Qualitative and Quantitative Requirements for Assessing Prognostic Markers in Prostate Cancer
Abstract
:1. Introduction
2. Experimental Section
| Parameter | No. of patients (%) | |
|---|---|---|
| Study cohort on TMA | Biochemical relapse among categories | |
| (n = 12,427) | ||
| Follow-up (mo) | ||
| n | 11,665 (93.9%) | 2769 (23.7%) |
| Mean | 48.9 | - |
| Median | 36.4 | - |
| Age (y) | ||
| ≤50 | 334 (2.7%) | 81 (24.3%) |
| 51–59 | 3061 (24.8%) | 705 (23%) |
| 60–69 | 7188 (58.2%) | 1610 (22.4%) |
| ≥70 | 1761 (14.3%) | 370 (21%) |
| Pretreatment prostate specific antigen (PSA) (ng/mL) | ||
| <4 | 1585 (12.9%) | 242 (15.3%) |
| 4–10 | 7480 (60.9%) | 1355 (18.1%) |
| 10–20 | 2412 (19.6%) | 737 (30.6%) |
| >20 | 812 (6.6%) | 397 (48.9%) |
| pT category (AJCC 2002) | ||
| pT2 | 8187 (66.2%) | 1095 (13.4%) |
| pT3a | 2660 (21.5%) | 817 (30.7%) |
| pT3b | 1465 (11.8%) | 796 (54.3%) |
| pT4 | 63 (0.5%) | 51 (81%) |
| Gleason grade | ||
| ≤3 + 3 | 2983 (24.1%) | 368 (12.3%) |
| 3 + 4 | 6945 (56.2%) | 1289 (18.6%) |
| 4 + 3 | 1848 (15%) | 788 (42.6%) |
| ≥4 + 4 | 584 (4.7%) | 311 (53.3%) |
| pN category | ||
| pN0 | 6970 (91%) | 1636 (23.5%) |
| pN+ | 693 (9%) | 393 (56.7%) |
| Surgical margin | ||
| Negative | 9990 (81.9%) | 1848 (18.5%) |
| Positive | 2211 (18.1%) | 853 (38.6%) |
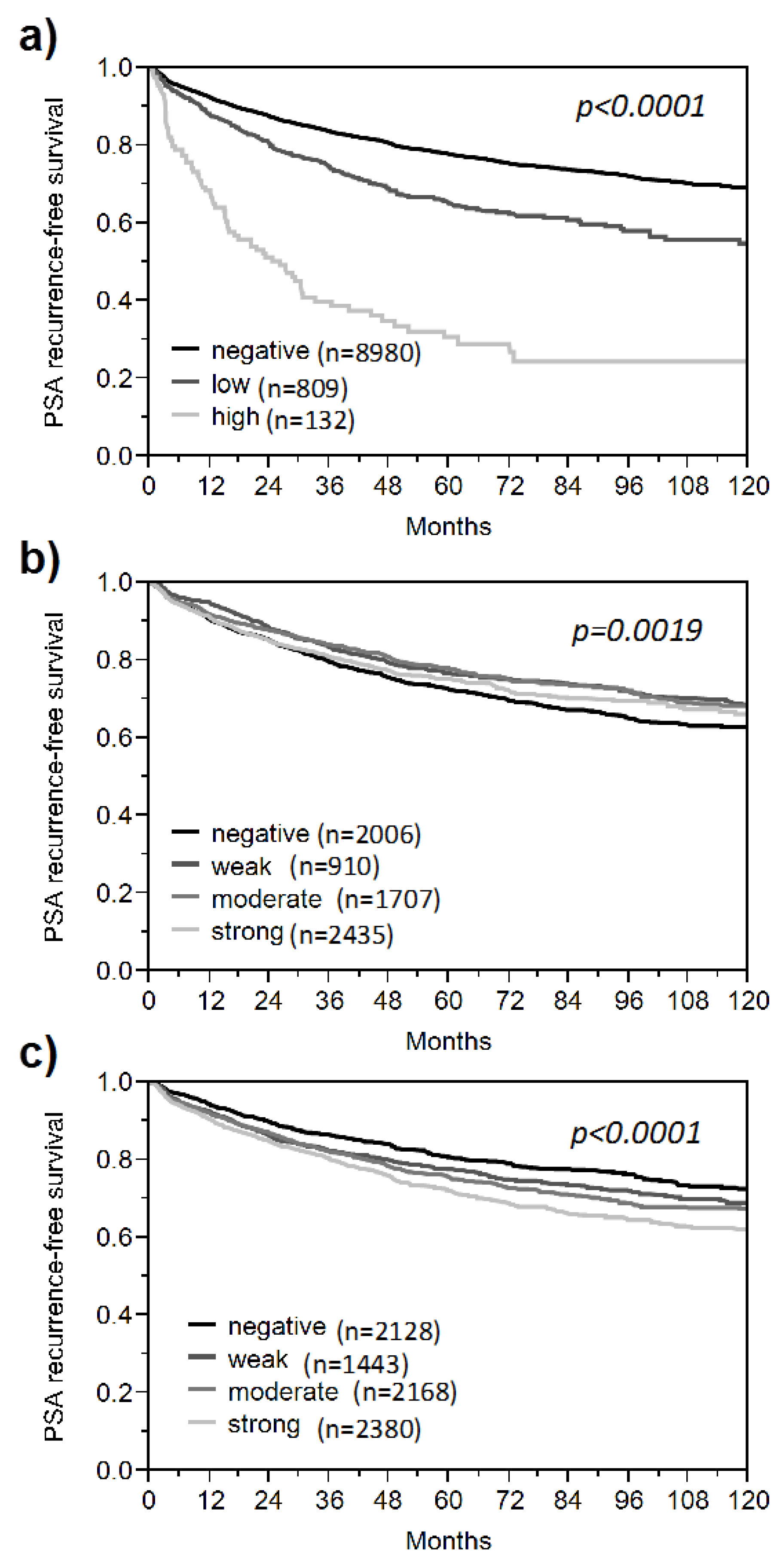
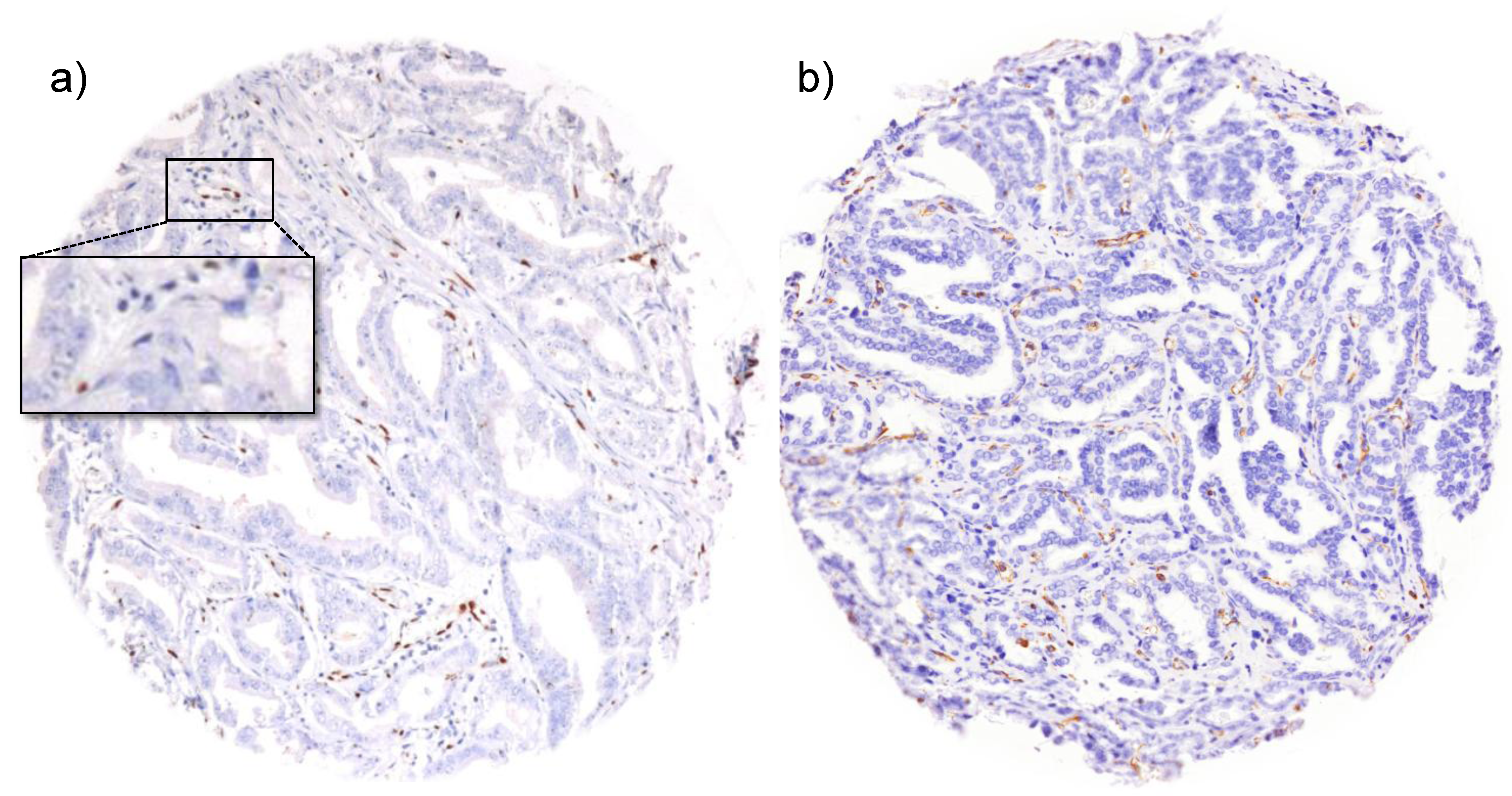
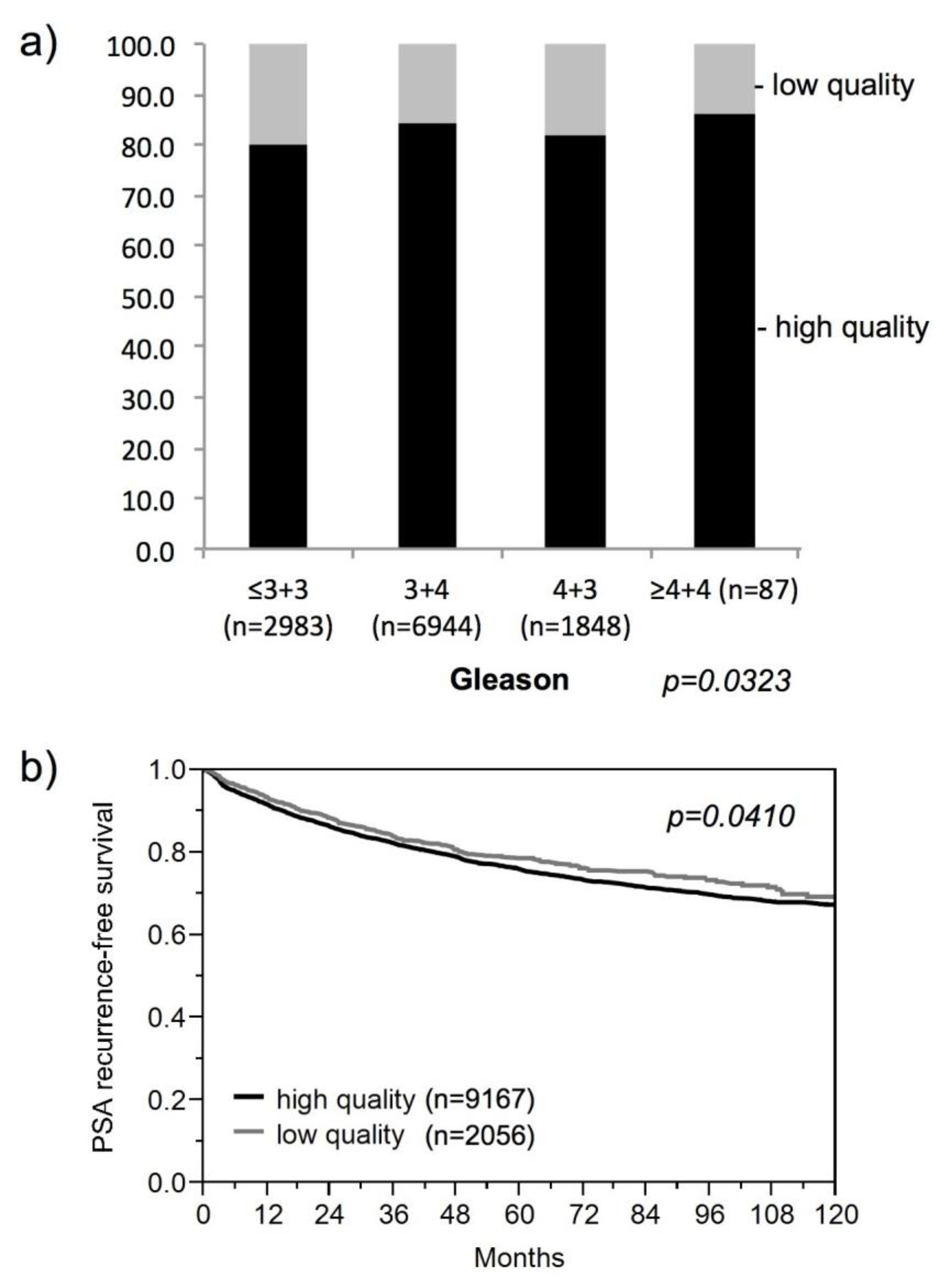
3. Results and Discussion
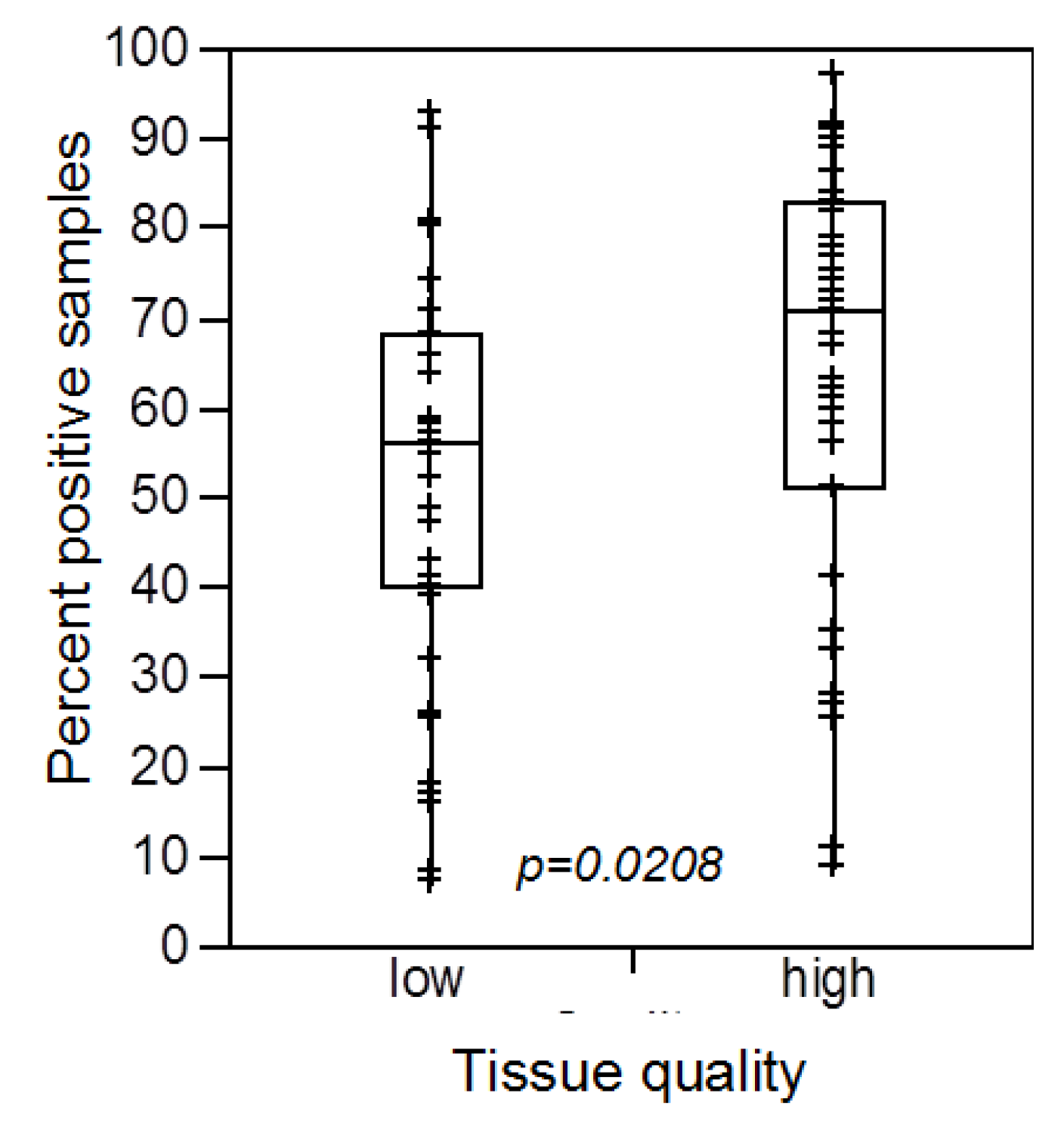
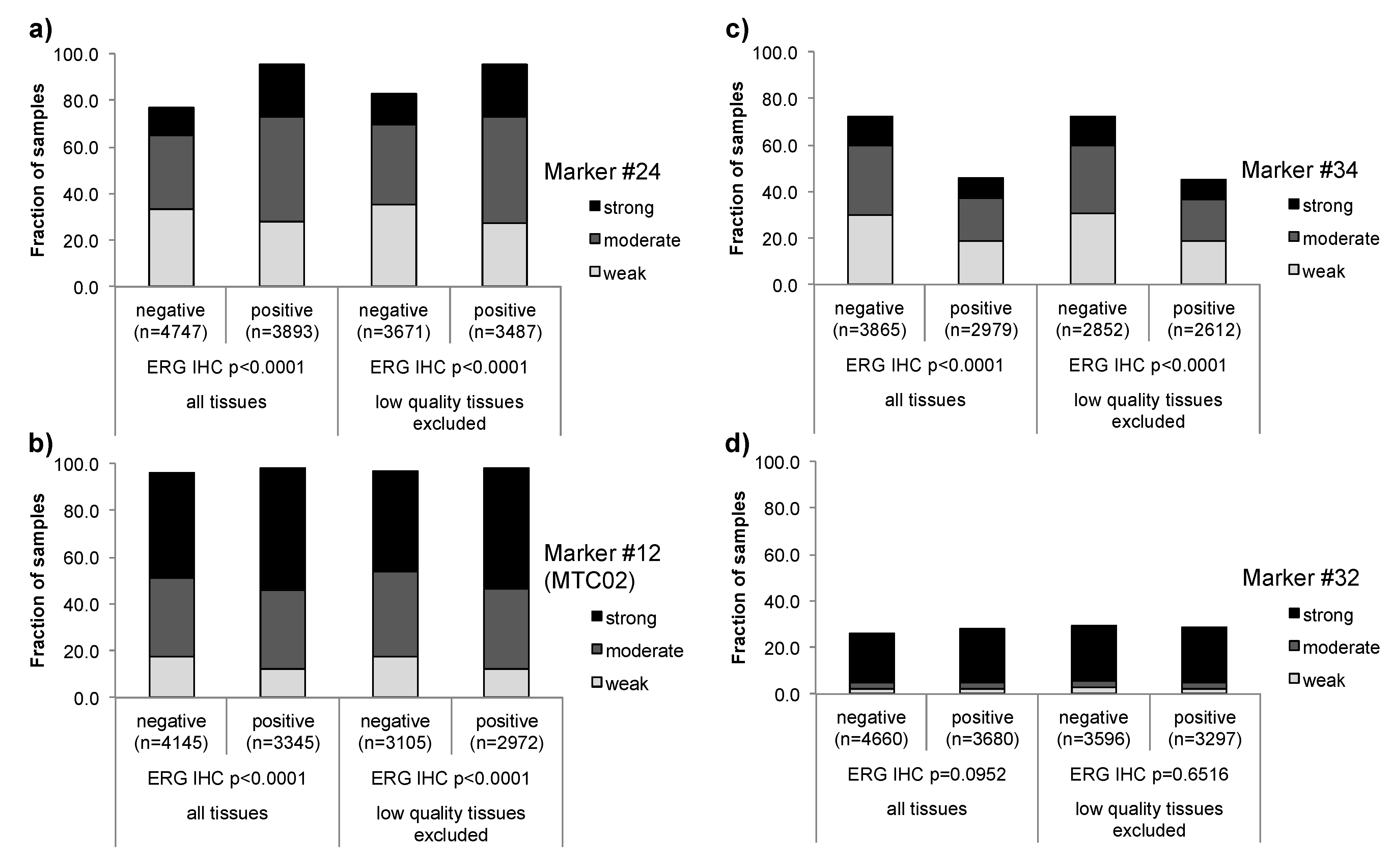
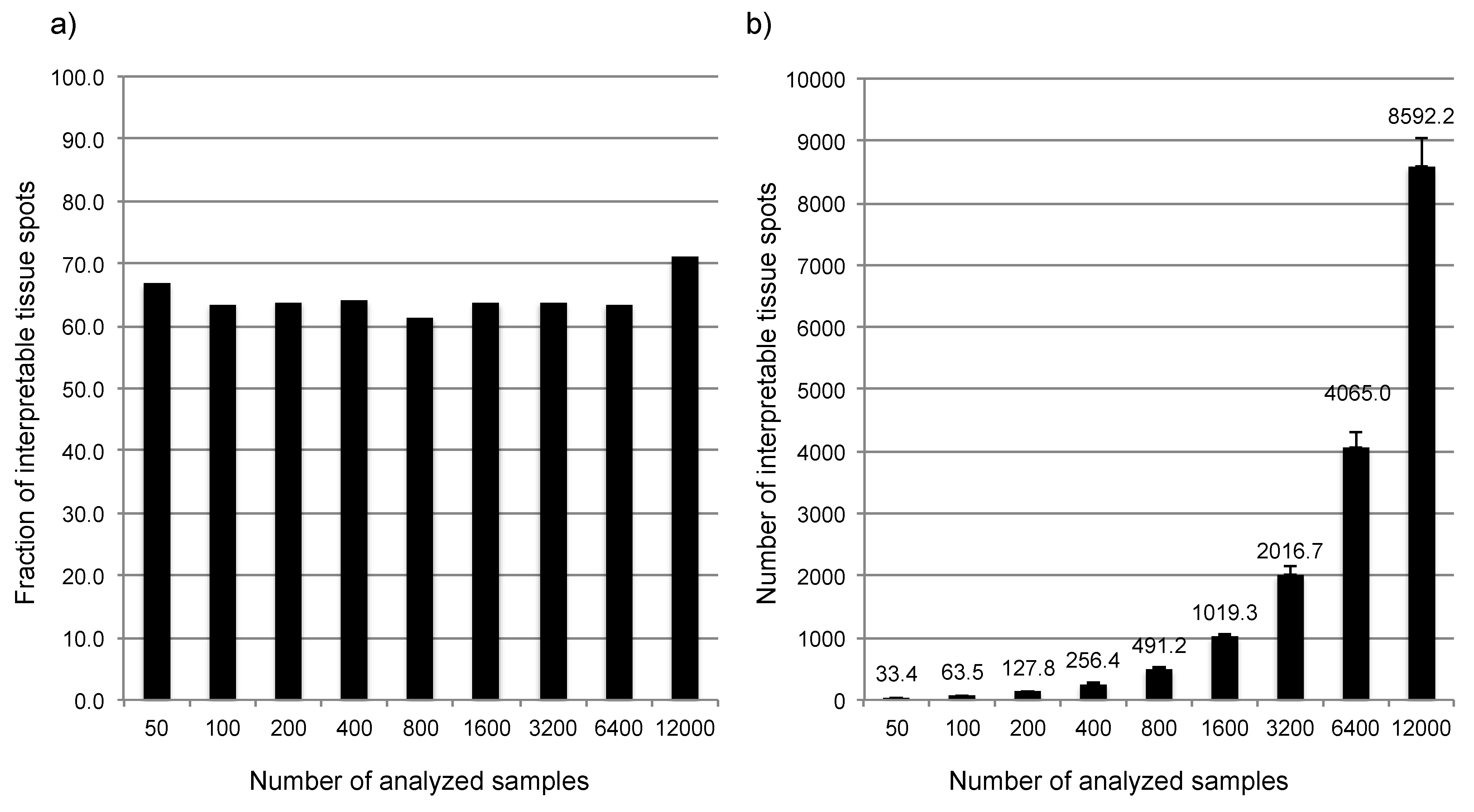
3.1. Impact of the Tissue Quality
| Marker | Low quality tissue | Number of analyzed tissue spots | Association strength | |||
|---|---|---|---|---|---|---|
| 1600 | 3200 | 6400 | 10,000 | |||
| Marker #32 | included | 0.1039 | 0.5243 | 0.1879 | 0.0952 | No association |
| excluded | 0.9431 | 0.6612 | 0.7653 | 0.6515 | ||
| Marker #13 | included | 0.0112 | <0.0001 | <0.0001 | <0.0001 | Weak |
| excluded | 0.012 | 0.0004 | <0.0001 | <0.0001 | ||
| Marker #12 (MTC02) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Weak |
| excluded | 0.0238 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #31 | included | 0.1307 | 0.0091 | <0.0001 | <0.0001 | Weak |
| excluded | 0.2414 | 0.0031 | <0.0001 | <0.0001 | ||
| Marker #37 | included | 0.0017 | <0.0001 | <0.0001 | <0.0001 | Weak |
| excluded | 0.1159 | 0.0012 | 0.0008 | <0.0001 | ||
| Marker #7 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #10 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #2 (CD10) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #21 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #27 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #39 (p53) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #4 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker#3 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #5 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #33 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #16 (NBS1) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #18 (AR) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #22 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #24 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #23 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #36 (KPNA2) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #35 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #30 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Moderate |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #6 (FOXP2) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #8 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #9 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #11 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #14 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #15 (LPCAT) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #19 (RBM3) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #26 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #26 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #25 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #29 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Strong |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #1 (CD147) | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Inverse |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #17 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Inverse |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #20 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Inverse |
| excluded | 0.0004 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #34 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Inverse |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker #38 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 | Inverse |
| excluded | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Marker | Low quality tissue | Number of analyzed tissue spots | |||
|---|---|---|---|---|---|
| 1600 | 3200 | 6400 | 10,000 | ||
| #2 (CD10) | included | 0.0937 | 0.0006 | <0.0001 | <0.0001 |
| excluded | 0.1215 | 0.0005 | <0.0001 | <0.0001 | |
| #3 | included | 0.0761 | 0.0946 | 0.0595 | 0.0037 |
| excluded | 0.1146 | 0.0761 | 0.0385 | 0.0043 | |
| #4 | included | 0.0810 | 0.1082 | 0.0151 | <0.0001 |
| excluded | 0.1059 | 0.0810 | 0.0060 | <0.0001 | |
| #18 (AR) | included | 0.0082 | 0.0206 | 0.0006 | <0.0001 |
| excluded | 0.0197 | 0.0082 | 0.0003 | <0.0001 | |
| #35 | included | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| excluded | 0.0230 | <0.0001 | <0.0001 | <0.0001 | |
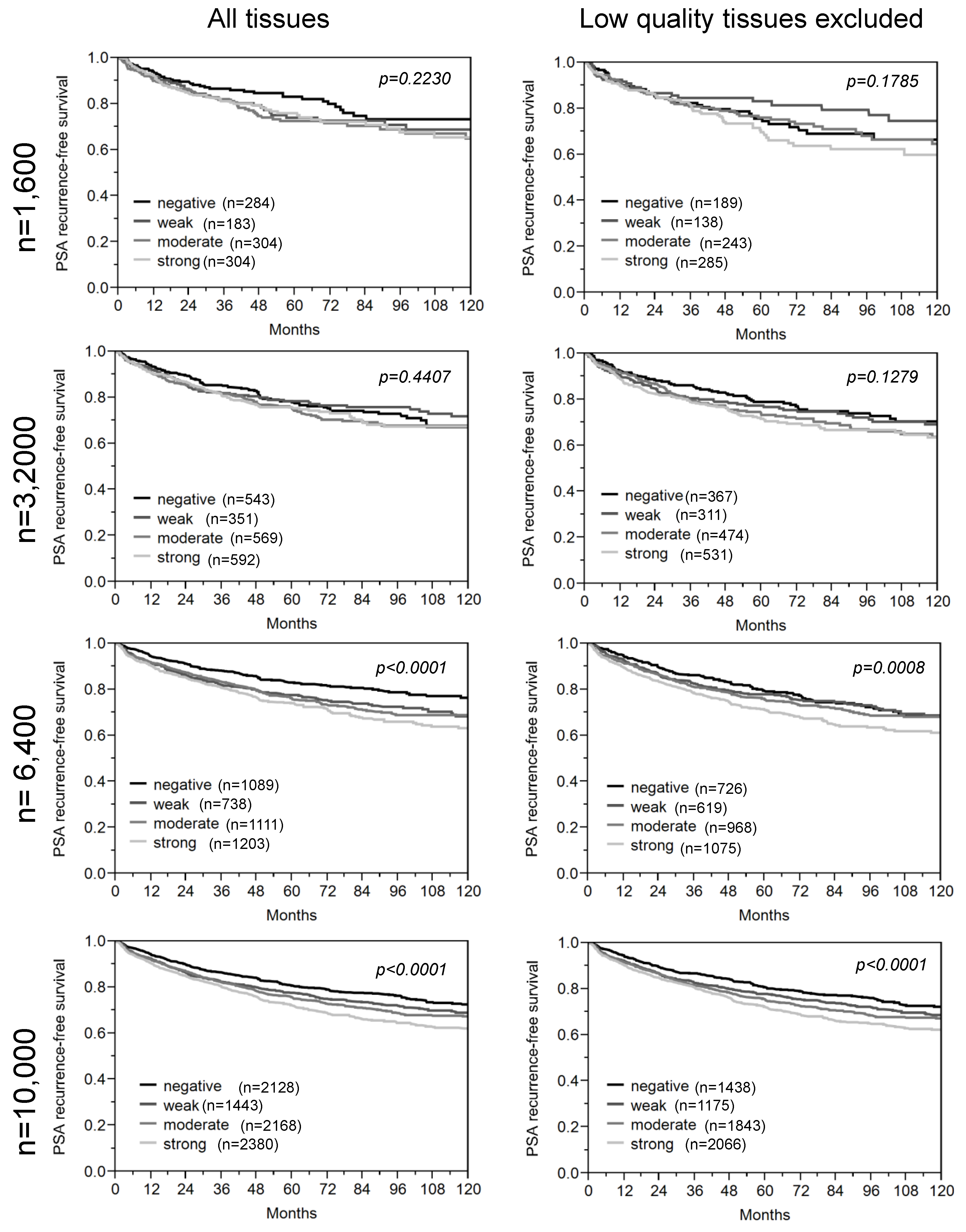
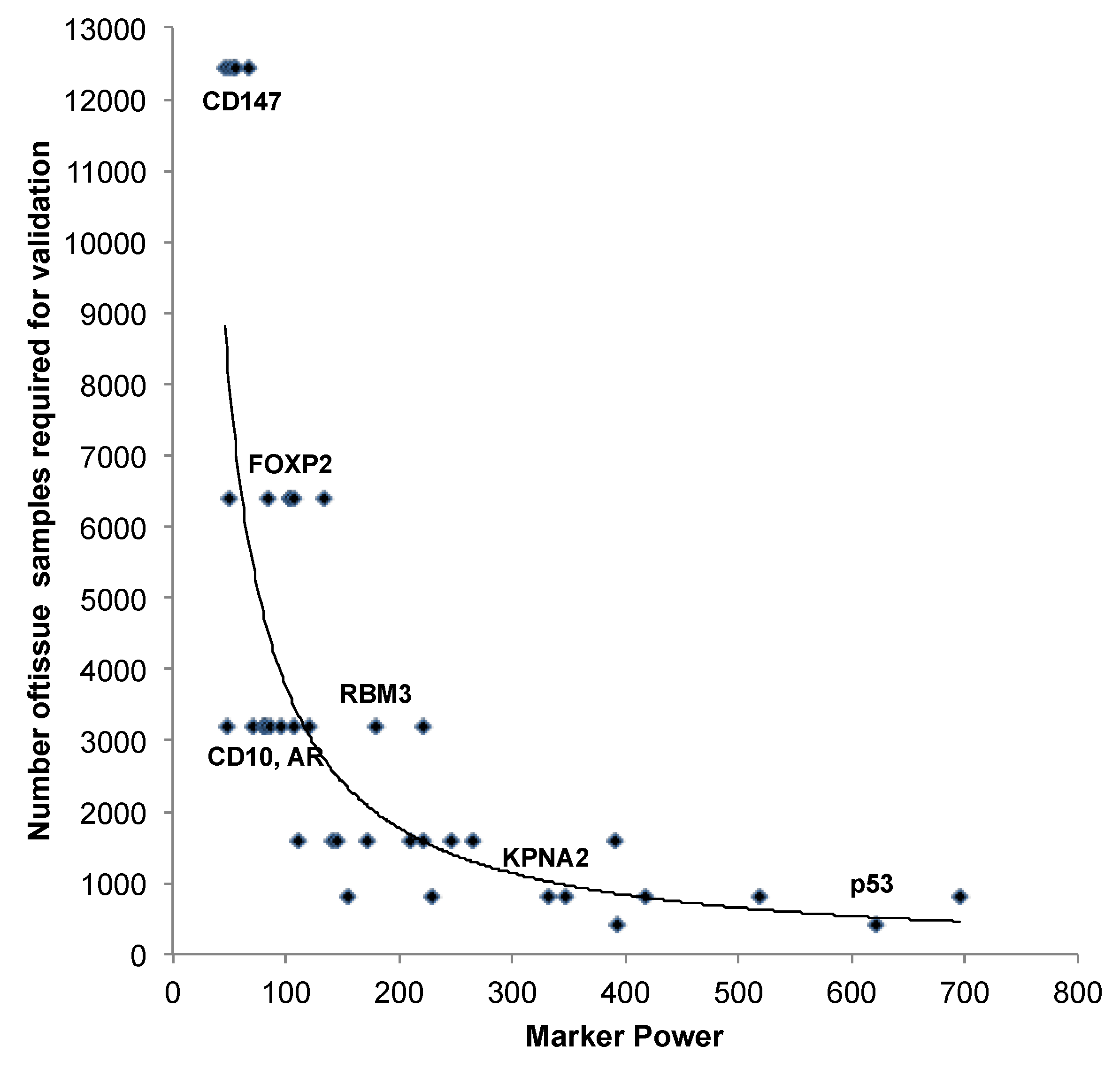
3.2. Impact of the Sample Size
| Marker | n analy-zable | Number of analyzed tissue spots | Marker Power | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 100 | 200 | 400 | 800 | 1600 | 3200 | 6400 | 12,427 | |||
| Marker #1 (CD147) | 7605 | 0.2417 | 0.4218 | 0.4550 | 0.4984 | 0.5396 | 0.3539 | 0.3428 | 0.0379 | 0.0019 | weak |
| Marker #4 | 8025 | 0.0863 | 0.0998 | 0.9252 | 0.4330 | 0.4648 | 0.5781 | 0.0571 | 0.0666 | 0.0002 | weak |
| Marker #3 | 7574 | 0.2276 | 0.0720 | 0.0673 | 0.7038 | 0.6825 | 0.0934 | 0.3195 | 0.0351 | 0.0016 | weak |
| Marker #5 | 9473 | 0.8553 | 0.2279 | 0.9171 | 0.5532 | 0.1026 | 0.3192 | 0.0797 | 0.0097 | 0.0016 | weak |
| Marker #6 (FOXP2) | 8284 | 0.7963 | 0.4436 | 0.0082 | 0.4058 | 0.4309 | 0.0776 | 0.0308 | 0.0011 | <0.0001 | weak |
| Marker #7 | 9485 | <0.0001 | 0.4999 | 0.0056 | 0.2196 | 0.0492 | 0.0464 | 0.0796 | 0.0007 | <0.0001 | moderate |
| Marker #8 | 8158 | 0.1175 | 0.5066 | 0.7638 | 0.1192 | 0.3055 | 0.0020 | 0.0548 | <0.0001 | <0.0001 | moderate |
| Marker #9 | 6494 | 0.0494 | 0.9606 | 0.5709 | 0.1780 | 0.0603 | 0.0421 | 0.0564 | <0.0001 | <0.0001 | moderate |
| Marker #33 | 9262 | 0.1088 | 0.2166 | 0.5932 | 0.6114 | 0.0017 | 0.0013 | 0.0105 | <0.0001 | <0.0001 | moderate |
| Marker #10 | 9516 | 0.6617 | 0.8651 | 0.0836 | 0.1074 | 0.1415 | 0.0489 | 0.0041 | 0.0034 | <0.0001 | weak |
| Marker #2 (CD10) | 8488 | 0.0722 | 0.5977 | 0.6268 | 0.2465 | 0.3409 | 0.5137 | 0.0001 | 0.0062 | 0.0012 | weak |
| Marker #11 | 9627 | 0.5518 | 0.1662 | 0.0652 | 0.4009 | 0.5480 | 0.1911 | 0.0009 | 0.0046 | <0.0001 | weak |
| Marker #12 (MTC02) | 8407 | 0.4313 | 0.9545 | 0.7656 | 0.9315 | 0.3693 | 0.2517 | 0.0013 | 0.0004 | <0.0001 | weak |
| Marker #14 | 8654 | 0.7139 | 0.0146 | 0.6741 | 0.4165 | 0.1473 | 0.4119 | 0.0094 | 0.0002 | <0.0001 | weak |
| Marker #16 (NBS1) | 8026 | 0.6506 | 0.0525 | 0.2071 | 0.7379 | 0.8382 | 0.2079 | 0.0004 | 0.0026 | <0.0001 | weak |
| Marker #13 | 9875 | 0.8336 | 0.3172 | 0.2557 | 0.6717 | 0.0790 | 0.0962 | 0.0026 | 0.0004 | <0.0001 | weak |
| Marker #15 (LPCAT) | 8762 | 0.7141 | 0.1713 | 0.5933 | 0.6978 | 0.2428 | 0.0449 | 0.0020 | 0.0003 | <0.0001 | weak |
| Marker #17 | 7275 | 0.7259 | 0.2301 | 0.2169 | 0.3562 | 0.3492 | 0.0661 | 0.0094 | <0.0001 | <0.0001 | moderate |
| Marker #18 (AR) | 7856 | 0.4576 | 0.0246 | 0.9451 | 0.6439 | 0.7573 | 0.3058 | <0.0001 | 0.0034 | <0.0001 | moderate |
| Marker #20 | 6638 | 0.7489 | 0.7507 | 0.2524 | 0.4930 | 0.9583 | 0.0863 | 0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #19 (RBM3) | 8303 | 0.4712 | 0.1035 | 0.4893 | 0.2494 | 0.0134 | 0.0658 | 0.0006 | <0.0001 | <0.0001 | moderate |
| Marker #21 | 9643 | 0.8921 | 0.6673 | 0.2718 | 0.0004 | 0.0086 | 0.0170 | <0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #28 | 6824 | 0.5465 | 0.0252 | 0.8696 | 0.6919 | 0.2038 | 0.0075 | 0.0026 | <0.0001 | <0.0001 | moderate |
| Marker #22 | 7117 | 0.2824 | 0.8367 | 0.5302 | 0.9432 | 0.0591 | 0.0095 | <0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #24 | 9756 | 0.5724 | 0.3132 | 0.8763 | 0.6290 | 0.0686 | 0.0016 | <0.0001 | 0.0002 | <0.0001 | moderate |
| Marker #23 | 9744 | 0.3087 | 0.0266 | 0.0008 | 0.1771 | 0.3495 | 0.0001 | 0.0004 | <0.0001 | <0.0001 | moderate |
| Marker #29 | 9403 | 0.8852 | 0.0612 | 0.1260 | 0.1830 | 0.8574 | 0.0028 | <0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #27 | 7588 | 0.0607 | 0.2364 | 0.2661 | 0.0050 | 0.4899 | 0.0001 | <0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #26 | 9633 | 0.0169 | 0.5727 | 0.1760 | 0.7661 | 0.0491 | 0.0003 | <0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #25 | 7677 | 0.8371 | 0.3814 | 0.7065 | 0.0233 | 0.9297 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | moderate |
| Marker #30 | 8174 | 0.3570 | 0.5619 | 0.3006 | 0.0028 | 0.1186 | 0.0002 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #31 | 10215 | 0.4072 | 0.4062 | 0.2342 | 0.3143 | 0.0040 | 0.0023 | 0.0002 | <0.0001 | <0.0001 | moderate |
| Marker #35 | 10216 | 0.8828 | 0.4813 | 0.1523 | 0.1304 | 0.0018 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #34 | 7670 | 0.0833 | 0.3010 | 0.2137 | 0.0703 | 0.0001 | 0.0002 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #37 | 7822 | 0.0044 | 0.0106 | 0.1209 | 0.0247 | 0.0002 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #32 | 9467 | 0.8994 | 0.3867 | 0.0006 | 0.0640 | <0.0001 | 0.0004 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #36 (KPNA2) | 7943 | 0.1490 | 0.3803 | 0.0483 | 0.0044 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #39 (p53) | 10946 | 0.2686 | 0.1962 | 0.0192 | <0.0001 | 0.0055 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | strong |
| Marker #38 | 9576 | 0.7878 | 0.3611 | 0.0091 | 0.0063 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | strong |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Graefen, M.; Ahyai, S.; Heuer, R.; Salomon, G.; Schlomm, T.; Isbarn, H.; Budaus, L.; Heinzer, H.; Huland, H. Active surveillance for prostate cancer. Urologe A 2008, 47, 261–269. [Google Scholar] [CrossRef]
- Cooperberg, M.R.; Simko, J.P.; Cowan, J.E.; Reid, J.E.; Djalilvand, A.; Bhatnagar, S.; Gutin, A.; Lanchbury, J.S.; Swanson, G.P.; Stone, S.; et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J. Clin. Oncol. 2013, 31, 1428–1434. [Google Scholar] [CrossRef]
- Cuzick, J.; Swanson, G.P.; Fisher, G.; Brothman, A.R.; Berney, D.M.; Reid, J.E.; Mesher, D.; Speights, V.O.; Stankiewicz, E.; Foster, C.S.; et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011, 12, 245–255. [Google Scholar] [CrossRef]
- Badani, K.; Thompson, D.J.; Buerki, C.; Davicioni, E.; Garrison, J.; Ghadessi, M.; Mitra, A.P.; Wood, P.J.; Hornberger, J. Impact of a genomic classifier of metastatic risk on postoperative treatment recommendations for prostate cancer patients: A report from the decide study group. Oncotarget 2013, 4, 600–609. [Google Scholar]
- Schlomm, T.; Erbersdobler, A.; Mirlacher, M.; Sauter, G. Molecular staging of prostate cancer in the year 2007. World J. Urol. 2007, 25, 19–30. [Google Scholar] [CrossRef]
- Zellweger, T.; Ninck, C.; Bloch, M.; Mirlacher, M.; Koivisto, P.A.; Helin, H.J.; Mihatsch, M.J.; Gasser, T.C.; Bubendorf, L. Expression patterns of potential therapeutic targets in prostate cancer. Int. J. Canc. 2005, 113, 619–628. [Google Scholar] [CrossRef]
- Schlomm, T.; Iwers, L.; Kirstein, P.; Jessen, B.; Kollermann, J.; Minner, S.; Passow-Drolet, A.; Mirlacher, M.; Milde-Langosch, K.; Graefen, M.; et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod. Pathol. 2008, 21, 1371–1379. [Google Scholar] [CrossRef]
- Vergis, R.; Corbishley, C.M.; Thomas, K.; Horwich, A.; Huddart, R.; Khoo, V.; Eeles, R.; Sydes, M.R.; Cooper, C.S.; Dearnaley, D.; Parker, C. Expression of Bcl-2, p53, and MDM2 in localized prostate cancer with respect to the outcome of radical radiotherapy dose escalation. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 35–41. [Google Scholar] [CrossRef]
- Kudahetti, S.; Fisher, G.; Ambroisine, L.; Foster, C.; Reuter, V.; Eastham, J.; Moller, H.; Kattan, M.W.; Cooper, C.S.; Scardino, P.; Cuzick, J.; Berney, D.M. P53 immunochemistry is an independent prognostic marker for outcome in conservatively treated prostate cancer. BJU Int. 2009, 104, 20–24. [Google Scholar] [CrossRef]
- Uzoaru, I.; Rubenstein, M.; Mirochnik, Y.; Slobodskoy, L.; Shaw, M.; Guinan, P. An evaluation of the markers p53 and Ki-67 for their predictive value in prostate cancer. J. Surg. Oncol. 1998, 67, 33–37. [Google Scholar] [CrossRef]
- Incognito, L.S.; Cazares, L.H.; Schellhammer, P.F.; Kuban, D.A.; van Dyk, E.O.; Moriarty, R.P.; Wright, G.L., Jr.; Somers, K.D. Overexpression of p53 in prostate carcinoma is associated with improved overall survival but not predictive of response to radiotherapy. Int. J. Oncol. 2000, 17, 761–769. [Google Scholar]
- Han, B.; Mehra, R.; Lonigro, R.J.; Wang, L.; Suleman, K.; Menon, A.; Palanisamy, N.; Tomlins, S.A.; Chinnaiyan, A.M.; Shah, R.B. Fluorescence in situ hybridization study shows association of PTEN deletion with erg rearrangement during prostate cancer progression. Mod. Pathol. 2009, 22, 1083–1093. [Google Scholar] [CrossRef]
- McCall, P.; Witton, C.J.; Grimsley, S.; Nielsen, K.V.; Edwards, J. Is pten loss associated with clinical outcome measures in human prostate cancer? Br. J. Canc. 2008, 99, 1296–1301. [Google Scholar] [CrossRef]
- Sircar, K.; Yoshimoto, M.; Monzon, F.A.; Koumakpayi, I.H.; Katz, R.L.; Khanna, A.; Alvarez, K.; Chen, G.; Darnel, A.D.; Aprikian, A.G.; et al. TEN genomic deletion is associated with p-akt and ar signalling in poorer outcome, hormone refractory prostate cancer. J. Pathol. 2009, 218, 505–513. [Google Scholar] [CrossRef]
- Yoshimoto, M.; Cunha, I.W.; Coudry, R.A.; Fonseca, F.P.; Torres, C.H.; Soares, F.A.; Squire, J.A. Fish analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br. J. Canc. 2007, 97, 678–685. [Google Scholar] [CrossRef]
- Reid, A.H.; Attard, G.; Ambroisine, L.; Fisher, G.; Kovacs, G.; Brewer, D.; Clark, J.; Flohr, P.; Edwards, S.; Berney, D.M.; et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br. J. Canc. 2010, 102, 678–684. [Google Scholar] [CrossRef]
- Koumakpayi, I.H.; Le Page, C.; Mes-Masson, A.M.; Saad, F. Hierarchical clustering of immunohistochemical analysis of the activated ErbB/PI3K/Akt/NF-kappaB signalling pathway and prognostic significance in prostate cancer. Br. J. Canc. 102, 1163–1173.
- Bedolla, R.; Prihoda, T.J.; Kreisberg, J.I.; Malik, S.N.; Krishnegowda, N.K.; Troyer, D.A.; Ghosh, P.M. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and akt activation. Clin. Canc. Res. 2007, 13, 3860–3867. [Google Scholar] [CrossRef]
- Osman, I.; Dai, J.; Mikhail, M.; Navarro, D.; Taneja, S.S.; Lee, P.; Christos, P.; Shen, R.; Nanus, D.M. Loss of neutral endopeptidase and activation of protein kinase b (AKT) is associated with prostate cancer progression. Cancer 2006, 107, 2628–2636. [Google Scholar] [CrossRef]
- McMenamin, M.E.; Soung, P.; Perera, S.; Kaplan, I.; Loda, M.; Sellers, W.R. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high gleason score and advanced stage. Canc. Res. 1999, 59, 4291–4296. [Google Scholar]
- Bertram, J.; Peacock, J.W.; Fazli, L.; Mui, A.L.; Chung, S.W.; Cox, M.E.; Monia, B.; Gleave, M.E.; Ong, C.J. Loss of PTEN is associated with progression to androgen independence. Prostate 2006, 66, 895–902. [Google Scholar] [CrossRef]
- Thomas, G.V.; Horvath, S.; Smith, B.L.; Crosby, K.; Lebel, L.A.; Schrage, M.; Said, J.; de Kernion, J.; Reiter, R.E.; Sawyers, C.L. Antibody-based profiling of the phosphoinositide 3-kinase pathway in clinical prostate cancer. Clin. Canc. Res. 2004, 10, 8351–8356. [Google Scholar] [CrossRef]
- Sauter, G.; Lee, J.; Bartlett, J.M.; Slamon, D.J.; Press, M.F. Guidelines for human epidermal growth factor receptor 2 testing: Biologic and methodologic considerations. J. Clin. Oncol. 2009, 27, 1323–1333. [Google Scholar] [CrossRef]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef]
- Schlomm, T.; Kirstein, P.; Lwers, L.; Daniel, B.; Steuber, T.; Walz, J.; Chun, F.H.K.; Haese, A.; Kollermann, J.; Graefen, M.; et al. Clinical significance of epidermal growth factor receptor protein overexpression and gene copy number gains in prostate cancer. Clin. Canc. Res. 2007, 13, 6579–6584. [Google Scholar] [CrossRef]
- Minner, S.; Jessen, B.; Stiedenroth, L.; Burandt, E.; Kollermann, J.; Mirlacher, M.; Erbersdobler, A.; Eichelberg, C.; Fisch, M.; Brummendorf, T.H.; et al. Low level HER2 overexpression is associated with rapid tumor cell proliferation and poor prognosis in prostate cancer. Clin. Canc. Res. 2010, 16, 1553–1560. [Google Scholar] [CrossRef]
- Kluth, M.; Harasimowicz, S.; Burkhardt, L.; Grupp, K.; Krohn, A.; Prien, K.; Gjoni, J.; Hass, T.; Galal, R.; Graefen, M.; et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int. J. Canc. 2014. [Google Scholar] [CrossRef]
- Muller, J.; Ehlers, A.; Burkhardt, L.; Sirma, H.; Steuber, T.; Graefen, M.; Sauter, G.; Minner, S.; Simon, R.; Schlomm, T.; Michl, U. Loss of p(ser2448)-mTOR expression is linked to adverse prognosis and tumor progression in erg-fusion-positive cancers. Int. J. Canc. 2013, 132, 1333–1340. [Google Scholar] [CrossRef]
- Fleischmann, A.; Schlomm, T.; Huland, H.; Kollermann, J.; Simon, P.; Mirlacher, M.; Salomon, G.; Chun, F.H.; Steuber, T.; Simon, R.; et al. Distinct subcellular expression patterns of neutral endopeptidase (CD10) in prostate cancer predict diverging clinical courses in surgically treated patients. Clin. Canc. Res. 2008, 14, 7838–7842. [Google Scholar] [CrossRef]
- Grupp, K.; Diebel, F.; Sirma, H.; Simon, R.; Breitmeyer, K.; Steurer, S.; Hube-Magg, C.; Prien, K.; Pham, T.; Weigand, P.; et al. SPINK1 expression is tightly linked to 6q15- and 5q21-deleted erg-fusion negative prostate cancers but unrelated to psa recurrence. Prostate 2013, 73, 1690–1698. [Google Scholar]
- Grupp, K.; Habermann, M.; Sirma, H.; Simon, R.; Steurer, S.; Hube-Magg, C.; Prien, K.; Burkhardt, L.; Jedrzejewska, K.; Salomon, G.; et al. High nuclear karyopherin alpha 2 expression is a strong and independent predictor of biochemical recurrence in prostate cancer patients treated by radical prostatectomy. Mod. Pathol. 2014, 27, 96–106. [Google Scholar] [CrossRef]
- Grupp, K.; Kohl, S.; Sirma, H.; Simon, R.; Steurer, S.; Becker, A.; Adam, M.; Izbicki, J.; Sauter, G.; Minner, S.; Schlomm, T.; Tsourlakis, M.C. Cysteine-rich secretory protein 3 overexpression is linked to a subset of PTEN-deleted ERG fusion-positive prostate cancers with early biochemical recurrence. Mod. Pathol. 2013, 26, 733–742. [Google Scholar] [CrossRef]
- Grupp, K.; Boumesli, R.; Tsourlakis, M.C.; Koop, C.; Wilczak, W.; Adam, M.; Sauter, G.; Simon, R.; Izbicki, J.R.; Graefen, M.; et al. The prognostic impact of high nijmegen breakage syndrome (NBS1) gene expression in erg negative prostate cancers lacking PTEN deletion is driven by kpna2 expression. Int. J. Canc. 2014. [Google Scholar] [CrossRef]
- Grupp, K.; Wilking, J.; Prien, K.; Hube-Magg, C.; Sirma, H.; Simon, R.; Steurer, S.; Budaus, L.; Haese, A.; Izbicki, J.; et al. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to erg activation and pten deletions. Eur. J. Canc. 2014, 50, 852–861. [Google Scholar] [CrossRef]
- Grupp, K.; Sanader, S.; Sirma, H.; Simon, R.; Koop, C.; Prien, K.; Hube-Magg, C.; Salomon, G.; Graefen, M.; Heinzer, H.; et al. High lysophosphatidylcholine acyltransferase 1 expression independently predicts high risk for biochemical recurrence in prostate cancers. Mol. Oncol. 2013, 7, 1001–1011. [Google Scholar] [CrossRef]
- Grupp, K.; Jedrzejewska, K.; Tsourlakis, M.C.; Koop, C.; Wilczak, W.; Adam, M.; Quaas, A.; Sauter, G.; Simon, R.; Izbicki, J.R.; et al. High mitochondria content is associated with prostate cancer disease progression. Mol. Canc. 2013, 12, 145. [Google Scholar] [CrossRef]
- Minner, S.; Wittmer, C.; Graefen, M.; Salomon, G.; Steuber, T.; Haese, A.; Huland, H.; Bokemeyer, C.; Yekebas, E.; Dierlamm, J.; et al. High level PSMA expression is associated with early psa recurrence in surgically treated prostate cancer. Prostate 2011, 71, 281–288. [Google Scholar] [CrossRef]
- Erbersdobler, A.; Isbarn, H.; Dix, K.; Steiner, I.; Schlomm, T.; Mirlacher, M.; Sauter, G.; Haese, A. Prognostic value of microvessel density in prostate cancer: A tissue microarray study. World J. Urol. 2010, 28, 687–692. [Google Scholar] [CrossRef]
- Fleischmann, A.; Schlomm, T.; Kollermann, J.; Sekulic, N.; Huland, H.; Mirlacher, M.; Sauter, G.; Simon, R.; Erbersdobler, A. Immunological microenvironment in prostate cancer: High mast cell densities are associated with favorable tumor characteristics and good prognosis. Prostate 2009, 69, 976–981. [Google Scholar] [CrossRef]
- Grupp, K.; Hohne, T.S.; Prien, K.; Hube-Magg, C.; Tsourlakis, M.C.; Sirma, H.; Pham, T.; Heinzer, H.; Graefen, M.; Michl, U.; et al. Reduced CD147 expression is linked to ERG fusion-positive prostate cancers but lacks substantial impact on psa recurrence in patients treated by radical prostatectomy. Exp. Mol. Pathol. 2013, 95, 227–234. [Google Scholar] [CrossRef]
- Minner, S.; de Silva, C.; Rink, M.; Dahlem, R.; Chun, F.; Fisch, M.; Hoppner, W.; Wagner, W.; Bokemeyer, C.; Terracciano, L.; et al. Reduced CD151 expression is related to advanced tumour stage in urothelial bladder cancer. Pathology 2012, 44, 448–452. [Google Scholar] [CrossRef]
- El Gammal, A.T.; Bruchmann, M.; Zustin, J.; Isbarn, H.; Hellwinkel, O.J.; Kollermann, J.; Sauter, G.; Simon, R.; Wilczak, W.; Schwarz, J.; et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin. Canc. Res. 2010, 16, 56–64. [Google Scholar] [CrossRef]
- Krohn, A.; Seidel, A.; Burkhardt, L.; Bachmann, F.; Grupp, K.; Becker, A.; Adam, M.; Graefen, M.; Huland, H.; Steurer, S.; et al. Recurrent deletion of 3p13 targets multiple tumor suppressor genes and defines a distinct subgroup of aggressive erg fusion positive prostate cancers. J. Pathol. 2013, 231, 130–141. [Google Scholar] [CrossRef]
- Burkhardt, L.; Fuchs, S.; Krohn, A.; Masser, S.; Mader, M.; Kluth, M.; Bachmann, F.; Huland, H.; Steuber, T.; Graefen, M.; et al. CHD1 is a 5q21 tumor suppressor required for erg rearrangement in prostate cancer. Canc. Res. 2013, 73, 2795–2805. [Google Scholar]
- Kluth, M.; Hesse, J.; Heinl, A.; Krohn, A.; Steurer, S.; Sirma, H.; Simon, R.; Schumacher, U.; Grupp, K.; Izbicki, J.; et al. Genomic deletion of MAP3K7 at 6q12–22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod. Pathol. 2013, 26, 975–983. [Google Scholar] [CrossRef]
- Krohn, A.; Diedler, T.; Burkhardt, L.; Mayer, P.S.; De Silva, C.; Meyer-Kornblum, M.; Kotschau, D.; Tennstedt, P.; Huang, J.; Gerhauser, C.; et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in erg fusion-positive and fusion-negative prostate cancer. Am. J. Pathol. 2012, 181, 401–412. [Google Scholar] [CrossRef]
- Minner, S.; Enodien, M.; Sirma, H.; Luebke, A.M.; Krohn, A.; Mayer, P.S.; Simon, R.; Tennstedt, P.; Muller, J.; Scholz, L.; et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin. Canc. Res. 2011, 17, 5878–5888. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Simon, R.; Feuerbach, L.; Schlangen, K.; Weichenhan, D.; Minner, S.; Wuttig, D.; Warnatz, H.J.; Stehr, H.; Rausch, T.; et al. Integrative genomic analyses reveal androgen-driven somatic alteration landscape in early-onset prostate cancer. Canc. Cell 2013, 23, 159–170. [Google Scholar] [CrossRef]
- Tapia, C.; Schraml, P.; Simon, R.; Al-Kuraya, K.S.; Maurer, R.; Mirlacher, M.; Novotny, H.; Spichtin, H.; Mihatsch, M.J.; Sauter, G. HER2 analysis in breast cancer: Reduced immunoreactivity in fish non-informative cancer biopsies. Int. J. Oncol. 2004, 25, 1551–1557. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Burdelski, C.; Matuszewska, A.; Kluth, M.; Koop, C.; Grupp, K.; Steurer, S.; Wittmer, C.; Minner, S.; Tsourlakis, M.C.; Sauter, G.; et al. Qualitative and Quantitative Requirements for Assessing Prognostic Markers in Prostate Cancer. Microarrays 2014, 3, 137-158. https://doi.org/10.3390/microarrays3020137
Burdelski C, Matuszewska A, Kluth M, Koop C, Grupp K, Steurer S, Wittmer C, Minner S, Tsourlakis MC, Sauter G, et al. Qualitative and Quantitative Requirements for Assessing Prognostic Markers in Prostate Cancer. Microarrays. 2014; 3(2):137-158. https://doi.org/10.3390/microarrays3020137
Chicago/Turabian StyleBurdelski, Christoph, Aleksandra Matuszewska, Martina Kluth, Christina Koop, Katharina Grupp, Stefan Steurer, Corinna Wittmer, Sarah Minner, Maria Christina Tsourlakis, Guido Sauter, and et al. 2014. "Qualitative and Quantitative Requirements for Assessing Prognostic Markers in Prostate Cancer" Microarrays 3, no. 2: 137-158. https://doi.org/10.3390/microarrays3020137
APA StyleBurdelski, C., Matuszewska, A., Kluth, M., Koop, C., Grupp, K., Steurer, S., Wittmer, C., Minner, S., Tsourlakis, M. C., Sauter, G., Schlomm, T., & Simon, R. (2014). Qualitative and Quantitative Requirements for Assessing Prognostic Markers in Prostate Cancer. Microarrays, 3(2), 137-158. https://doi.org/10.3390/microarrays3020137