Application of Tissue Microarray Technology to Stem Cell Research
Abstract
:1. Introduction
2. Experimental Section
2.1. Cell Culture for CMA Preparation
2.2. CMA Construction
2.3. Immunofluorescence (IF) Analysis
3. Results
3.1. CMA Technology to Screen for Bonafide Pluripotent Stem Cells and Evaluation of Population Heterogeneity within the Lines
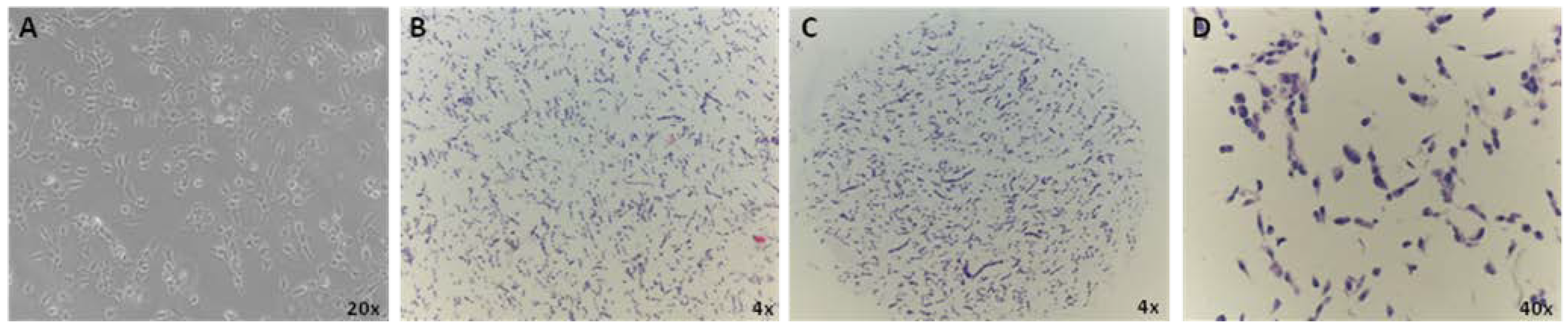
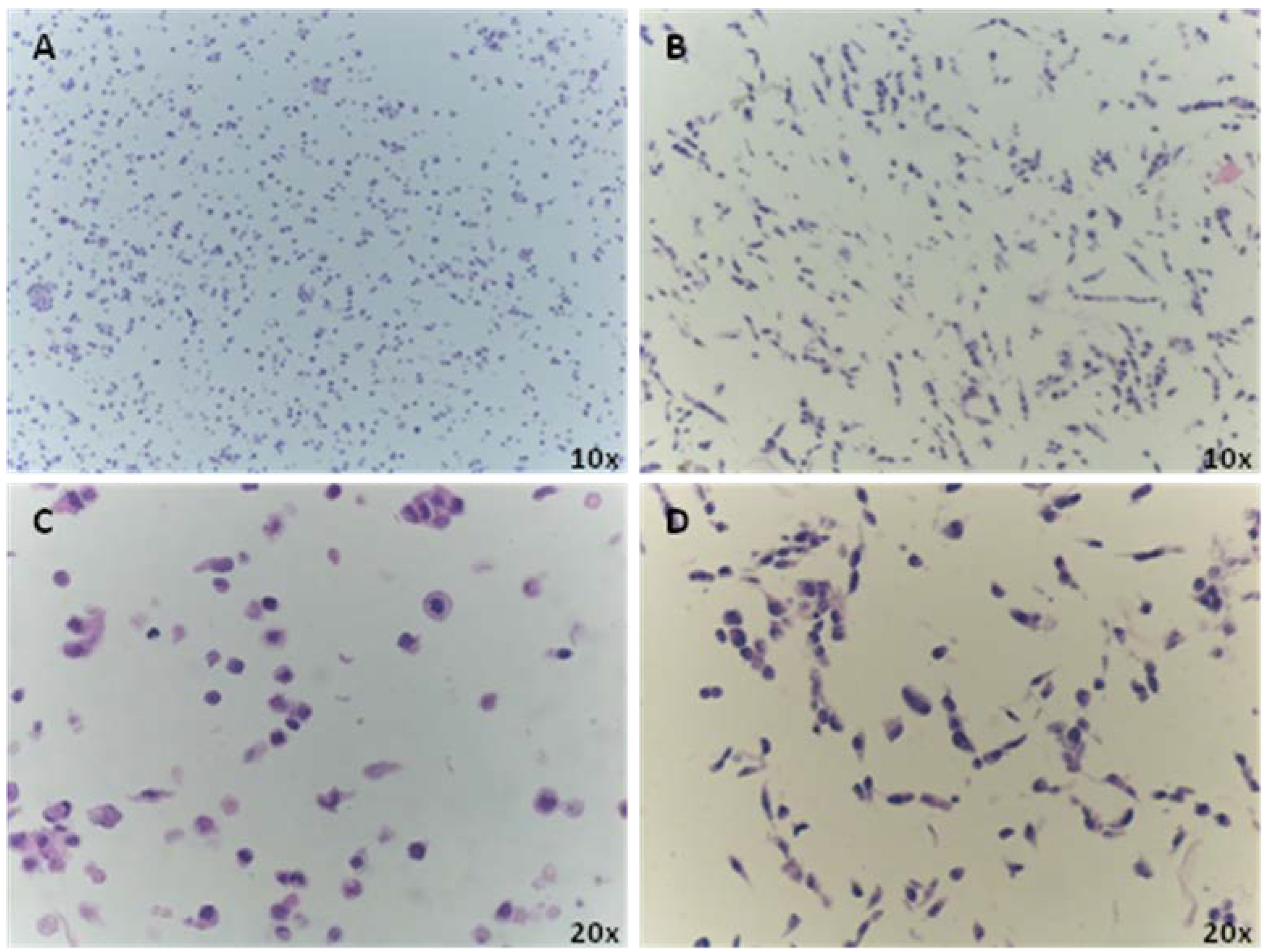
3.2. Immunocharacterization of AF22 iPS-Derived Cells by CMA Technology versus Cover Slip

4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Warford, A.; Howat, W.; McCafferty, J. Expression profiling by high-throughput immunohistochemistry. J. Immunol. Methods 2004, 290, 81–92. [Google Scholar]
- Takikita, M.; Chung, J.Y.; Hewitt, S.M. Tissue microarrays enabling high-throughput molecular pathology. Curr. Opin. Biotechnol. 2007, 18, 318–325. [Google Scholar] [CrossRef]
- Kallioniemi, O.P.; Wagner, U.; Kononen, J.; Sauter, G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum. Mol. Genet. 2001, 10, 657–662. [Google Scholar] [CrossRef]
- Torhorst, J.; Bucher, C.; Kononen, J.; Haas, P.; Zuber, M.; Köchli, O.R.; Mross, F.; Dieterich, H.; Moch, H.; Mihatsch, M.; Kallioniemi, O.P.; Sauter, G. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am. J. Pathol. 2001, 159, 2249–2256. [Google Scholar] [CrossRef]
- Gillett, C.E.; Springall, R.J.; Barnes, D.M.; Hanby, A.M. Multiple tissue core arrays in histopathology research: a validation study. J. Pathol. 2000, 192, 549–553. [Google Scholar]
- Burandt, E.; Schreiber, M.; Stein, A.; Minner, S.; Clauditz, T.S.; Bokemeyer, C.; Jänicke, F.; Fisch, M.; Izbicki, J.R.; Knecht, R.; et al. Continuous tissue microarray based identification of cancers with homogeneous target expression for successful targeted therapy in clinical routine practice. Gene. Chromosome. Canc. 2014, 53, 228–239. [Google Scholar] [CrossRef]
- Waterworth, A.; Hanby, A.; Speirs, V. A novel cell array technique for high-throughput, cell-based analysis. In Vitro Cell Dev. Biol. Anim. 2005, 41, 185–187. [Google Scholar] [CrossRef]
- Kim, M.S.; Kuppireddy, S.V.; Sakamuri, S.; Singal, M.; Getnet, D.; Harsha, H.C.; Goel, R.; Balakrishnan, L.; Jacob, H.K.; Kashyap, M.K.; et al. Rapid characterization of candidate biomarkers for pancreatic cancer using cell microarrays (CMAs). J. Proteome Res. 2012, 11, 5556–5563. [Google Scholar] [CrossRef]
- Takahashi, K.; Okita, K.; Nakagawa, M.; Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007, 2, 3081–3089. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Phillips, B.W.; Crook, J.M. Pluripotent human stem cells: A novel tool in drug discovery. BioDrugs 2010, 24, 99–108. [Google Scholar]
- Andersson, A.C.; Strömberg, S.; Bäckvall, H.; Kampf, C.; Uhlen, M.; Wester, K.; Pontén, F. Analysis of protein expression in cell microarrays: A tool for antibody-based proteomics. J. Histochem. Cytochem. 2006, 54, 1413–1423. [Google Scholar]
- Falk, A.; Koch, P.; Kesavan, J.; Takashima, Y.; Ladewig, J.; Alexander, M.; Wiskow, O.; Tailor, J.; Trotter, M.; Pollard, S.; et al. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One 2012, 7, e29597. [Google Scholar] [CrossRef]
- Koch, P.; Opitz, T.; Steinbeck, J.A.; Ladewig, J.; Brüstle, O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. USA 2009, 106, 3225–3230. [Google Scholar]
- Marchenko, S.; Flanagan, L. Immunocytochemistry: human neural stem cells. J. Vis. Exp. 2007, 7, 267. [Google Scholar]
- Orlandi, R.; Cattaneo, M.; Troglio, F.; Campiglio, M.; Biunno, I.; Ménard, S. Production of a monoclonal antibody directed against the recombinant SEL1L protein. Int. J. Biol. Markers 2002, 17, 104–111. [Google Scholar]
- Jensen, J.B.; Parmar, M. Strengths and limitations of the neurosphere culture system. Mol Neurobiol 2006, 34, 153–61. [Google Scholar] [CrossRef]
- Ferrer, B.; Bermudo, R.; Thomson, T.; Nayach, I.; Soler, M.; Sánchez, M.; Castillo, M.; Calvo, J.; Campo, E.; Fernández, P.L. Paraffin-Embedded Cell Line Microarray (PECLIMA): Development and Validation of a High-Throughput Method for Antigen Profiling of Cell Lines. Pathobiology 2005, 72, 225–232. [Google Scholar] [CrossRef]
- Yamazoe, H.; Iwata, H. Cell microarray for screening feeder cells for differentiation of embryonic stem cells. J. Biosci. Bioeng. 2005, 100, 292–296. [Google Scholar] [CrossRef]
- Kiskinis, E.; Eggan, K. Progress toward the clinical application of patient-specific pluripotent stem cells. J. Clin. Invest. 2010, 120, 51–59. [Google Scholar] [CrossRef]
- Choong, C.; Rao, M.S. Human embryonic stem cells. Neurosurg. Clin. N. Am. 2007, 18, 1–14. [Google Scholar] [CrossRef]
- Cardano, M.; Diaferia, G.R.; Falavigna, M.; Spinelli, C.C.; Sessa, F.; De Blasio, P.; Biunno, I. Cell and tissue microarray technologies for protein and nucleic acid expression profiling. J. Histochem. Cytochem. 2013, 61, 116–124. [Google Scholar] [CrossRef]
- Pilla, D.; Bosisio, F.M.; Marotta, R.; Faggi, S.; Forlani, P.; Falavigna, M.; Biunno, I.; Martella, E.; De Blasio, P.; Borghesi, S.; Cattoretti, G. Tissue microarray design and construction for scientific, industrial and diagnostic use. J. Pathol. Inform. 2012, 3, 42. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
La Spada, A.; Rainoldi, B.; De Blasio, A.; Biunno, I. Application of Tissue Microarray Technology to Stem Cell Research. Microarrays 2014, 3, 159-167. https://doi.org/10.3390/microarrays3030159
La Spada A, Rainoldi B, De Blasio A, Biunno I. Application of Tissue Microarray Technology to Stem Cell Research. Microarrays. 2014; 3(3):159-167. https://doi.org/10.3390/microarrays3030159
Chicago/Turabian StyleLa Spada, Alberto, Barnaba Rainoldi, Andrea De Blasio, and Ida Biunno. 2014. "Application of Tissue Microarray Technology to Stem Cell Research" Microarrays 3, no. 3: 159-167. https://doi.org/10.3390/microarrays3030159




