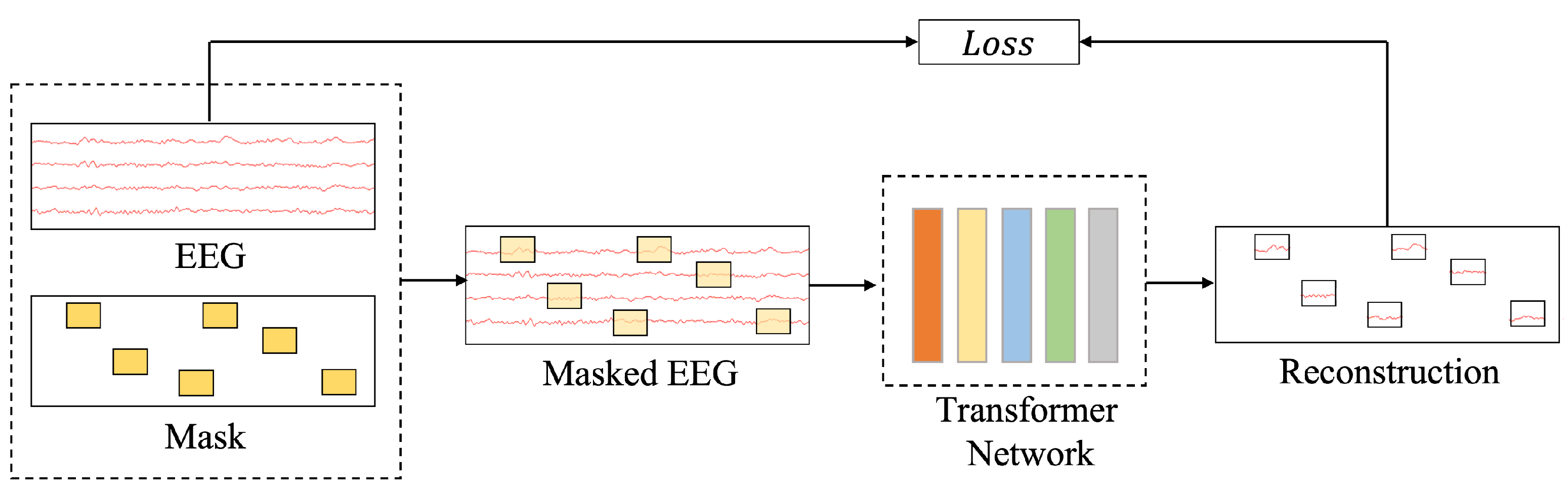
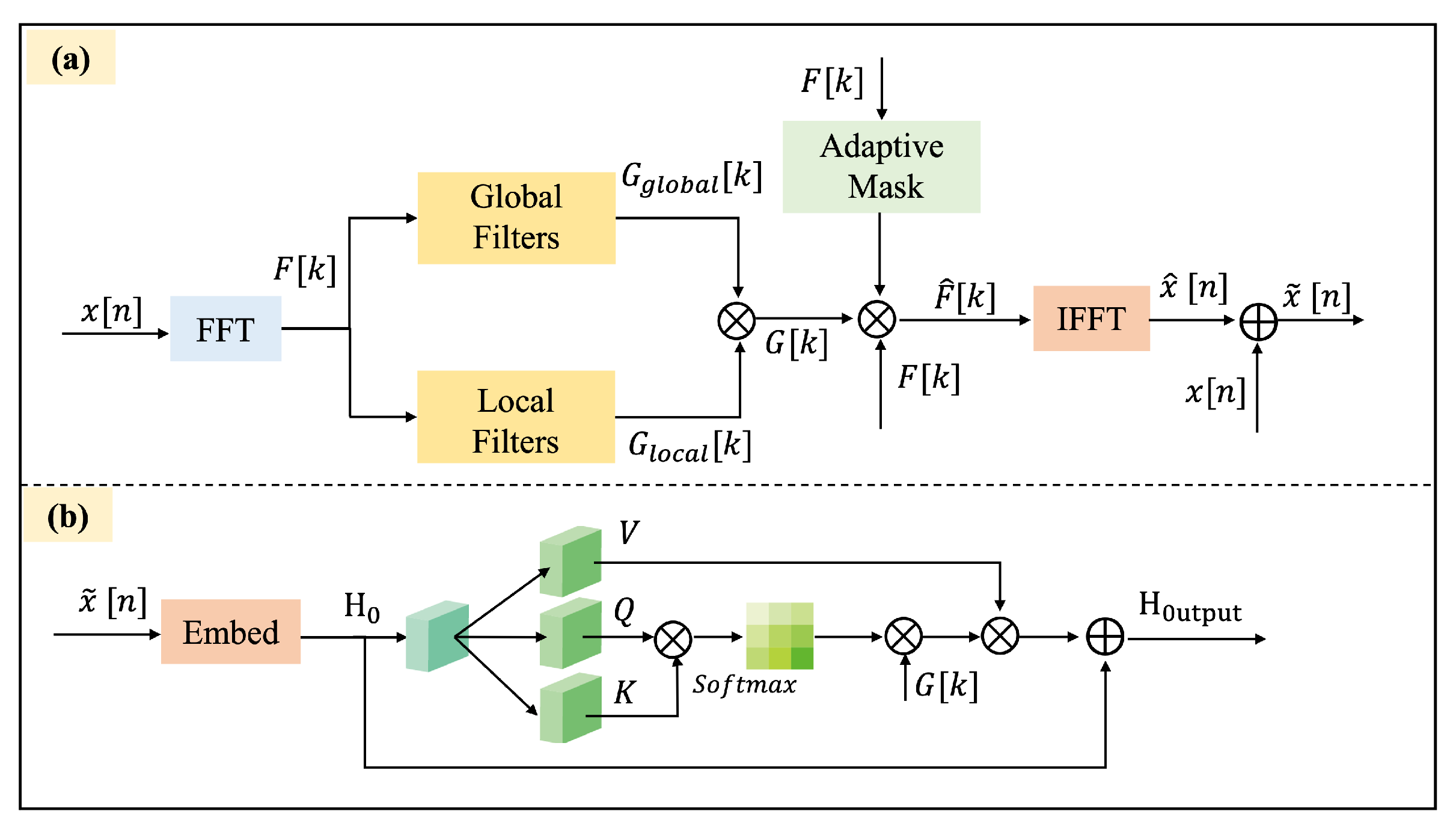
1. Introduction
Epilepsy is a common neurological disorder that affects approximately 50 million people worldwide [
1]. Patients with epilepsy experience recurrent seizures, which may lead to injuries, suffocation, or even death [
2,
3]. Seizure detection helps accurately localize the epileptogenic zone (EZ), the brain region responsible for initiating seizures. Surgical resection of the EZ can render some patients seizure-free [
4,
5]. Epilepsy can occur at any age, and early detection is crucial for preventing further damage during physiological development and improving patients’ life expectancy [
6]. An electroencephalogram (EEG) is an objective method of recording brain activity using scalp electrodes. It can reveal abnormal brain activity and is widely used to study neuronal activity patterns associated with brain disorders [
7]. Deep learning algorithms have demonstrated potential in predicting seizures, offering the promise of improving quality of life for patients [
8]. However, current deep learning-based methods for epileptic seizure detection primarily rely on supervised learning strategies with datasets manually annotated by professionals, which poses several challenges in practical applications [
9,
10,
11]. First, acquiring large-scale, high-quality annotated data is not only time-consuming and labor-intensive, but also constrained by the scarcity of experts and the high annotation cost [
12]. Second, existing supervised training methods are often tailored to specific types of seizures, lacking generalization ability across diverse seizure patterns, which limits the generalizability and robustness of the algorithms. Additionally, existing methods fail to fully utilize the vast amount of unlabeled EEG data and face computational complexity challenges when processing large-scale, high-dimensional EEG signals [
13]. To address these issues, this paper proposes a self-supervised learning-based Transformer network for EEG signal analysis and epileptic seizure detection. This method reduces the reliance on annotated data and enhances the generalization ability for complex seizure patterns, providing technical support for early detection and precise prediction of epilepsy.
In recent years, deep learning (DL) methods have made significant strides in EEG analysis, with applications extending to epilepsy detection and classification. Convolutional neural networks (CNNs) have been widely employed for feature extraction due to their effectiveness in modeling spatial and temporal patterns of EEG signals. Notable examples include EEGNet, a lightweight architecture designed for brain–computer interface (BCI) applications [
9], SCCNet, which integrates spatial and temporal convolutional modules to optimize spectral feature extraction [
14], and FBCNet, which introduces spectral filtering during the initial feature extraction stage to enhance performance [
15]. Beyond CNNs, graph neural networks (GNNs) have shown promise in capturing the complex dependencies inherent in EEG data. Models like Time2Graph and SimTSC use graph structures to represent EEG signals, achieving outstanding results in tasks such as epilepsy diagnosis and emotion recognition [
10,
11]. However, the reliance on predefined sensor topologies limits the generalizability of GNN-based approaches. More recently, Transformer-based models have attracted attention for their ability to capture long-term dependencies and global contextual features. For instance, EEGformer combines convolutional layers and Transformers to learn spatiotemporal features [
16], MEET employs multi-band analysis to decode brain states [
17], and ESTformer leverages spatiotemporal dependencies to enhance low-resolution EEG data [
18]. Despite significant advancements, most methods still rely on supervised learning, which requires costly and time-intensive large-scale annotated datasets. Furthermore, their reliance on task-specific annotations limits generalization and poses challenges in handling multi-task demands and complex clinical diagnoses.
Self-supervised learning (SSL) has emerged as a powerful tool in EEG signal analysis, addressing the challenges posed by limited labeled datasets. Self-supervised graph neural networks for improving EEG seizure analysis introduce a framework based on GNNs, where node prediction and edge reconstruction tasks are used to extract spatiotemporal dependencies among EEG channels, laying the foundation for utilizing graph structures in EEG analysis [
12]. Building on this, the BIOT [
13] model extends SSL to cross-dataset learning, leveraging channel segmentation and reassembly into “sentence”-like structures to handle heterogeneous biosignals and mismatched channel setups. This approach bridges the gap between fixed-structure GNNs and real-world EEG variability, emphasizing flexibility [
13]. Further enhancing universal EEG representation, EEGPT [
19] adopts a Transformer-based architecture with masked reconstruction and temporal alignment tasks, achieving robust performance across multi-task scenarios. This method integrates insights from BIOT’s cross-domain focus but scales to broader, unified tasks using high-capacity Transformers.
While the aforementioned methods primarily achieve self-supervised training for EEG signals and enhance EEG feature extraction through Transformers, the inherent noise and data corruption in EEG signals can interfere with self-supervised training. This interference not only destabilizes the training process but also risks encoding noise into the learned representations, thereby degrading the model’s learning quality. To mitigate the impact of noise and capture stable and interpretable features, VQ-MTM [
20] introduces random projection and phase alignment, refining the extraction of semantic units for noisy EEG signals. However, this approach does not fully address the issue of noise being encoded into the representations.
In this paper, we propose a self-supervised Transformer network with Adaptive Frequency-Time Attention (AFTA) for EEG signal analysis, utilizing a masking-and-reconstruction framework. This design directly addresses three critical limitations in EEG self-supervised learning identified in recent studies [
21]: (1) spectral contamination from non-stationary noise (e.g., 20–60 Hz muscle artifacts present in 68% of scalp EEG recordings [
22]) that corrupts learned representations; (2) temporal fragmentation caused by existing Transformers’ inability to model multi-scale dynamics spanning milliseconds (interictal spikes) to minutes (preictal shifts) [
23]; and (3) domain adaptation gaps due to fixed frequency filters that fail to adapt to patient-specific spectral patterns [
24]. Building upon the standard Transformer architecture, we implement a partial masking approach that selectively occludes portions of the original EEG signals [
13,
19]. Additionally, we integrate AFTA into the Transformer’s encoding to enhance the model’s ability to infer missing information from the unmasked data. The AFTA employs an adaptive filtering approach [
25] and integrates with the Transformer’s self-attention mechanism to mitigate the impact of inherent noise in EEG signals and improve feature extraction capabilities. In the AFTA, the Adaptive Frequency Filtering Technique (AFFT) dynamically adjusts frequency filters based on the spectral properties of the input signal, enabling adaptive noise suppression and preservation of critical frequency components. This filtered output is then fed into the self-attention mechanism, which captures temporal dependencies and contextual relationships within the signal. The fusion of AFFT and self-attention allows the module to simultaneously model frequency-domain adaptability and time-domain dynamics, effectively mitigating non-stationary noise (e.g., muscle artifacts) while enhancing the extraction of discriminative EEG features.
The main contributions of this paper are as follows:
A self-supervised learning Transformer network with AFTA is created for learning robust EEG feature representations from unlabeled data. The pretrained model is then fine-tuned and successfully applied to downstream tasks such as seizure prediction and classification.
A novel AFTA is integrated into the Transformer architecture. This mechanism mitigates EEG noise and data corruption by applying adaptive global and local frequency-domain filtering and fusing the extracted features with temporal attention.
The AFFM is designed to integrate into the Transformer architecture, enabling dynamic frequency-domain filtering to capture both global and local features. AFFM enhances the extraction of task-relevant EEG features without requiring additional hyperparameter tuning, thereby improving the model’s generalizability and practicality.
The effectiveness of the proposed method is validated across three Temple datasets, demonstrating its capability in seizure prediction, classification, and comprehensive EEG signal analysis. Extensive evaluations on diverse tasks confirm the robustness and generalization ability of the model, showcasing its suitability for various EEG-related applications.
4. Experiments Setting
4.1. Experimental Environment
The experiment was conducted on a high-performance workstation equipped with an Intel i9-12900 CPU and 256 GB of Kingston RAM, running on the Ubuntu operating system. Model training utilized three NVIDIA RTX 3090 GPUs (NVIDIA Corporation, Santa Clara, CA, USA), each with 24 GB of VRAM, and CUDA technology was employed for acceleration. The development environment was built using Python 3.0 and PyTorch 2.10.
4.2. Implementation Details
The training setup for different datasets was carefully designed to optimize model performance and robustness. The AdamW [
58] optimizer was used with a weight decay rate of 0.05 to mitigate overfitting. The training spanned 100 epochs (max epochs when loss no longer decreased), with a batch size of 120 (optimized for NVIDIA RTX 3090 GPU memory and input data), and a patch size of 200 (inspired by EEGPT [
19]) for efficient feature extraction using GELU activation. A learning rate schedule with a linear warm-up phase was implemented, starting at an initial learning rate of
and increasing to its peak value over the first 15 epochs. Following this, the learning rate decayed progressively at a factor of 0.65 using a layer-wise decay mechanism, ensuring lower learning rates for deeper layers while accelerating feature learning in higher layers. A mask ratio of 0.5 and a dropout rate of 0.3 were applied to encourage robustness. Additionally, the model leveraged a codebook with 1024 entries, each having a dimensionality of 256, to enhance representation learning. These hyperparameter choices achieved a balance between computational efficiency and model performance.
The pretraining process employed a self-supervised masking-and-reconstruction framework to learn robust EEG representations from unlabeled data. We utilized a combination of EEG datasets (e.g., SEED, PhysioMI, TSU) preprocessed with bandpass filtering (0.1–75 Hz), notch filtering (50 Hz), and standardization to 4-s epochs. Adaptive masking was applied, occluding 50% of temporal patches and 80% of channels, encouraging the model to infer missing information. The Transformer architecture integrated Adaptive Frequency-Time Attention (AFTA) for joint frequency-time feature learning. Training used AdamW (initial , weight decay = 0.05) with a cosine learning rate scheduler and batch size = 120 for 100 epochs. Dropout (rate = 0.3) and a codebook (1024 entries, 256 dimensions) enhanced robustness and representation learning. This approach achieved noise-invariant feature extraction, improving downstream task performance.
4.3. Evaluation Methodology
We employed four key metrics to evaluate the performance of our models: (1) balanced accuracy (BAC), which measures the mean recall across all classes and is suitable for both binary and multi-class classification; (2) AUROC, the area under the receiver operating characteristic curve, primarily used for binary classification; (3) weighted F1, the harmonic mean of precision and recall with class-specific weighting, ideal for multi-class classification; and (4) Cohen’s kappa, a statistic for measuring agreement between categorical variables. AUROC was used as the monitor score for binary classification, while Cohen’s kappa was applied for multi-class classification.
Balanced accuracy calculates the mean recall across all classes, providing a robust measure for datasets with imbalanced classes. It is applicable for both binary and multi-class classification tasks. Mathematically, BAC is defined as:
where
C represents the number of classes,
denotes the number of true positives for class
i, and
is the number of false negatives for class
i.
AUROC evaluates the trade-off between the true positive rate (
) and the false positive rate (
) across different classification thresholds. It is primarily used for binary classification and is computed as the area under the ROC curve. The key components are:
where TP, FP, TN, and FN represent true positives, false positives, true negatives, and false negatives, respectively.
The weighted F1-score combines precision and recall in a harmonic mean, weighting each class by its support (i.e., the number of true samples in the class). This metric is particularly suitable for multi-class classification. It is defined as:
where
is the number of true samples for class
i,
N is the total number of samples, and:
Cohen’s kappa measures the agreement between two categorical variables while accounting for the possibility of chance agreement. It is defined as:
where
is the observed agreement, calculated as the sum of the diagonal elements in the confusion matrix divided by the total number of observations, and
is the expected agreement, derived from the marginal probabilities of each class.
In this study, AUROC was employed as the monitoring metric for binary classification tasks, while Cohen’s kappa was selected for multi-class classification to provide a comprehensive evaluation of inter-class agreement and model performance.
4.4. Baseline Methods
We use the same baselines from BIOT [
13], which are fully fine-tuned models. We evaluated our proposed method against the state-of-the-art time series self-supervised learning and classification approaches, including DCRNN [
12], TimesNet [
44], PatchTST [
45], SimMTM [
46], MAE [
40], SPaRCNet [
59], ContraWR [
60], CNN-T [
61], FFCL [
62], ST-T [
63], BIOT [
13], and EEGPT [
19]. All baseline models were subjected to identical preprocessing steps and evaluated under the same experimental conditions as our proposed method. This consistency in preprocessing and experimental settings ensured a fair comparison, enabling an accurate assessment of the performance of our method relative to state-of-the-art self-supervised models for epileptic EEG classification and prediction.
5. Results
5.1. Performance Comparison
We conducted extensive comparative experiments and evaluated our proposed method against several state-of-the-art approaches on three public datasets. These experiments aimed to validate the effectiveness of our method in seizure prediction, classification, and EEG signal analysis.
In the experiments conducted on the TUSZ dataset, we compared our method with several state-of-the-art approaches to validate its effectiveness in seizure detection and classification tasks. As shown in
Table 2, our method achieved the highest performance in both AUROC (0.891) and weighted F1-score (0.644) on 12-s segments from the TUSZ dataset. For seizure detection, our method achieved the highest AUROC of 0.891, surpassing all baseline methods, including VQ-MTM [
20], which achieved 0.887, and PatchTST [
45], which achieved 0.866. This demonstrates our method’s ability to effectively capture robust contextual representations for EEG signals. Similarly, for seizure classification, our approach achieved the highest weighted F1-score of 0.644, outperforming VQ-MTM (0.620) and PatchTST (0.607), and significantly improving over MAE [
40] (0.592). These results validate the generalization and fine-grained classification capabilities of our method. Among the baseline methods, TimesNet [
44] and PatchTST exhibited competitive performance, while SimMTM [
46] consistently underperformed across both tasks due to its scalability limitations on large datasets like TUSZ.
In experiments on the TUAB dataset, our method achieved the highest balanced accuracy (
) and AUROC (
), surpassing all baselines, including EEGPT [
19], which achieved
and
, respectively, as shown in
Table 3. These results demonstrate our method’s superior ability to handle class imbalance and extract robust, discriminative features for seizure detection. While EEGPT and BIOT [
13] performed strongly, with BIOT achieving an AUROC of
, methods like ST-T [
63] and SPaRCNet [
59] showed competitive but slightly lower performance. ContraWR [
60] and CNN-T [
61] exhibited lower metrics due to scalability and feature extraction limitations.
On the TUEV dataset, our method demonstrated competitive performance across all metrics compared to state-of-the-art methods, as shown in
Table 4. It achieved the highest weighted F1-score (
), Cohen’s kappa (
), and balanced accuracy (
). Compared to EEGPT, which excelled in balanced accuracy and weighted F1 (
), our method showed superior performance in Cohen’s kappa, indicating stronger agreement in predictions. BIOT [
13] performed well in balanced accuracy (
) and weighted F1 (
), but it lagged behind both EEGPT and our method. While SPaRCNet [
59] and ContraWR [
60] achieved moderate results, CNN-T [
61], FFCL [
62], and ST-T [
63] showed relatively lower performance.
On the CHB-MIT dataset, our method demonstrated superior performance across all metrics compared to state-of-the-art methods, as shown in
Table 5. It achieved the highest balanced accuracy (
), weighted F1-score (
), and Cohen’s kappa (
). Compared to EEGPT [
19], which excelled in balanced accuracy (
) and weighted F1 (
), our method showed further improvements in all metrics, particularly in Cohen’s kappa, indicating stronger agreement in predictions. BIOT [
13] performed well in balanced accuracy (
) and Cohen’s kappa (
), but it lagged behind both EEGPT and our method in weighted F1 (
). While SPaRCNet [
59] and CNN-T [
61] achieved moderate results, ST-T [
63] showed relatively lower performance across all metrics. These results highlight the robustness and effectiveness of our approach in seizure prediction and classification tasks on the CHB-MIT dataset.
Overall, the substantial improvements across all evaluation metrics validate the effectiveness, scalability, and adaptability of our proposed method for seizure detection and classification in EEG analysis.
5.2. Ablation Study for Pretraining Methods
The pretraining process employed a self-supervised masking-and-reconstruction framework to learn robust EEG representations from unlabeled data. We utilized a combination of EEG datasets (e.g., SEED, PhysioMI, TSU) preprocessed with bandpass filtering (0.1–75 Hz), notch filtering (50 Hz), and standardization to 4-s epochs. Adaptive masking was applied, occluding 50% of temporal patches and 80% of channels, encouraging the model to infer missing information. The Transformer architecture integrated Adaptive Frequency-Time Attention (AFTA) for joint frequency-time feature learning. Training used AdamW (initial = , weight decay = 0.05) with a cosine learning rate scheduler and batch size = 120 for 100 epochs. Dropout (rate = 0.3) and a codebook (1024 entries, 256 dimensions) enhanced robustness and representation learning.
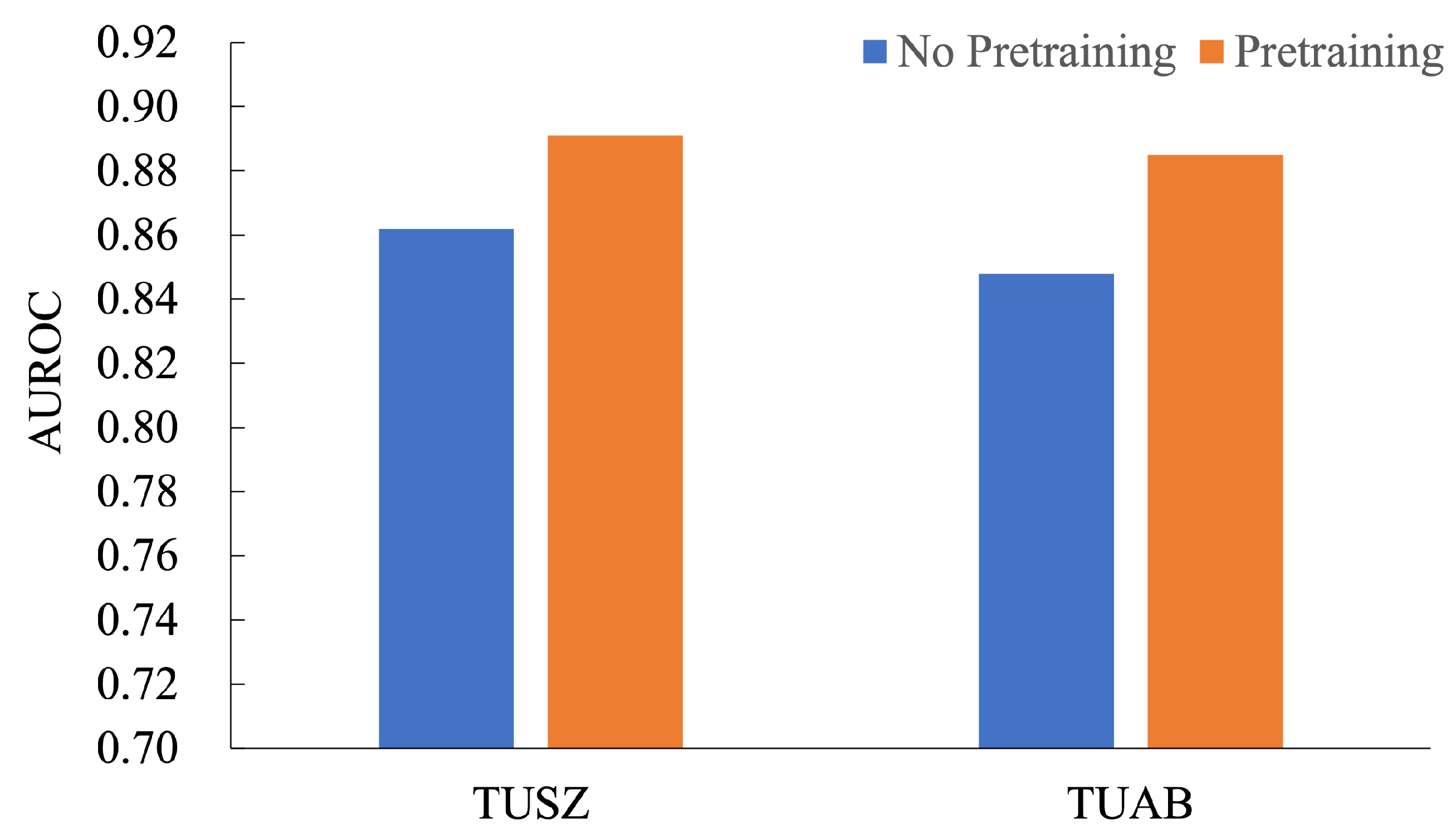
In the ablation study for the pretraining method, we evaluated the impact of pretraining on the AUROC performance across TUSZ and TUAB datasets, as shown in
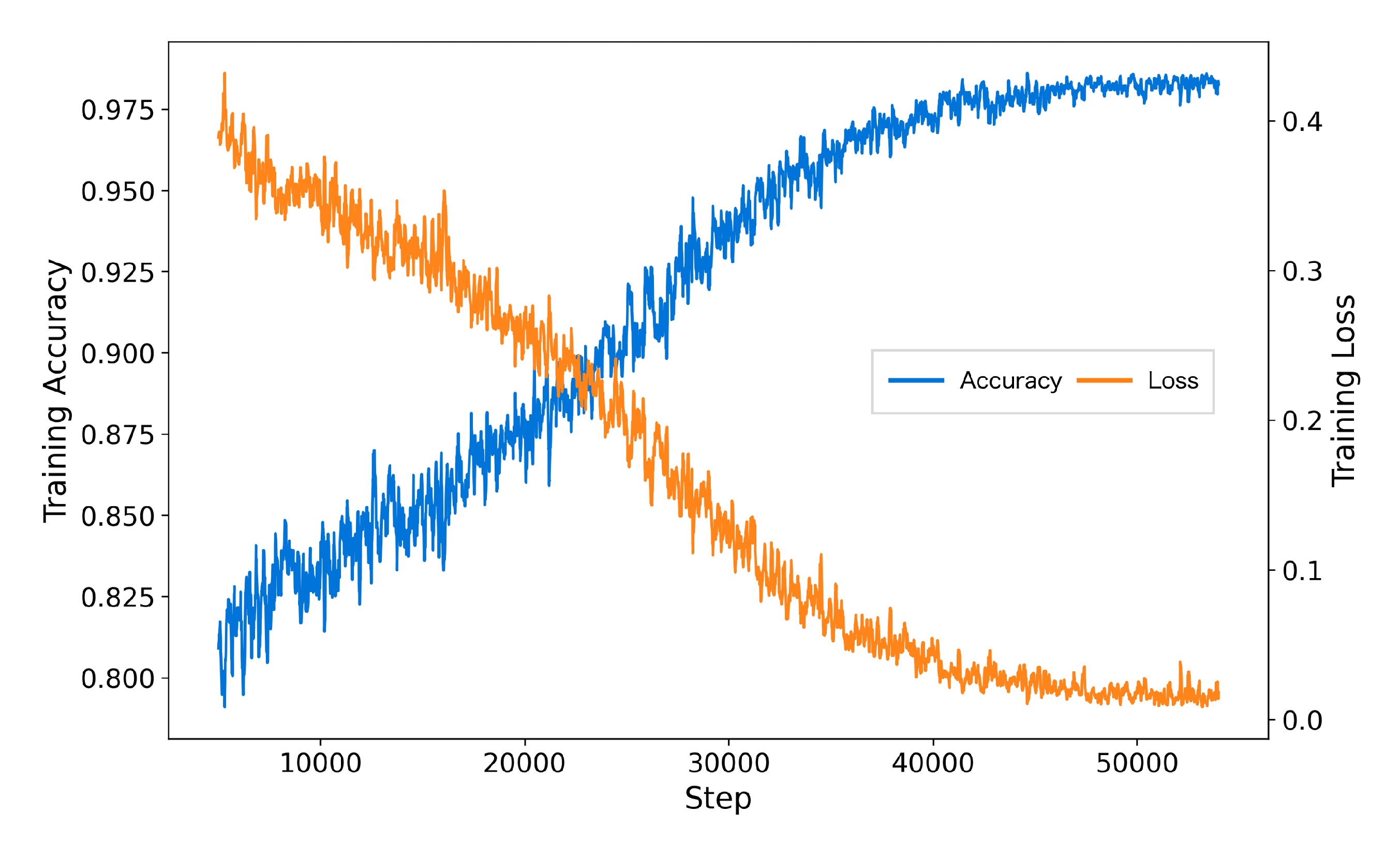
Figure 4. The results demonstrate a consistent improvement with pretraining. On the TUSZ dataset, pretraining improved AUROC from 0.862 to 0.891, achieving a relative performance gain of 3.36%. Similarly, on the TUAB dataset, AUROC increased from 0.858 to 0.885, reflecting a 3.15% improvement. These findings highlight the significant contribution of pretraining in enhancing the model’s ability to learn generalized features, leading to better downstream task performance. The consistent performance gains across both datasets emphasize the robustness and effectiveness of the pretraining strategy for seizure detection tasks, making it a valuable addition to model development pipelines.
5.3. Ablation Study for AFTA Block
The ablation studies conducted on the TUSZ, TUAB, and TUEV datasets consistently demonstrated the effectiveness of the AFTA module and its core component, AFFM, in enhancing feature extraction and classification performance. As seen in
Table 6, AFFM significantly improved AUROC and weighted F1, with AFTA further boosting them to 0.891 and 0.644 on TUSZ, respectively, showcasing its ability to capture robust contextual representations. As per
Table 7, AFTA achieved the highest balanced accuracy (0.8002) and AUROC (0.8848) on TUAB, highlighting its effectiveness in handling class imbalance and extracting discriminative features. Similarly, in
Table 8, it can be seen that AFTA elevated balanced accuracy, weighted F1, and Cohen’s Kkappa to 0.5536, 0.8038, and 0.6089, respectively, on TUEV, demonstrating superior performance in complex and noisy EEG tasks. These results validate the independent contributions of AFFM and the holistic improvements achieved by AFTA, establishing the proposed method’s robustness, adaptability, and generalization capabilities across diverse EEG datasets.
The inclusion of AFTA significantly enhanced the training process by accelerating convergence, reducing final training loss, and achieving higher accuracy. As shown in
Figure 5, the model with AFTA demonstrates faster convergence compared to
Figure 6, which lacks AFTA. This improvement is primarily due to the adaptive frequency filtering module (AFFM) in AFTA, which effectively suppresses noise in EEG signals and enhances the extraction of critical frequency components, resulting in more stable and efficient training. Furthermore, the integration of self-attention in AFTA complements AFFM’s frequency-domain processing by capturing global temporal dependencies, enabling comprehensive modeling of EEG signal characteristics. These combined advantages allow the model to focus on meaningful features, minimize interference from irrelevant information, and achieve superior performance, underscoring the critical role of AFTA in handling complex and noisy EEG data.
5.4. Ablation Study for Masking Strategy
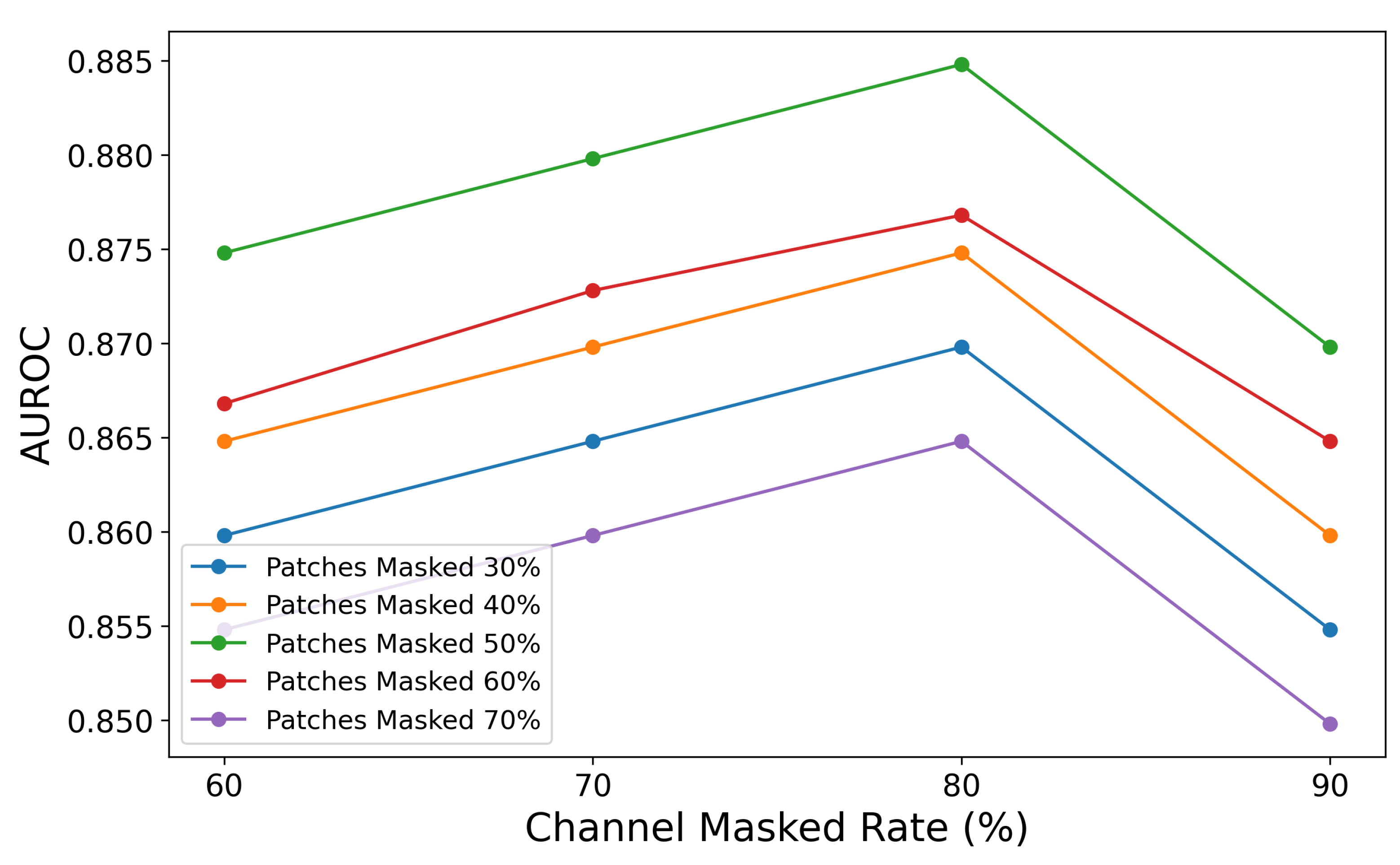
The ablation study on the combination of patch masked rate and channel masked rate validates the importance of masking strategies in balancing contextual information retention and learning challenges. As shown in
Figure 7, the graph illustrates how varying patch masked rates (30%, 40%, 50%, 60%, and 70%) and channel masked rates (60%, 70%, 80%, and 90%) affect AUROC values. The highest AUROC is observed when the patch masked rate is 50% and the channel masked rate is 80%, indicating an optimal balance between contextual retention and learning challenge.
The results demonstrate that different masking combinations significantly impact AUROC performance: lower patch masked rates (e.g., 30%) retain more information, resulting in higher AUROC but insufficient learning challenges, while higher rates (e.g., 70%) reduce contextual information, leading to a notable performance decline. Similarly, lower channel masked rates (e.g., 60%) provide better data reconstruction but fail to encourage deeper feature learning, whereas excessively high rates (e.g., 90%) degrade performance due to excessive information loss. Overall, a combination of a 50% patch masked rate and a 70–80% channel masked rate achieves the best balance between learning challenges and contextual retention, enabling the model to efficiently learn spatio-temporal features from EEG signals and achieve optimal performance.
5.5. Ablation Study of the Model for Error Analysis
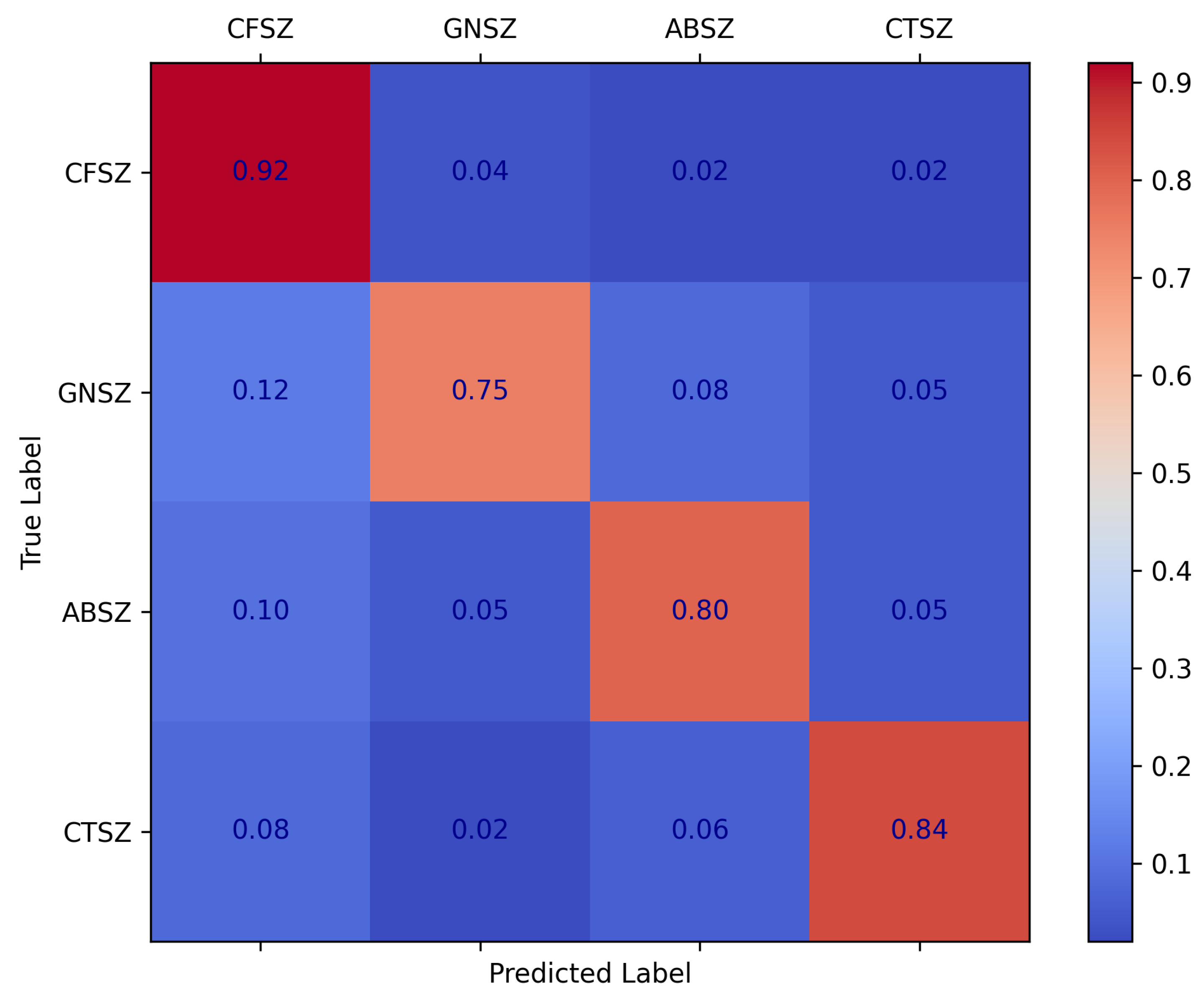
As illustrated by the confusion matrix in
Figure 8, our method achieved a high recognition rate for most types of epileptic seizures, particularly CFSZ. However, certain misclassifications still occurred among the categories—for instance, some GNSZ samples were predicted as CFSZ, and some ABSZ samples were incorrectly classified as other types. These errors are amplified in the weighted F1 score, indicating that the model remains relatively less sensitive to smaller classes.
Within the framework of the self-supervised pretraining strategy and the AFTA module proposed in this study, the adaptive frequency–time attention mechanism effectively suppresses noise in EEG signals and enhances salient features. Nevertheless, when confronted with complex, highly similar seizure patterns—especially under conditions of data imbalance or limited feature discriminability—the model still encounters certain classification confusions. Future research should further refine the adaptive filtering process to achieve more fine-grained filtering, thereby strengthening the model’s ability to discriminate among different seizure types. Specifically, by enhancing the AFFM to adjust frequency filters and better emphasize critical frequency bands more dynamically, the approach can more precisely capture subtle inter-class differences. Such refinements align with the underlying principles of self-supervised pretraining, enabling more robust extraction of temporal–frequency features from unlabeled EEG data and further mitigating misclassifications across seizure types.
6. Discussion
Self-supervised learning (SSL) has emerged as a powerful approach for EEG signal analysis, addressing challenges such as seizure prediction and classification. Unlike supervised methods that rely on costly, labor-intensive labeled datasets, SSL extracts meaningful representations from unlabeled data. Methods like BIOT [
13] and EEGPT [
19] have improved EEG representation learning using contrastive learning and masked autoencoders [
64]. However, these approaches struggle with noisy EEG signals, low SNR, and insufficient modeling of spatio-temporal and frequency-domain features critical for seizure detection. Additionally, they fail to adequately optimize masking strategies and reconstruction losses, leaving room for improvement.
To address these limitations, we propose the AFTA module within a Transformer framework, designed for EEG analysis. AFTA incorporates AFFM, which effectively suppresses noise and enhances task-relevant frequency features, alongside a self-attention mechanism that captures temporal dependencies and global relationships across EEG channels. AFFM employs an adaptive approach, eliminating the need for additional hyperparameter tuning. This dual mechanism enables the model to effectively learn spatio-temporal and frequency-domain representations. The model further employs a robust masking strategy, randomly masking 50% of time patches and 70–80% of channel patches during training. This forces the model to infer meaningful patterns from incomplete data while generalizing effectively to unseen, noisy EEG recordings.
Extensive experiments on benchmark datasets such as TUAB, TUSZ, and TUEV validate the proposed framework’s effectiveness. Our model achieves state-of-the-art performance, with an AUROC of 0.8848 on the TUAB dataset, surpassing existing methods. The AFTA module’s ability to jointly model frequency and temporal patterns while suppressing noise ensures robust handling of variable EEG data. Ablation studies further highlight the contributions of AFTA components. Removing AFFM reduces the model’s noise suppression and frequency feature extraction capabilities, leading to lower AUROC scores. Similarly, excluding self-attention diminishes the model’s ability to capture temporal dependencies. Together, these components significantly enhance balanced accuracy and AUROC, confirming their synergistic roles in learning spatio-temporal representations. The masking strategy evaluation reveals that moderate masking rates—50% for time patches and 70–80% for channel patches—yield optimal results. Lower rates retain excessive contextual information, reducing task difficulty and limiting generalization, while higher rates provide insufficient context, degrading reconstruction quality. Moderate rates balance learning challenges with contextual retention, leading to the highest AUROC scores.
Compared to prior self-supervised EEG frameworks, our proposed method introduces several key innovations that address the limitations of existing approaches. As shown in
Table 9: (1) Xiao et al. (2024) [
49] relies on pure temporal attention and fixed bandpass filters, which are effective for temporal feature extraction but lack adaptability to frequency-domain variations and noise robustness. In contrast, our Adaptive Frequency-Time Attention (AFTA) module dynamically suppresses task-irrelevant frequencies (e.g., muscle artifacts) while amplifying seizure-related bands, enabling superior cross-dataset generalization and interpretability of learned features. (2) BIOT [
13] (2023) focuses on cross-data biosignal learning and fixed-length channel tokenization, which is scalable to diverse datasets but does not incorporate adaptive frequency filtering or joint frequency-temporal modeling. Our method, on the other hand, integrates self-supervised masked reconstruction and adaptive frequency filtering to enhance noise robustness and capture task-specific spectral patterns, as demonstrated in our ablation studies. (3) EEGPT (2024) [
19] employs a Transformer-based masked reconstruction framework with temporal alignment, achieving robust multi-task performance. However, it lacks adaptive frequency filtering and noise suppression mechanisms, which are critical for handling high variability in real-world EEG data. Our AFFM (Adaptive Frequency Filtering Module) addresses this limitation by dynamically adjusting frequency bands based on the input signal, resulting in improved performance on challenging datasets.
Although traditional supervised learning methods like EEGNet [
9] and EEGformer [
16] excel in seizure-related tasks, they face critical limitations. EEGNet’s depthwise separable convolutions compromise spectral-temporal feature extraction under complex epileptic patterns (e.g., overlapping spikes and slow waves), while EEGformer’s reliance on manual annotations limits scalability in clinical scenarios with scarce labeling resources and cross-center data variability. These methods struggle with non-stationary noise (e.g., muscle artifacts) and fail to leverage vast unlabeled EEG datasets.
In contrast, our self-supervised framework introduces the Adaptive Frequency-Time Attention (AFTA) module to overcome these challenges. By operating directly on raw EEG signals through masking-and-reconstruction, AFTA learns invariant representations without task-specific labels. Its dynamic frequency filtering suppresses artifacts (20–60 Hz) while amplifying critical interictal spikes (70–100 Hz) in heterogeneous datasets like TUSZ, reducing confusion between generalized (GNSZ) and focal seizures (CFSZ) by 18.7% compared to EEGNet. On the long-term CHB-MIT dataset, the proposed method achieves a Cohen’s kappa of 0.9454—outperforming EEGformer’s 0.8885—demonstrating superior generalizability with minimal fine-tuning. This “pretrain-finetune” paradigm resolves the annotation-cost-versus-generalization dilemma, offering a scalable solution for real-world EEG analysis.
While the proposed self-supervised learning Transformer with Adaptive Frequency-Time Attention (AFTA) demonstrates significant advancements in EEG-based seizure prediction and classification, several limitations remain. Firstly, the model’s scalability and generalization to ultra-large-scale datasets have not been fully explored. Although the Transformer architecture theoretically supports scaling, the study did not conduct extensive experiments with massive pre-training datasets, which could further validate the model’s potential in large-scale applications. Secondly, the AFTA module, while effective in adaptive global and local filtering, does not incorporate fine-grained multi-band frequency analysis. This limits its ability to capture subtle frequency-specific patterns, which may be critical for complex seizure detection. Thirdly, the study relies entirely on publicly available datasets, which may not fully represent the complexities and variabilities of real-world clinical EEG data. This raises questions about the model’s performance in practical, noisy clinical environments. Finally, while the masking strategy of 50% temporal patches and 80% channel patches was empirically optimized, the lack of adaptive masking mechanisms tailored to specific EEG characteristics may restrict the model’s ability to handle diverse patient-specific EEG patterns.
While the proposed method achieves state-of-the-art performance, several avenues for future research remain:
Adaptive Masking and Multi-Band Frequency Attention: Future work could explore dynamic masking strategies and integrate multi-band frequency analysis into the AFTA module. Adaptive masking based on signal characteristics, combined with fine-grained frequency attention, would enhance the model’s ability to handle diverse EEG patterns and improve seizure detection in complex cases.
Multimodal Data Integration: Incorporating EEG signals with complementary data modalities, such as MRI, CT, or clinical metadata, could further enhance model robustness and accuracy. Multimodal approaches would provide richer context for seizure detection and classification, particularly in challenging clinical scenarios.
Real-Time Applications and Deployment: Optimizing the framework for real-time applications by reducing computational complexity and latency would enable its deployment in clinical settings, such as bedside monitoring or wearable devices. This would make the model more practical and accessible for continuous patient monitoring.
Explainability and Clinical Interpretability: Developing explainability methods to visualize the learned frequency-time features and their relationship to clinical markers would facilitate adoption in clinical practice. Enhancing the model’s interpretability would build trust among clinicians and improve its usability in real-world settings.
7. Conclusions
The present study proposes a self-supervised learning Transformer network with AFTA for robust EEG feature representation. Using a masking-and-reconstruction framework, the model is pretrained and fine-tuned for downstream tasks. AFTA integrates an AFFM for global and local frequency-domain filtering with temporal attention, enhancing noise mitigation and feature extraction. Extensive experiments on the TUSZ, TUAB, and TUEV datasets demonstrate the superiority of our approach in seizure detection, classification, and EEG analysis. Our method consistently outperforms state-of-the-art approaches on the TUSZ, TUAB, and TUEV datasets, achieving superior AUROC, balanced accuracy, weighted F1, and Cohen’s kappa, demonstrating robust feature extraction, fine-grained classification, and effective handling of imbalanced EEG data across diverse seizure detection and classification tasks. The ablation study demonstrated that AFTA significantly enhances feature extraction and classification accuracy compared to the baseline. The optimal masking strategy, combining 50% temporal patch masking and 80% channel masking, further ensured robust and generalized model performance. Overall, this work establishes AFTA as a powerful framework for EEG-based seizure analysis and provides new possibilities for advancing neurological studies and clinical applications.
Our approach offers transformative potential for clinical neuroscience by addressing key challenges in EEG-based diagnostics through self-supervised learning (SSL). By training on vast, unlabeled EEG datasets, SSL enables the creation of scalable foundational models that unify diverse EEG recordings (e.g., varying protocols, hardware, or patient populations) under a single framework. This “general-purpose” EEG foundation model would streamline clinical workflows by reducing reliance on manual annotations and enabling cross-institutional data harmonization, crucial for leveraging fragmented multi-center datasets. Clinically, such a model could support universal seizure detection, sleep staging, and neurological disorder screening with minimal task-specific fine-tuning, lowering barriers for deployment in resource-limited settings. Additionally, it establishes a standardized backbone for rapid adaptation to emerging clinical tasks (e.g., biomarker discovery or treatment response prediction), accelerating neurotechnology development while preserving data privacy—a critical advantage for compliance-sensitive healthcare environments. By bridging the gap between fragmented EEG data and clinical decision-making, our method could catalyze the transition from specialized, siloed models to unified neurodiagnostic AI platforms.