Use of a Pan–Genomic DNA Microarray in Determination of the Phylogenetic Relatedness among Cronobacter spp. and Its Use as a Data Mining Tool to Understand Cronobacter Biology
Abstract
:1. Introduction
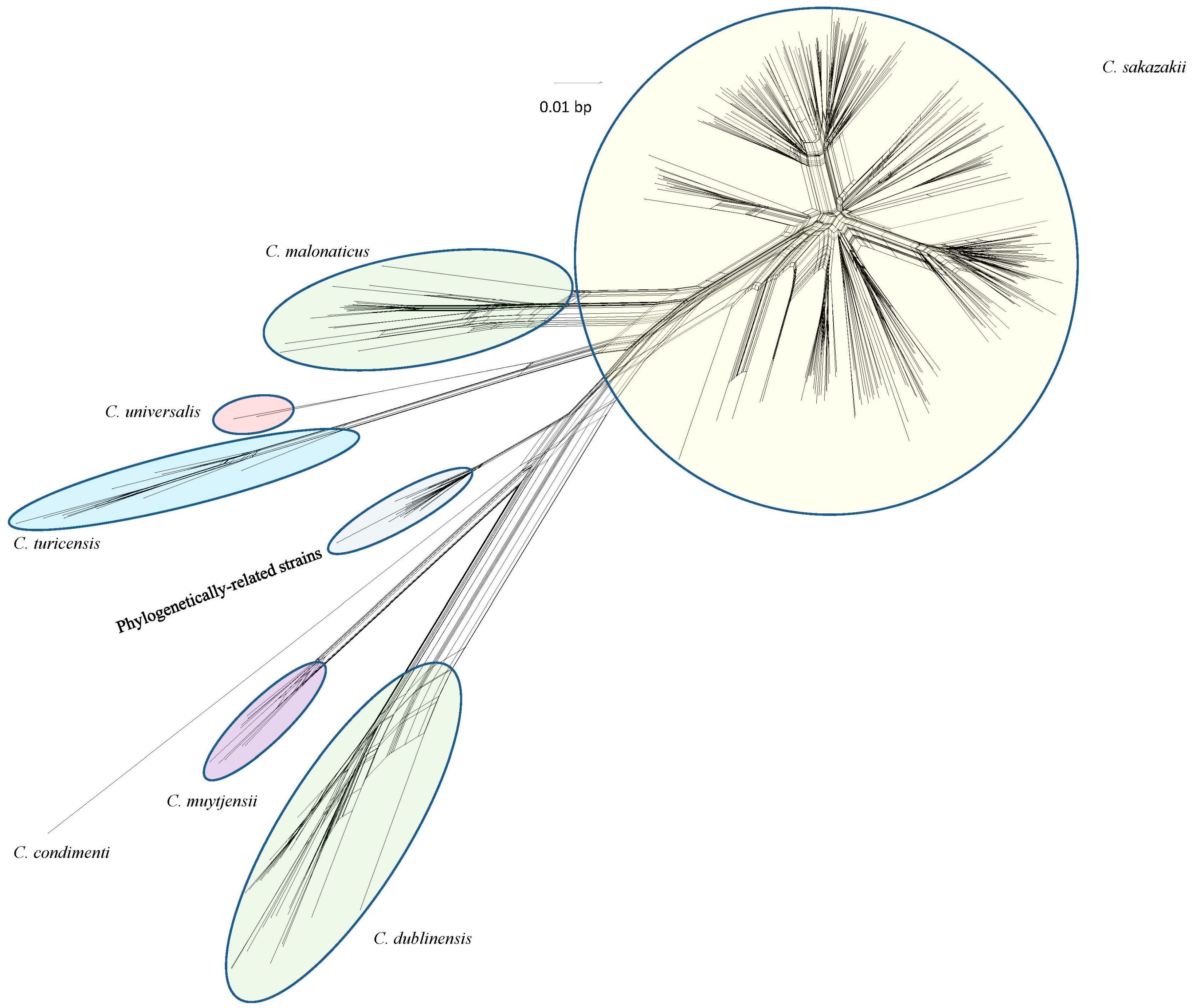
2. Previous Work in the Development of a DNA Microarray for Cronobacter Species Identity
3. Recent Work Describing the Use of the Cronobacter Microarray to Data Mine Specific Alleles
4. Future Directions
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yan, Q.Q.; Condell1, O.; Power, K.; Butler, F.; Tall, B.D.; Fanning, S. Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: A review of our current understanding of the biology of this bacterium. J. Appl. Microbiol. 2012, 113, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, Z.W.; Al Mousa, W.; Elbetieha, A.; Al Nabulsi, A.; Tall, B.D. Cronobacter spp.—Opportunistic food borne pathogen. A review of its virulence and environmental adaptive traits. J. Med. Microbiol. 2014, 63, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Tall, B.D.; Chen, Y.; Yan, Q.Q.; Gopinath, G.R.; Grim, C.J.; Jarvis, K.G.; Fanning, S.; Lampel, K.A. Cronobacter: An emergent pathogen causing meningitis to neonates through their feeds. Sci. Prog. 2014, 97, 154–172. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.E.; Mahon, B.E.; Greene, S.A.; Rounds, J.; Cronquist, A.; Wymore, K.; Boothe, E.; Lathrop, S.; Palmer, A.; Bowen, A. Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg. Infect. Dis. 2014, 20, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Jason, J. Prevention of invasive Cronobacter infections in young infants fed powdered infant formulas. Pediatrics 2012, 130, e1076–e1084. [Google Scholar] [CrossRef] [PubMed]
- Osaili, T.; Forsythe, S. Desiccation resistance and persistence of Cronobacter species in infant formula. Intern. J. Food Microbiol. 2009, 136, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, J.; You, C.; Liu, Z. Diversity of Cronobacter spp. isolates from the vegetables in the middle-east coastline of China. World J. Microbiol. Biotechnol. 2016, 32, 90. [Google Scholar] [CrossRef] [PubMed]
- El-Sharoud, W.M.; El-Din, M.Z.; Ziada, D.M.; Ahmed, S.F.; Klena, J.D. Surveillance and genotyping of Enterobacter sakazakii suggest its potential transmission from milk powder into imitation recombined soft cheese. J. Appl. Microbiol. 2008, 105, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. Morb. Mortal. Wkly. Rep. 2002, 51, 297–300. [Google Scholar]
- Noriega, F.R.; Kotloff, K.L.; Martin, M.A.; Schwalbe, R.S. Nosocomial bacteremia caused by Enterobacter sakazakii and Leuconostoc mesenteroides resulting from extrinsic contamination of infant formula. Pediatr. Infect. Dis. J. 1990, 9, 447–449. [Google Scholar] [PubMed]
- Yan, Q.; Fanning, S. Strategies for the identification and tracking of Cronobacter species: An opportunistic pathogen of concern to neonatal health. Front. Pediatr. 2015, 3, 38. [Google Scholar] [CrossRef] [PubMed]
- Bowen, A.B.; Braden, C.R. Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 2006, 12, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Gosney, M.A.; Martin, M.V.; Wright, A.E.; Gallagher, M. Enterobacter sakazakii in the mouths of stroke patients and its association with aspiration pneumonia. Eur. J. Intern. Med. 2006, 17, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Holý, O.; Petrželová, J.; Hanulík, V.; Chromá, M.; Matoušková, I.; Forsythe, S.J. Epidemiology of Cronobacter spp. isolates from patients admitted to the Olomouc University Hospital (Czech Republic). Epidemiol. Mikrobiol. Imunol. 2014, 63, 69–72. [Google Scholar] [PubMed]
- Alsonosi, A.; Hariri, S.; Kajsík, M.; Oriešková, M.; Hanulík, V.; Röderová, M.; Petrželová, J.; Kollárová, H.; Drahovská, H.; Forsythe, S.; et al. The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Iversen, C.; Mullane, N.; McCardell, B.; Tall, B.D.; Lehner, A.; Fanning, S.; Stephan, R.; Joosten, H. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. System. Evol. Microbiol. 2008, 58, 1442–1447. [Google Scholar]
- Joseph, S.; Cetinkaya, E.; Drahovska, H.; Levican, A.; Figueras, M.J.; Forsythe, S.J. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2012, 62, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Tall, B.D.; Gangiredla, J.; Gopinath, G.R.; Yan, Q.Q.; Chase, H.R.; Lee, B.; Hwang, S.; Trach, L.; Park, E.; Yoo, Y.J.; et al. Development of a custom-designed, pan genomic DNA microarray to characterize strain-level diversity among Cronobacter spp. Front. Pediatr. 2015, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kotewicz, M.L.; Power, K.A.; Gopinath, G.; Franco, A.A.; Jarvis, K.G.; Yan, Q.Q.; Jackson, S.A.; Sathyamoorthy, V.; Hu, L.; et al. Pan-genome analysis of the emerging foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence driven by niche adaptation. BMC Genom. 2013, 14, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Jackson, S.A.; Gangiredla, J.; Wang, W.; Liua, H.; Tall, B.D.; Jean-Gilles Beaubrun, J.; Jay-Russell, M.; Vellidis, G.; Elkins, C.A. Genomic evidence reveals numerous re-introduction events of Salmonella enterica serovar Newport in irrigation ponds in Suwannee watershed. Appl. Environ. Microbiol. 2015, 81, 8243–8253. [Google Scholar] [CrossRef]
- Jackson, S.A.; Patel, I.R.; Barnaba, T.; LeClerc, J.E.; Cebula, T.A. Investigating the global diversity of Escherichia coli using a multi-genome DNA microarray platform with novel gene prediction strategies. BMC Genom. 2011, 12, 349. [Google Scholar] [CrossRef] [PubMed]
- Laksanalamai, P.; Jackson, S.A.; Mammel, M.K.; Datta, A.R. High density microarray analysis reveals new insights into genetic footprints of Listeria monocytogenes strains involved in listeriosis outbreaks. PLoS ONE 2012, 7, e32896. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Grim, C.J.; Gopinath, G.R.; Mammel, M.K.; Sathyamoorthy, V.; Trach, L.H.; Chase, H.R.; Fanning, S.; Tall, B.D. Re-examination of the taxonomic status of Enterobacter helveticus sp. nov., Enterobacter pulveris sp. nov., and Enterobacter turicensis sp. nov. as members of Cronobacter: Proposal of two new genera Siccibacter gen. nov. and Franconibacter gen. nov. and descriptions of Siccibacter turicensis sp. nov., Franconibacter helveticus sp. nov., and Franconibacter pulveris sp. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 3402–3410. [Google Scholar] [PubMed]
- Yan, Q.Q.; Wang, J.; Gangiredla, J.; Cao, Y.; Martins, M.; Gopinath, G.R.; Stephan, R.; Lampel, K.; Tall, B.D.; Fanning, S. Comparative genotypic and phenotypic analysis of Cronobacter species cultured from four powdered infant formula production facilities: Indication of patho-adaptation along the food chain. Appl. Environ. Microbiol. 2015, 81, 4388–4402. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.A.; Hu, L.; Grim, C.J.; Gopinath, G.; Sathyamoorthy, V.; Jarvis, K.G.; Lee, C.; Sadowski, J.; Kim, J.; Kothary, M.H.; et al. Characterization of Putative Virulence Genes Encoded on the related RepFIB Plasmids Harbored by Cronobacter spp. Appl. Environ. Microbiol. 2011, 77, 3255–3267. [Google Scholar] [CrossRef] [PubMed]
- Cronobacter GenomeTrakr Project 258403: FDA-CFSAN. Available online: https://www.ncbi.nlm.nih.gov/bioproject/258403 (accessed on 28 February 2017).
- Kothary, M.H.; Gopinath, G.R.; Gangiredla, J.; Rallabhandi, P.V.; Harrison, L.M.; Yan, Q.Q.; Chase, H.R.; Lee, B.; Park, E.; Yoo, Y.; et al. Analysis and Characterization of Proteins Associated with Outer Membrane Vesicles Secreted by Cronobacter spp. Front. Microbiol. 2017, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Eshwar, A.K.; Tall, B.D.; Gangiredla, J.; Gopinath, G.R.; Patel, I.R.; Neuhauss, S.C.F.; Stephan, R.; Lehner, A. Linking Genomo- and Pathotype: Exploiting the Zebrafish Embryo Model to Investigate the Divergent Virulence Potential among Cronobacter spp. PLoS ONE 2016, 11, e0158428. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.; Xicohtencatl-Cortes, J.; González-Pedrajo, B.; Bobadilla, M.; Eslava, C.; Rosas, I. Virulence traits in Cronobacter species isolated from different sources. Can. J. Microbiol. 2011, 57, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Lehner, A.; Tischler, P.; Rattei, T. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J. Bacteriol. 2011, 193, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Kucerova, E.; Clifton, S.W.; Xia, X.Q.; Long, F.; Porwollik, S.; Fulton, L.; Fronick, C.; Minx, P.; Kyung, K.; Warren, W.; et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS ONE 2010, 5, e9556. [Google Scholar] [CrossRef] [PubMed]
- Power, K.A.; Yan, Q.; Fox, E.M.; Cooney, S.; Fanning, S. Genome sequence of Cronobacter sakazakii SP291, a persistent thermotolerant isolate derived from a factory producing powdered infant formula. Genome Announc. 2013, 1, e0008213. [Google Scholar] [CrossRef] [PubMed]
- Moine, D.; Kassam, M.; Baert, L.; Tang, Y.; Barretto, C.; Ngom Bru, C.; Klijn, A.; Descombes, P. Fully Closed Genome Sequences of Five Type Strains of the Genus Cronobacter and One Cronobacter sakazakii Strain. Genome Announc. 2016, 4, e00142-16. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, J.; Li, C.; Xie, T.; Ling, N.; Wu, Q.; Ye, Y. The driving force of prophages and CRISPR-Cas system in the evolution of Cronobacter sakazakii. Sci. Rep. 2017, 7, 40206. [Google Scholar] [CrossRef] [PubMed]
- Farmer, J.J., III. My 40-year history with Cronobacter/Enterobacter sakazakii—Lessons learned, myths debunked, and recommendations. Front. Pediatr. 2015, 3, 84.35. [Google Scholar] [CrossRef] [PubMed]
| Species and Strain | Number of Alleles from Each Strain | Number Present (% Present) b | Genome NCBI c Accession Numbers | Plasmid Name, NCBI Accession Numbers |
|---|---|---|---|---|
| C. sakazakii ATCC BAA-894 | 2035 | 1904 (93.5) | NC_009778 | pESA3, NC_009780.1; pESA2, NC_009779.1 |
| 4.01C | 139 | 132 (94.9) | AJLB00000000.1 | pESA3-like, AJLB00000000.1 |
| 2151 | 201 | 171 (85.0) | AJKT01000000.1 | pESA3- and pCSA2151, AJKT01000000.1 |
| Es35 | 202 | 188 (93.0) | AJLC00000000.1 | pESA3- and pCTU3-like, AJLC00000000.1) |
| Es764 | 304 | 266 (87.5) | AJLA00000000.1 | pESA3-like, AJLA00000000.1 |
| C. turicensis LMG23827T | 4402 | 4039 (91.7) | NC_013282.2 | pCTU1, NC_013283.1; pCTU2, NC_013284.1; pCTU3, NC_013285 |
| C. malonaticus | ||||
| LMG23826T | 1582 | 1434 (90.6) | AJKV01000000.1 | pCMA1, NZ_CP013941.1/CP013941.1; pCMA2, NZ_CP013942.1/CP013942.1 |
| CDC2193-01 | 257 | JXTD00000000.1 | pCTU1-like, JXTD00000000.1 | |
| C. dublinensis | ||||
| C. dublinensis subsp.d dublinensis LMG23823T | 781 | 745 (95.4) | AJKZ01000000.1 | pCTU1-like, AJKZ01000000.1 |
| C. dublinensis subsp. lausannensis LMG23824T | 2580 | 2386 (92.5) | AJKY01000000.1 | pCTU1-like, AJKY01000000.1 |
| C. muytjensii ATCC 51329T | 1754 | 1708 (97.3) | AJKU01000000.1 | No Plasmid |
| C. universalis NCTC9529T | 1315 | 1201 (91.3) | AJKW01000000.1 | pEAS3-like, AJKW01000000.1 |
| C. condimenti LMG26250T | 2611 | 2498 (95.7) | CAKW00000000.1 | pCTU1-like, CAKW00000000.1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tall, B.D.; Gangiredla, J.; Grim, C.J.; Patel, I.R.; Jackson, S.A.; Mammel, M.K.; Kothary, M.H.; Sathyamoorthy, V.; Carter, L.; Fanning, S.; et al. Use of a Pan–Genomic DNA Microarray in Determination of the Phylogenetic Relatedness among Cronobacter spp. and Its Use as a Data Mining Tool to Understand Cronobacter Biology. Microarrays 2017, 6, 6. https://doi.org/10.3390/microarrays6010006
Tall BD, Gangiredla J, Grim CJ, Patel IR, Jackson SA, Mammel MK, Kothary MH, Sathyamoorthy V, Carter L, Fanning S, et al. Use of a Pan–Genomic DNA Microarray in Determination of the Phylogenetic Relatedness among Cronobacter spp. and Its Use as a Data Mining Tool to Understand Cronobacter Biology. Microarrays. 2017; 6(1):6. https://doi.org/10.3390/microarrays6010006
Chicago/Turabian StyleTall, Ben D., Jayanthi Gangiredla, Christopher J. Grim, Isha R. Patel, Scott A. Jackson, Mark K. Mammel, Mahendra H. Kothary, Venugopal Sathyamoorthy, Laurenda Carter, Séamus Fanning, and et al. 2017. "Use of a Pan–Genomic DNA Microarray in Determination of the Phylogenetic Relatedness among Cronobacter spp. and Its Use as a Data Mining Tool to Understand Cronobacter Biology" Microarrays 6, no. 1: 6. https://doi.org/10.3390/microarrays6010006
APA StyleTall, B. D., Gangiredla, J., Grim, C. J., Patel, I. R., Jackson, S. A., Mammel, M. K., Kothary, M. H., Sathyamoorthy, V., Carter, L., Fanning, S., Iversen, C., Pagotto, F., Stephan, R., Lehner, A., Farber, J., Yan, Q. Q., & Gopinath, G. R. (2017). Use of a Pan–Genomic DNA Microarray in Determination of the Phylogenetic Relatedness among Cronobacter spp. and Its Use as a Data Mining Tool to Understand Cronobacter Biology. Microarrays, 6(1), 6. https://doi.org/10.3390/microarrays6010006








