Protein Profiling Gastric Cancer and Neighboring Control Tissues Using High-Content Antibody Microarrays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Isolation and Labeling
2.2. Microarray Production
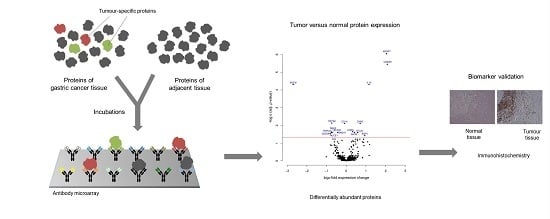
2.3. Microarray Incubation, Scanning, and Image Processing
2.4. Data Analysis and Sample Classification
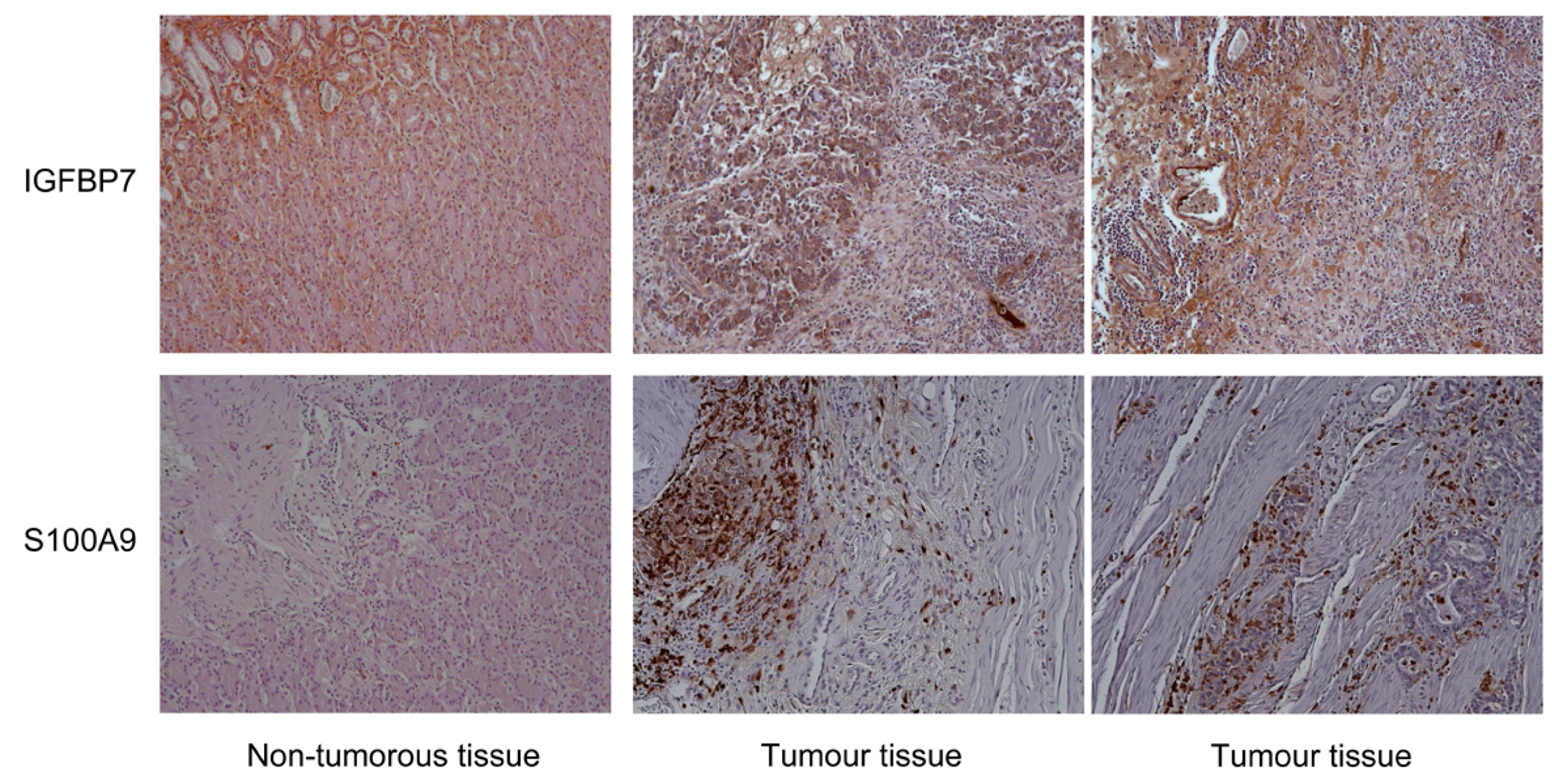
2.5. Immunohistochemistry
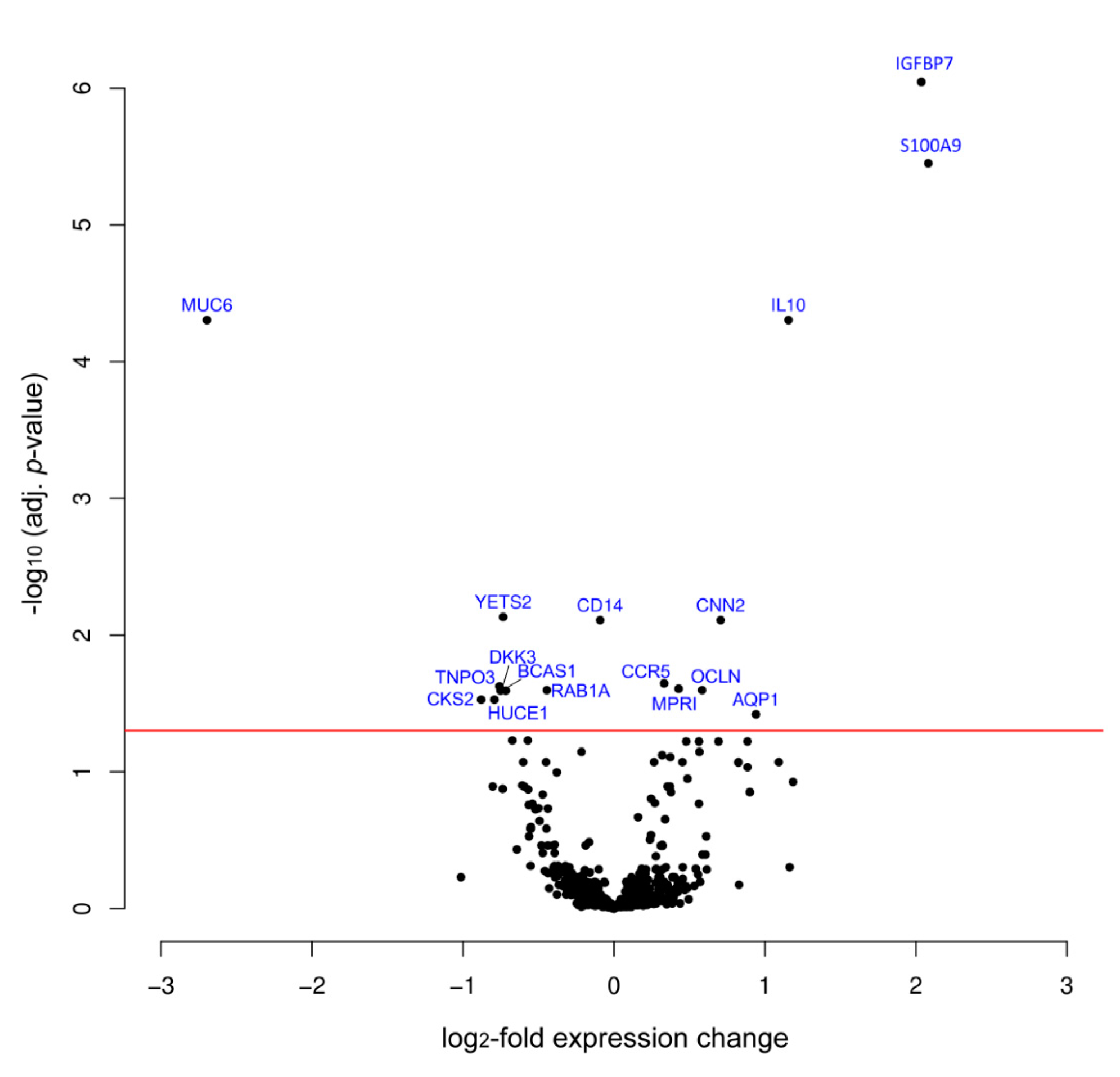
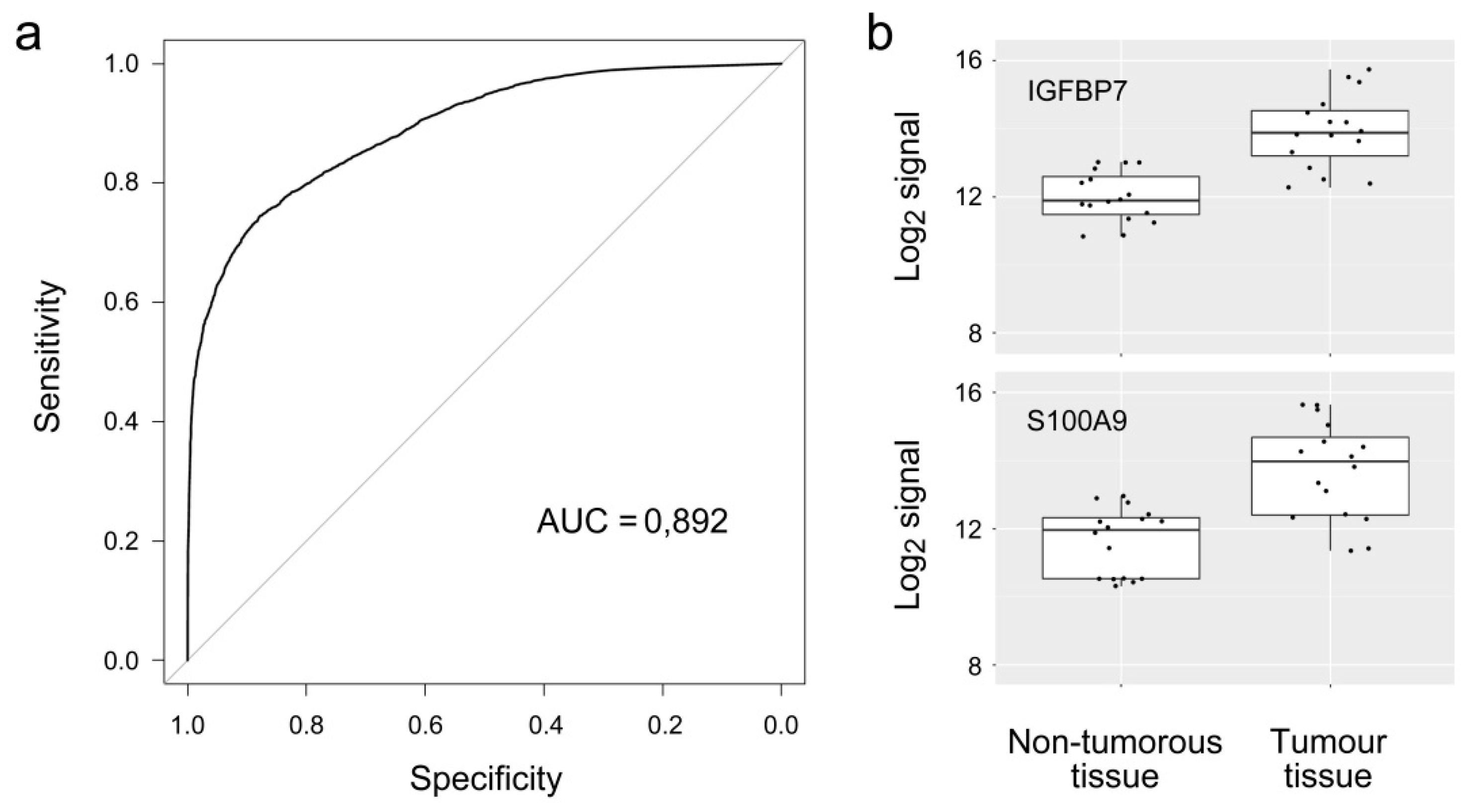
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Karimi, P.; Islami, F.; Anandasabapathy, S.; Freedman, N.D.; Kamangar, F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomark. Prev. 2014, 23, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Dicken, B.J.; Bigam, D.L.; Cass, C.; Mackey, J.R.; Joy, A.A.; Hamilton, S.M. Gastric adenocarcinoma: Review and considerations for future directions. Ann. Surg. 2005, 241, 27–39. [Google Scholar] [PubMed]
- Sanford, M. Trastuzumab: A review of its use in HER2-positive advanced gastric cancer. Drugs 2013, 73, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; de Braud, F.; da Prat, V.; Perrone, F.; Pierotti, M.A.; Gariboldi, M.; Fanetti, G.; Biondani, P.; Pellegrinelli, A.; Bossi, I.; et al. A review on biomarkers for prediction of treatment outcome in gastric cancer. Anticancer Res. 2013, 33, 1257–1266. [Google Scholar] [PubMed]
- Brenner, H.; Rothenbacher, D.; Arndt, V. Epidemiology of stomach cancer. Methods Mol. Biol. 2009, 472, 467–477. [Google Scholar] [PubMed]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [PubMed]
- Flejou, J.F. Who classification of digestive tumors: The fourth edition. Ann. Pathol. 2011, 31, S27–S31. [Google Scholar] [PubMed]
- Zheng, L.; Wang, L.; Ajani, J.; Xie, K. Molecular basis of gastric cancer development and progression. Gastric Cancer 2004, 7, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.Y.; Yuen, S.T.; Chu, K.M.; Mathy, J.A.; Li, R.; Chan, A.S.; Law, S.; Wong, J.; Chen, X.; So, S. Expression profiling identifies chemokine (C-C motif) ligand 18 as an independent prognostic indicator in gastric cancer. Gastroenterology 2004, 127, 457–469. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar]
- Betzen, C.; Alhamdani, M.S.; Lueong, S.; Schroder, C.; Stang, A.; Hoheisel, J.D. Clinical proteomics: Promises, challenges and limitations of affinity arrays. Proteom. Clin. Appl. 2015, 9, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Borrebaeck, C.A.; Wingren, C. Antibody array generation and use. Methods Mol. Biol. 2014, 1131, 563–571. [Google Scholar] [PubMed]
- Brennan, D.J.; O’Connor, D.P.; Rexhepaj, E.; Ponten, F.; Gallagher, W.M. Antibody-based proteomics: Fast-tracking molecular diagnostics in oncology. Nat. Rev. Cancer 2010, 10, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Hoheisel, J.D.; Alhamdani, M.S.; Schroder, C. Affinity-based microarrays for proteomic analysis of cancer tissues. Proteom. Clin. Appl. 2013, 7, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Schroder, C.; Jacob, A.; Tonack, S.; Radon, T.P.; Sill, M.; Zucknick, M.; Ruffer, S.; Costello, E.; Neoptolemos, J.P.; Crnogorac-Jurcevic, T.; et al. Dual-color proteomic profiling of complex samples with a microarray of 810 cancer-related antibodies. Mol. Cell. Proteom. 2010, 9, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Wingren, C.; Sandstrom, A.; Segersvard, R.; Carlsson, A.; Andersson, R.; Lohr, M.; Borrebaeck, C.A. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012, 72, 2481–2490. [Google Scholar] [CrossRef] [PubMed]
- Ingvarsson, J.; Wingren, C.; Carlsson, A.; Ellmark, P.; Wahren, B.; Engstrom, G.; Harmenberg, U.; Krogh, M.; Peterson, C.; Borrebaeck, C.A. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics 2008, 8, 2211–2219. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Jacak, J.; Schirwitz, C.; Stadler, V.; Michel, G.; Marme, N.; Schutz, G.J.; Hoheisel, J.D.; Knemeyer, J.P. Single-molecule detection on a protein-array assay platform for the exposure of a tuberculosis antigen. J. Proteome Res. 2011, 10, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Alhamdani, M.S.; Schroder, C.; Werner, J.; Giese, N.; Bauer, A.; Hoheisel, J.D. Single-step procedure for the isolation of proteins at near-native conditions from mammalian tissue for proteomic analysis on antibody microarrays. J. Proteome Res. 2010, 9, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K.; Michaud, J.; Scott, H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Alhamdani, M.S.; Youns, M.; Buchholz, M.; Gress, T.M.; Beckers, M.C.; Marechal, D.; Bauer, A.; Schroder, C.; Hoheisel, J.D. Immunoassay-based proteome profiling of 24 pancreatic cancer cell lines. J. Proteom. 2012, 75, 3747–3759. [Google Scholar] [CrossRef] [PubMed]
- Nijaguna, M.B.; Schroder, C.; Patil, V.; Shwetha, S.D.; Hegde, A.S.; Chandramouli, B.A.; Arivazhagan, A.; Santosh, V.; Hoheisel, J.D.; Somasundaram, K. Definition of a serum marker panel for glioblastoma discrimination and identification of interleukin 1beta in the microglial secretome as a novel mediator of endothelial cell survival induced by C-reactive protein. J. Proteom. 2015, 128, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Botla, S.K.; Gholami, A.M.; Malekpour, M.; Moskalev, E.A.; Fallah, M.; Jandaghi, P.; Aghajani, A.; Bondar, I.S.; Omranipour, R.; Malekpour, F.; et al. Diagnostic values of GHSR DNA methylation pattern in breast cancer. Breast Cancer Res. Treat. 2012, 135, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Kondo, Y.; Rosner, G.L.; Xiao, L.; Hernandez, N.S.; Vilaythong, J.; Houlihan, P.S.; Krouse, R.S.; Prasad, A.R.; Einspahr, J.G.; et al. Mgmt promoter methylation and field defect in sporadic colorectal cancer. J. Natl. Cancer Inst. 2005, 97, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Wolff, E.M.; Chihara, Y.; Pan, F.; Weisenberger, D.J.; Siegmund, K.D.; Sugano, K.; Kawashima, K.; Laird, P.W.; Jones, P.A.; Liang, G. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010, 70, 8169–8178. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Inokuchi, M.; Takagi, Y.; Otsuki, S.; Fujimori, Y.; Yanaka, Y.; Kobayashi, K.; Higuchi, K.; Kojima, K.; Kawano, T. Relationship between expression of IGFBP7 and clinicopathological variables in gastric cancer. J. Clin. Pathol. 2015, 68, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, Z.; Zhang, W.; Yan, B.; Gu, Q.; Jiao, J.; Yue, X. Decreased expression of IGFBP7 was a poor prognosis predictor for gastric cancer patients. Tumour Biol. 2014, 35, 8875–8881. [Google Scholar] [CrossRef] [PubMed]
- Rupp, C.; Scherzer, M.; Rudisch, A.; Unger, C.; Haslinger, C.; Schweifer, N.; Artaker, M.; Nivarthi, H.; Moriggl, R.; Hengstschlager, M.; et al. IGFBP7, a novel tumor stroma marker, with growth-promoting effects in colon cancer through a paracrine tumor-stroma interaction. Oncogene 2015, 34, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kang, H.J.; Lee, H.; Lee, S.T.; Yu, M.H.; Kim, H.; Lee, C. Identification of S100A8 and S100A9 as serological markers for colorectal cancer. J. Proteome Res. 2009, 8, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Zhang, L.H.; Jia, Y.N.; Zhong, X.Y.; Liu, Y.Q.; Cheng, X.J.; Wang, X.H.; Xing, X.F.; Hu, Y.; Li, Y.A.; et al. Presence of S100A9-positive inflammatory cells in cancer tissues correlates with an early stage cancer and a better prognosis in patients with gastric cancer. BMC Cancer 2012, 12, 316. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, H.; Zhu, X.; Ma, Y.; Liu, Q.; Xue, Y.; Chu, H.; Wu, W.; Wang, J.; Zou, H. S100A9 promotes human lung fibroblast cells activation through receptor for advanced glycation end-product-mediated extracellular-regulated kinase 1/2, mitogen-activated protein-kinase and nuclear factor-κB-dependent pathways. Clin. Exp. Immunol. 2013, 173, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.H.; Moon, H.J.; Park, H.J.; Choi, J.H.; Park do, Y. S100A8 and S100A9 promotes invasion and migration through p38 mitogen-activated protein kinase-dependent NF-κB activation in gastric cancer cells. Mol. Cells 2013, 35, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.J.; Argyris, P.P.; Zou, X.; Ross, K.F.; Herzberg, M.C. S100A8/A9 regulates MMP-2 expression and invasion and migration by carcinoma cells. Int. J. Biochem. Cell Biol. 2014, 55, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Terai, M.; Tamura, Y.; Alexeev, V.; Mastrangelo, M.J.; Selvan, S.R. Interleukin 10 in the tumor microenvironment: A target for anticancer immunotherapy. Immunol. Res. 2011, 51, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Szkaradkiewicz, A.; Karpinski, T.M.; Drews, M.; Borejsza-Wysocki, M.; Majewski, P.; Andrzejewska, E. Natural killer cell cytotoxicity and immunosuppressive cytokines (IL-10, TGF-β1) in patients with gastric cancer. J. Biomed. Biotech. 2010, 2010, 901564. [Google Scholar] [CrossRef] [PubMed]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The master regulator of immunity to infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Huang, R.R.; Wen, D.R.; Paul, E.; Wunsch, P.H.; Cochran, A.J. IL-10 expression by primary tumor cells correlates with melanoma progression from radial to vertical growth phase and development of metastatic competence. Mod. Pathol. 2011, 24, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Leir, S.H.; Harris, A. MUC6 mucin expression inhibits tumor cell invasion. Exp. Cell Res. 2011, 317, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.G.; He, Z.Y.; Wang, Q. Transcriptome profiling of the cancer and normal tissues from gastric cancer patients by deep sequencing. Tumour Biol. 2014, 35, 7423–7427. [Google Scholar] [CrossRef] [PubMed]
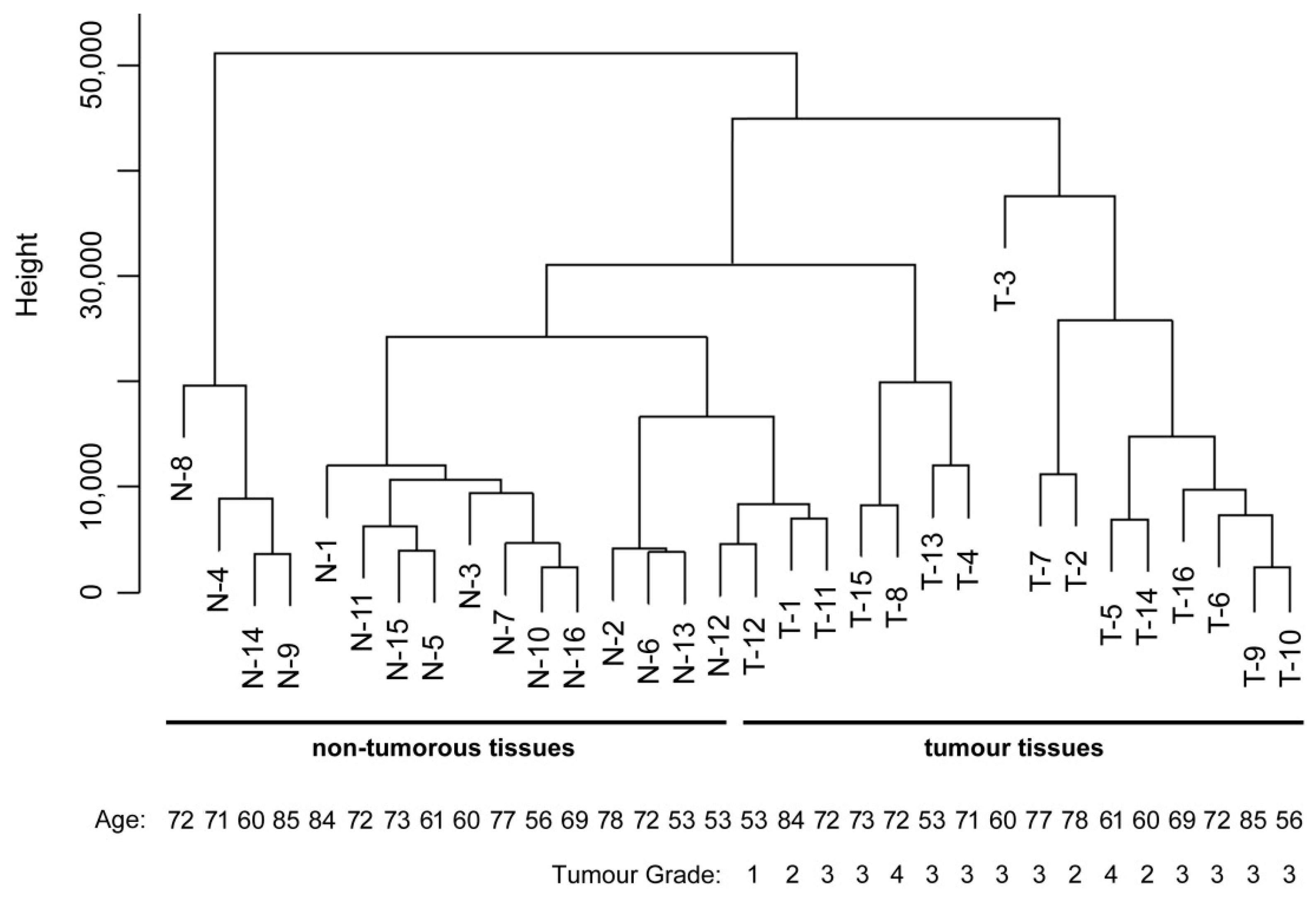
| Sample Number | Patient Gender | Patient Age | Location in Stomach | Tumour Grade | Lauren Classification | Number of Positive Lymph Nodes |
|---|---|---|---|---|---|---|
| 1 | Male | 84 | Corpus | 2 | Intestinal | 1 |
| 2 | Male | 78 | Cardia | 2 | Intestinal | 0 |
| 3 | Male | 60 | Cardia | 3 | Intestinal | 7 |
| 4 | Male | 71 | Cardia | 3 | Diffuse | 5 |
| 5 | Male | 61 | Antrum | 4 | Mixed | 8 |
| 6 | Female | 72 | Antrum | 3 | Intestinal | 2 |
| 7 | Female | 77 | Antrum | 3 | Intestinal | 0 |
| 8 | Male | 72 | Antrum | 4 | Mixed | 0 |
| 9 | Male | 85 | Antrum | 3 | Mixed | 0 |
| 10 | Male | 56 | Corpus | 3 | Diffuse | 1 |
| 11 | Male | 72 | Cardia | 3 | Intestinal | 21 |
| 12 | Male | 53 | Corpus | 1 | Intestinal | 0 |
| 13 | Male | 53 | Spread | 3 | Intestinal | 12 |
| 14 | Male | 60 | Spread | 2 | Intestinal | 10 |
| 15 | Male | 73 | Corpus | 3 | Intestinal | 12 |
| 16 | Female | 69 | Spread | 3 | Intestinal | 16 |
| Protein | Variation log2FC | Adj. p-Value | Swissprot No. | Full Protein Name |
|---|---|---|---|---|
| Proteins with most significant and strongest variation in expression | ||||
| IGFBP7 | 2.04 | 8.99 × 10−7 | Q16270 | Insulin-like growth factor-binding protein 7 |
| S100A9 | 2.08 | 3.54 × 10−6 | P06702 | S100 calcium binding protein A9 |
| IL10 | 1.16 | 4.96 × 10−5 | P22301 | Interleukin 10 |
| MUC6 | −2.70 | 4.96 × 10−5 | Q6W4X9 | Mucin 6 |
| Other proteins of higher abundance in tumor | ||||
| CNN2 | 0.71 | 7.77 × 10−3 | Q99439 | Calponin-2 |
| CCR5 | 0.33 | 2.26 × 10−2 | P51681 | C-C chemokine receptor type 5 |
| MPRI | 0.43 | 2.41 × 10−4 | P11717 | Cation-independent mannose-6-phosphate receptor |
| OCLN | 0.58 | 2.53 × 10−2 | Q16625 | Occludin |
| AQP1 | 0.94 | 3.80 × 10−2 | P29972 | Aquaporin 1 |
| Other proteins of lower abundance in tumor | ||||
| YETS2 | −0.73 | 7.37 × 10−3 | Q9ULM3 | YEATS domain-containing protein 2 |
| CD14 | -0.09 | 1.07 × 10−2 | P08571 | Monocyte differentiation antigen CD14 |
| TNPO3 | −0.75 | 2.35 × 10−2 | Q9Y5L0 | Transportin-3 |
| BCAS1 | −0.72 | 2.53 × 10−2 | O75363 | Breast carcinoma-amplified sequence 1 |
| RAB1A | −0.44 | 2.53 × 10−2 | P62820 | Ras-related protein Rab-1A |
| DKK3 | −0.74 | 2.53 × 10−2 | Q9UBP4 | Dickkopf-related protein 3 |
| CKS2 | −0.88 | 2.97× 10−2 | P33552 | Cyclin-dependent kinases regulatory subunit 2 |
| HUCE1 | -0.79 | 2.97× 10−2 | O43159 | Cerebral protein 1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sill, M.; Schröder, C.; Shen, Y.; Marzoq, A.; Komel, R.; Hoheisel, J.D.; Nienhüser, H.; Schmidt, T.; Kastelic, D. Protein Profiling Gastric Cancer and Neighboring Control Tissues Using High-Content Antibody Microarrays. Microarrays 2016, 5, 19. https://doi.org/10.3390/microarrays5030019
Sill M, Schröder C, Shen Y, Marzoq A, Komel R, Hoheisel JD, Nienhüser H, Schmidt T, Kastelic D. Protein Profiling Gastric Cancer and Neighboring Control Tissues Using High-Content Antibody Microarrays. Microarrays. 2016; 5(3):19. https://doi.org/10.3390/microarrays5030019
Chicago/Turabian StyleSill, Martin, Christoph Schröder, Ying Shen, Aseel Marzoq, Radovan Komel, Jörg D. Hoheisel, Henrik Nienhüser, Thomas Schmidt, and Damjana Kastelic. 2016. "Protein Profiling Gastric Cancer and Neighboring Control Tissues Using High-Content Antibody Microarrays" Microarrays 5, no. 3: 19. https://doi.org/10.3390/microarrays5030019
APA StyleSill, M., Schröder, C., Shen, Y., Marzoq, A., Komel, R., Hoheisel, J. D., Nienhüser, H., Schmidt, T., & Kastelic, D. (2016). Protein Profiling Gastric Cancer and Neighboring Control Tissues Using High-Content Antibody Microarrays. Microarrays, 5(3), 19. https://doi.org/10.3390/microarrays5030019