.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Park, S.C.; Ji, C.Y.; Kim, H.S.; Lee, H.S.; Kwak, S.S. Metabolic engineering of carotenoids in transgenic sweetpotato. Breed. Sci. 2017, 67, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Wang, W.; Kang, L.; Kim, S.E.; Lee, C.J.; Park, S.C.; Park, W.S.; Ahn, M.J.; Kwak, S.S. Metabolic engineering of low molecular weight antioxidants in sweetpotao. Plant Biotechnol. Rep. 2020, 14, 193–205. [Google Scholar] [CrossRef]
- Mohanraj, R.; Sivasankar, S. Sweet potato (Ipomoea batatas [L.] Lam)—A valuable medicinal food: A review. J. Med. Food 2014, 17, 733–741. [Google Scholar] [CrossRef]
- Liu, Q. Improvement for agronomically important traits by gene engineering in sweetpotato. Breed. Sci. 2017, 67, 15–26. [Google Scholar] [CrossRef]
- Lu, S.; Van Eck, J.; Zhou, X. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef]
- Tzuri, G.; Zhou, X.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 2015, 82, 267–279. [Google Scholar] [CrossRef]
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’Ar, U.; Tzuri, G.; Zheng, Y.; Mazourek, M.; Gepstein, S.; Zhou, X.; et al. Distinct mechanisms of the ORANGE protein in controlling carotenoid flux. Plant Physiol. 2017, 173, 376–389. [Google Scholar] [CrossRef]
- Lopez, A.B.; Van Eck, J.; Conlin, B.J.; Paolillo, D.J.; O’Neill, J.; Li, L. Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. J. Exp. Bot. 2008, 59, 213–223. [Google Scholar] [CrossRef]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Owsiany, K.; Sheeja, T.; Zhou, X.; Rodriguez, C.; Li, Y.; Welsch, R.; Chayut, N.; Yang, Y.; Thannhauser, T.W.; et al. A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis. Plant Physiol. 2015, 169, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kim, S.H.; Park, S.; Lee, H.U.; Lee, J.S.; Park, W.S.; Ahn, M.J.; Kim, Y.H.; Jeong, J.C.; Lee, H.S.; et al. Enhanced accumulation of carotenoids in sweetpotato plants overexpressing IbOr-Ins gene in purple-fleshed sweetpotato cultivar. Plant Physiol. Biochem. 2015, 86, 82–90. [Google Scholar] [CrossRef]
- Wang, Z.; Ke, Q.; Kim, M.D.; Kim, S.H.; Ji, C.Y.; Jeong, J.C.; Lee, H.S.; Park, W.S.; Ahn, M.J.; Li, H.; et al. Transgenic alfalfa plants expressing the sweetpotato Orange gene exhibit enhanced abiotic stress tolerance. PLoS ONE 2015, 10, e0126050. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Ji, C.Y.; Lee, C.J.; Kim, S.E.; Park, S.C.; Kwak, S.S. Orange: A target gene for regulating carotenoid homeostasis and increasing plant tolerance to environmental stress in marginal lands. J. Exp. Bot. 2018, 69, 3393–3400. [Google Scholar] [CrossRef]
- Park, S.; Kim, H.S.; Jung, Y.J.; Kim, S.H.; Ji, C.Y.; Wang, Z.; Jeong, J.C.; Lee, H.S.; Lee, S.Y.; Kwak, S.S. Orange protein has a role in phytoene synthase stabilization in sweetpotato. Sci. Rep. 2016, 6, 33563. [Google Scholar] [CrossRef]
- Kang, L.; Kim, H.S.; Kwon, Y.S.; Ke, Q.; Ji, C.Y.; Park, S.C.; Lee, H.S.; Deng, X.; Kwak, S.S. IbOr regulates photosynthesis under heat stress by stabilizing IbPsbP in sweetpotato. Front. Plant Sci. 2017, 8, 989. [Google Scholar] [CrossRef]
- Park, S.C.; Kang, L.; Park, W.S.; Ahn, M.J.; Kwak, S.S.; Kim, H.S. Carotenoid cleavage dioxygenase 4 (CCD4) cleaves β-carotene and interacts with IbOr in sweetpotato. Plant Biotechnol. Rep. 2020, 14, 737–742. [Google Scholar] [CrossRef]
- Yazdani, M.; Sun, Z.; Yuan, H.; Zeng, S.; Thannhauser, T.W.; Vrebalov, J.; Ma, Q.; Xu, Y.; Fei, Z.; Van Eck, J.; et al. Ectopic expression of ORANGE promotes carotenoid accumulation and fruit development in tomato. Plant Biotechnol. J. 2018, 17, 33–49. [Google Scholar] [CrossRef]
- Ellison, S.L.; Luby, C.H.; Corak, K.E.; Coe, K.M.; Senalik, D.; Iorizzo, M.; Goldman, I.L.; Simon, P.W.; Dawson, J.C. Carotenoid presence is associated with the or gene in domesticated carrot. Genetics 2018, 210, 1497–1508. [Google Scholar] [CrossRef]
- Kim, S.E.; Kim, H.S.; Wang, Z.; Ke, Q.; Lee, C.J.; Park, S.U.; Lim, Y.H.; Park, W.S.; Ahn, M.J.; Kwak, S.S. A single amino acid change at position 96 (Arg to His) of the sweetpotato Orange protein leads to carotenoid overaccumulation. Plant Cell Rep. 2019, 38, 1393–1402. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.J.; Lee, H.S.; Kwak, S.S. Down-regulation of β-carotene hydroxylase increases β-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweetpotato. Phytochemistry 2012, 74, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.J.; Jeong, J.C.; Lee, H.S.; Kwak, S.S. Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol. Biochem. 2013, 70, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, C.; Simon, G.; Rock, E.; Amouroux, P.; Rémésy, C. Genetic variability influences carotenoid, vitamin, phenolic and mineral content in white, yellow, purple, orange, and dark-orange carrot cultivars. J. Am. Soc. Hortic. Sci. 2004, 129, 523–529. [Google Scholar] [CrossRef]
- Takahata, Y.; Noda, T.; Nagata, T. HPLC determination of β-carotene content of sweet-potato cultivars and its relationship with color values. Jpn. J. Breed. 1993, 43, 421–427. [Google Scholar] [CrossRef]
- Teow, C.C.; Truong, V.D.; McFeeters, R.F.; Thompson, R.L.; Pecota, K.V.; Yencho, G.C. Antioxidant activities, phenolic, and β-carotene contents of sweetpotato genotypes with varying flesh colours. Food Chem. 2007, 103, 829–838. [Google Scholar] [CrossRef]
- Zhao, C.; Craig, J.C.; Petzold, H.E.; Dickerman, A.W.; Beers, E.P. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root hypocotyl. Plant Physiol. 2005, 138, 803–818. [Google Scholar] [CrossRef]
- Kim, J.E.; Rensing, K.H.; Douglas, C.J.; Cheng, K.M. Chromoplasts ultrastructure and estimated carotene content in root secondary phloem of different carrot varieties. Planta 2010, 231, 549–558. [Google Scholar] [CrossRef]
- Lu, P.J.; Wang, C.Y.; Yin, T.T.; Zhong, S.L.; Grierson, D.; Chen, K.S.; Xu, C.J. Cytological and molecular characterization of carotenoid accumulation in normal and high-lycopene mutant oranges. Sci. Rep. 2017, 7, 761. [Google Scholar] [CrossRef]
- Perrin, F.; Hartmann, L.; Dubois-Laurent, C.; Welsch, R.; Huet, S.; Hamama, L.; Briard, M.; Peltier, D.; Gagné, S.; Geoffriau, E. Carotenoid gene expression explains the difference of carotenoid accumulation in carrot root tissues. Planta 2017, 245, 737–747. [Google Scholar] [CrossRef]
- Jeffery, J.; Holzenburg, A.; King, S. Physical barriers to carotenoid bioaccessibility. Ultrastructure survey of chromoplast and cell wall morphology in nine carotenoid-containing fruits and vegetables. Food Chem. 2012, 133, 1471–1477. [Google Scholar] [CrossRef] [PubMed]
- Pogson, B.; McDonald, K.A.; Truong, M.; Britton, G.; DellaPenna, D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 1996, 8, 1627–1639. [Google Scholar] [PubMed]
- Kim, J.; Smith, J.J.; Tian, L.; Dellapenna, D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009, 50, 463–479. [Google Scholar] [CrossRef]
- Beisel, K.G.; Jahnke, S.; Hofmann, D.; Köppchen, S.; Schurr, U.; Matsubara, S. Continuous turnover of carotenes and chlorophyll a in mature leaves of Arabidopsis revealed by 14CO2 pulse-chase labeling. Plant Physiol. 2010, 152, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- Dhami, N.; Cazzonelli, C.I. Environmental impacts on carotenoid metabolism in leaves. Plant Growth Regul. 2020, 7, 1–23. [Google Scholar] [CrossRef]
- Beltran, J.C.; Stange, C. Apocarotenoids: A new carotenoid-derived pathway. Subcell. Biochem. 2016, 79, 239–272. [Google Scholar] [PubMed]
- Sun, T.; Tadmor, Y.; Li, L. Pathways for carotenoid biosynthesis, degradation, and storage. In Methods in Molecular Biology; Rodríguez-Concepción, M., Welsch, R., Eds.; Humana: New York, NY, USA, 2020; Volume 2083, pp. 3–23. [Google Scholar]
- Hermanns, A.S.; Zhou, X.; Xu, Q.; Tadmor, Y.; Li, L. Carotenoid pigment accumulation in horticultural plants. Hortic. Plant J. 2020. [Google Scholar] [CrossRef]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef]
- Aksoy, L.; Kolay, E.; Ağılönüa, Y.; Aslan, Z.; Kargıoğlu, M. Free radical scavenging activity, total phenolic content, total antioxidant status, and total oxidant status of endemic Thermopsis Turc. Saudi J. Biol. Sci. 2013, 20, 235–239. [Google Scholar] [CrossRef]
- Xiong, L.M.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Gouveia, C.S.S.; Ganança, J.F.T.; Slaski, J.J.; Lebot, V.; De Carvalho, M.Â.A.P. Abscisic acid phytohormone estimation in tubers and shoots of Ipomoea batatas subjected to long drought stress using competitive immunological assay. Physiol. Plant. 2020. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-E.; Lee, C.-J.; Park, S.-U.; Lim, Y.-H.; Park, W.S.; Kim, H.-J.; Ahn, M.-J.; Kwak, S.-S.; Kim, H.S. Overexpression of the Golden SNP-Carrying Orange Gene Enhances Carotenoid Accumulation and Heat Stress Tolerance in Sweetpotato Plants. Antioxidants 2021, 10, 51. https://doi.org/10.3390/antiox10010051
Kim S-E, Lee C-J, Park S-U, Lim Y-H, Park WS, Kim H-J, Ahn M-J, Kwak S-S, Kim HS. Overexpression of the Golden SNP-Carrying Orange Gene Enhances Carotenoid Accumulation and Heat Stress Tolerance in Sweetpotato Plants. Antioxidants. 2021; 10(1):51. https://doi.org/10.3390/antiox10010051
Chicago/Turabian StyleKim, So-Eun, Chan-Ju Lee, Sul-U Park, Ye-Hoon Lim, Woo Sung Park, Hye-Jin Kim, Mi-Jeong Ahn, Sang-Soo Kwak, and Ho Soo Kim. 2021. "Overexpression of the Golden SNP-Carrying Orange Gene Enhances Carotenoid Accumulation and Heat Stress Tolerance in Sweetpotato Plants" Antioxidants 10, no. 1: 51. https://doi.org/10.3390/antiox10010051
APA StyleKim, S.-E., Lee, C.-J., Park, S.-U., Lim, Y.-H., Park, W. S., Kim, H.-J., Ahn, M.-J., Kwak, S.-S., & Kim, H. S. (2021). Overexpression of the Golden SNP-Carrying Orange Gene Enhances Carotenoid Accumulation and Heat Stress Tolerance in Sweetpotato Plants. Antioxidants, 10(1), 51. https://doi.org/10.3390/antiox10010051






