Brain Tumor Segmentation Network with Multi-View Ensemble Discrimination and Kernel-Sharing Dilated Convolution
Abstract
1. Introduction
1.1. Motivation
1.2. Contributions
2. Related Work
2.1. Multi-View Fusion
2.2. Multi-Scale Receptive-Field Feature Extraction Model
3. Method
3.1. ACSF Convolution
3.2. Hierarchical Multi-View Fusion Module Based on ACSF Convolution
3.3. Kernel-Sharing Dilated Convolution
4. Results
4.1. Datasets and Evaluation Indicators
4.2. Implementation Details
4.3. Experimental Results and Analysis
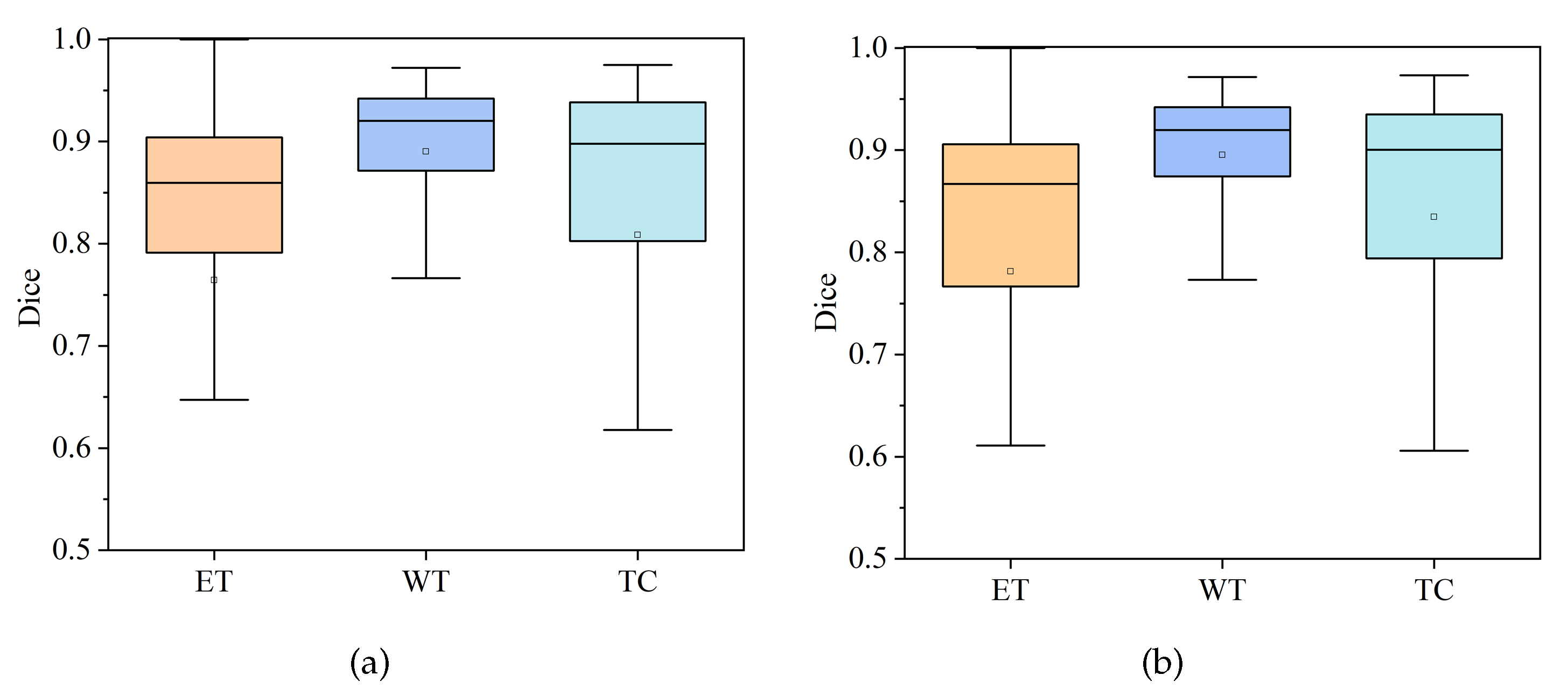
4.3.1. Comparison of HDC-Net and MVKS-Net Boxplots
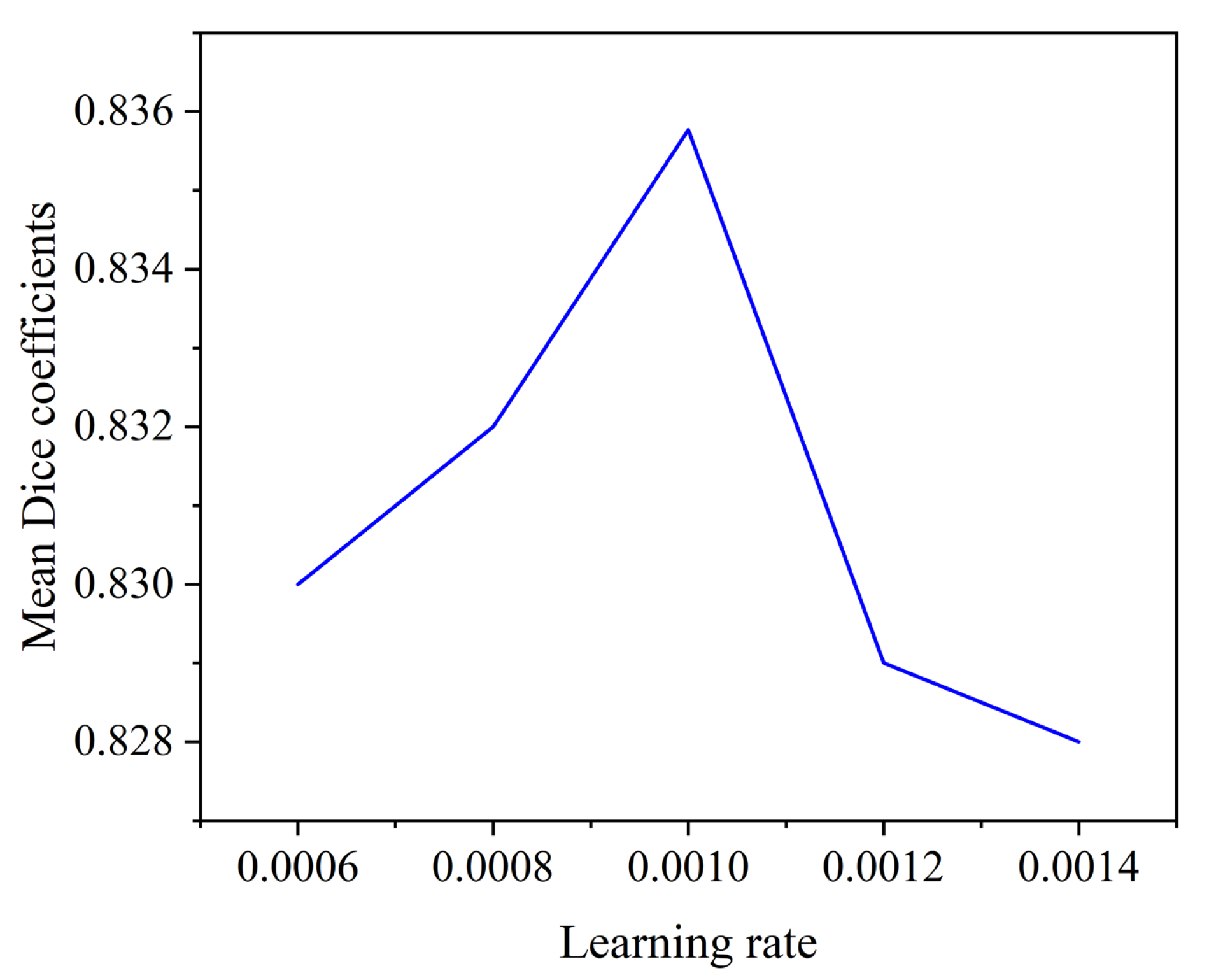
4.3.2. Effect of Initial Learning Rate on Segmentation Performance
4.3.3. Ablation Experimental Analysis
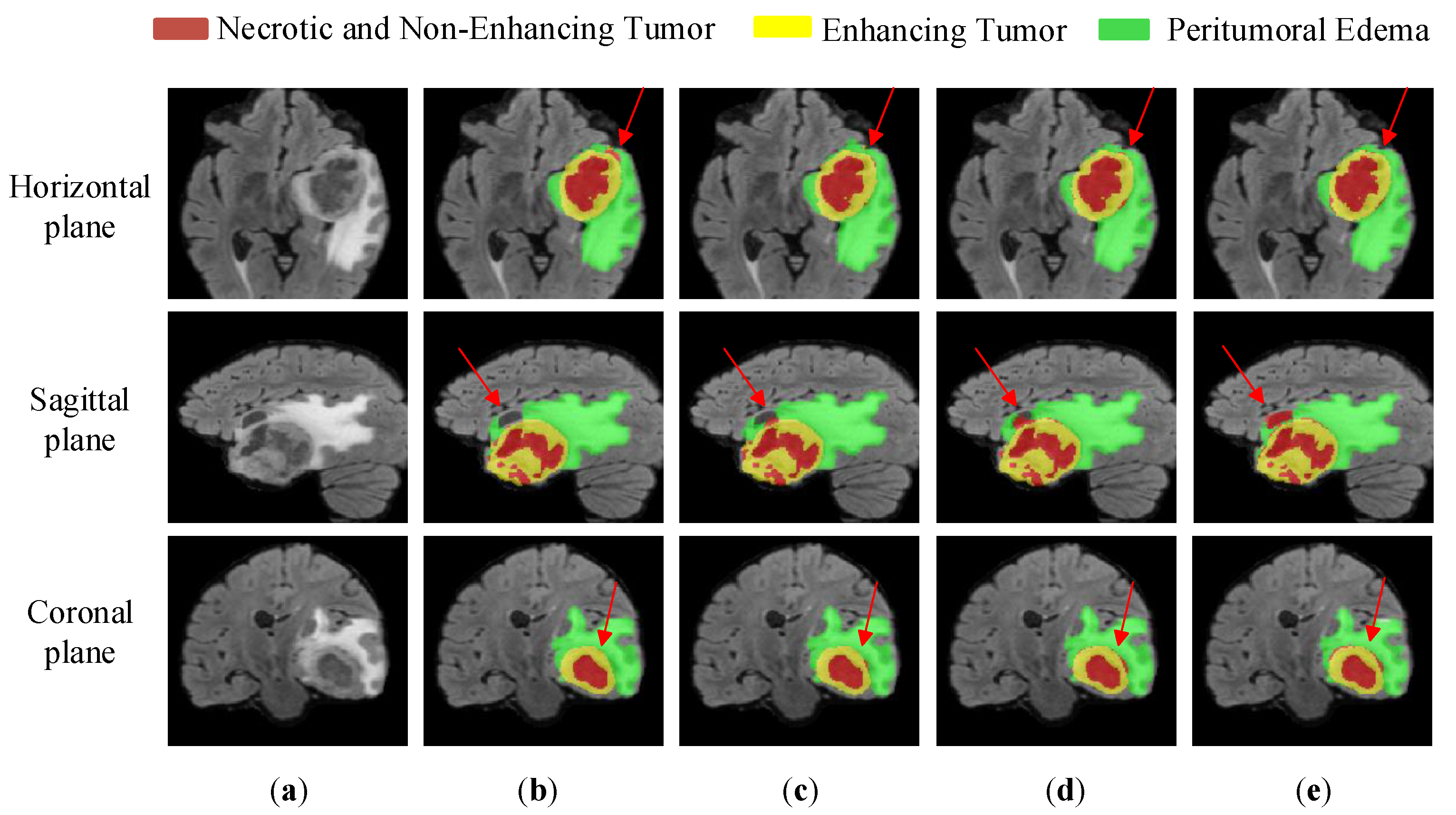
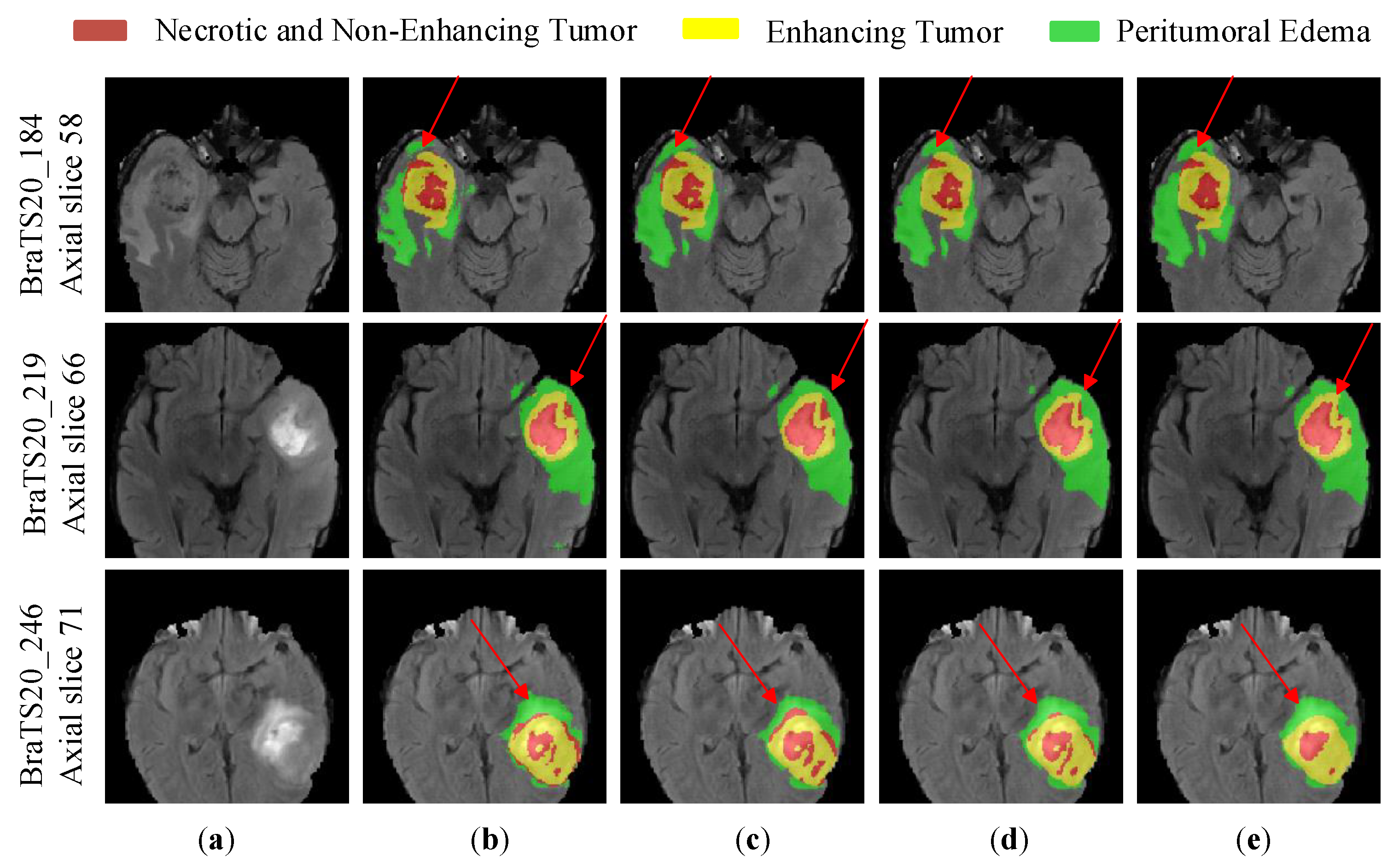
4.3.4. Comparative Experiments and Analysis
5. Conclusions
6. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rahman, T.; Islam, M.S. MRI brain tumor detection and classification using parallel deep convolutional neural networks. Meas. Sensors 2023, 26, 100694. [Google Scholar] [CrossRef]
- Yu, B.; Xie, H.; Xu, Z. PN-GCN: Positive-negative graph convolution neural network in information system to classification. Inf. Sci. 2023, 632, 411–423. [Google Scholar] [CrossRef]
- Karim, A.M.; Kaya, H.; Alcan, V.; Sen, B.; Hadimlioglu, I.A. New optimized deep learning application for COVID-19 detection in chest X-ray images. Symmetry 2022, 14, 1003. [Google Scholar] [CrossRef]
- Osborne, A.; Dorville, J.; Romano, P. Upsampling Monte Carlo Neutron Transport Simulation Tallies using a Convolutional Neural Network. Energy 2023, 13, 100247. [Google Scholar] [CrossRef]
- Fawzi, A.; Achuthan, A.; Belaton, B. Brain image segmentation in recent years: A narrative review. Brain Sci. 2021, 11, 1055. [Google Scholar] [CrossRef]
- Wang, P.; Chung, A.C. Relax and focus on brain tumor segmentation. Med. Image Anal. 2022, 75, 102259. [Google Scholar] [CrossRef]
- Fang, L.; Wang, X. Brain tumor segmentation based on the dual-path network of multi-modal MRI images. J. Pattern Recognit. Soc. 2022, 124, 108434. [Google Scholar] [CrossRef]
- Zhuang, Y.; Liu, H.; Song, E.; Hung, C.C. A 3D Cross-Modality Feature Interaction Network with Volumetric Feature Alignment for Brain Tumor and Tissue Segmentation. IEEE J. Biomed. Health Inform. 2022, 27, 75–86. [Google Scholar] [CrossRef]
- Ding, Y.; Zheng, W.; Geng, J.; Qin, Z.; Choo, K.K.R.; Qin, Z.; Hou, X. MVFusFra: A multi-view dynamic fusion framework for multimodal brain tumor segmentation. IEEE Biomed. Health Inform. 2021, 26, 1570–1581. [Google Scholar] [CrossRef]
- Lahoti, R.; Vengalil, S.K.; Venkategowda, P.B.; Sinha, N.; Reddy, V.V. Whole Tumor Segmentation from Brain MR images using Multi-view 2D Convolutional Neural Network. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 4111–4114. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation based on cascaded convolutional neural networks with uncertainty estimation. Front. Comput. Neurosci. 2019, 13, 56. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2018, 43, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Y.; Chen, W.; Chang, Y.; Gu, H.; Yu, B. MSMANet: A multi-scale mesh aggregation network for brain tumor segmentation. Appl. Soft Comput. 2021, 110, 107733. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Q.; Jia, W.; Lu, Y.; Li, Y.; He, X. See more than once: Kernel-sharing atrous convolution for semantic segmentation. Neurocomputing 2021, 443, 26–34. [Google Scholar] [CrossRef]
- Wang, J.; Gao, J.; Ren, J.; Luan, Z.; Yu, Z.; Zhao, Y.; Zhao, Y. DFP-ResUNet: Convolutional neural network with a dilated convolutional feature pyramid for multimodal brain tumor segmentation. Comput. Methods Programs Biomed. 2021, 208, 106208. [Google Scholar] [CrossRef]
- Zhou, Z.; He, Z.; Jia, Y. AFPNet: A 3D fully convolutional neural network with atrous-convolution feature pyramid for brain tumor segmentation via MRI images. Neurocomputing 2020, 402, 235–244. [Google Scholar] [CrossRef]
- Ahmad, P.; Jin, H.; Qamar, S.; Zheng, R.; Saeed, A. RD 2 A: Densely connected residual networks using ASPP for brain tumor segmentation. Multimed. Tools Appl. 2021, 80, 27069–27094. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 3rd International Workshop, BrainLes 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 14 September 2017; Springer: Berlin/Heidelberg, Germany, 2018; pp. 178–190. [Google Scholar]
- Hu, K.; Gan, Q.; Zhang, Y.; Deng, S.; Xiao, F.; Huang, W.; Cao, C.; Gao, X. Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 2019, 7, 92615–92629. [Google Scholar] [CrossRef]
- Pan, X.; Phan, T.L.; Adel, M.; Fossati, C.; Gaidon, T.; Wojak, J.; Guedj, E. Multi-View Separable Pyramid Network for AD Prediction at MCI Stage by 18F-FDG Brain PET Imaging. IEEE Trans. Med. Imaging 2021, 40, 81–92. [Google Scholar] [CrossRef]
- Liang, S.; Thung, K.H.; Nie, D.; Zhang, Y.; Shen, D. Multi-view spatial aggregation framework for joint localization and segmentation of organs at risk in head and neck CT images. IEEE Trans. Med. Imaging 2020, 39, 2794–2805. [Google Scholar] [CrossRef]
- Zhao, C.; Dewey, B.E.; Pham, D.L.; Calabresi, P.A.; Reich, D.S.; Prince, J.L. SMORE: A self-supervised anti-aliasing and super-resolution algorithm for MRI using deep learning. IEEE Trans. Med. Imaging 2020, 40, 805–817. [Google Scholar] [CrossRef]
- Yang, J.; Huang, X.; He, Y.; Xu, J.; Yang, C.; Xu, G.; Ni, B. Reinventing 2D convolutions for 3D images. IEEE J. Biomed. Health Informatics 2021, 25, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yang, C.; Zeng, M.; Wang, X. TransConver: Transformer and convolution parallel network for developing automatic brain tumor segmentation in MRI images. Quant. Imaging Med. Surg. 2022, 12, 2397. [Google Scholar] [CrossRef] [PubMed]
- Punn, N.S.; Agarwal, S. Multi-modality encoded fusion with 3D inception U-net and decoder model for brain tumor segmentation. Multimed. Tools Appl. 2021, 80, 30305–30320. [Google Scholar] [CrossRef]
- Hussain, S.; Anwar, S.M.; Majid, M. Segmentation of glioma tumors in brain using deep convolutional neural network. Neurocomputing 2018, 282, 248–261. [Google Scholar] [CrossRef]
- Khened, M.; Kollerathu, V.A.; Krishnamurthi, G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 2019, 51, 21–45. [Google Scholar] [CrossRef]
- Asgari Taghanaki, S.; Abhishek, K.; Cohen, J.P.; Cohen-Adad, J.; Hamarneh, G. Deep semantic segmentation of natural and medical images: A review. Artif. Intell. Rev. 2021, 54, 137–178. [Google Scholar] [CrossRef]
- Nuechterlein, N.; Mehta, S. 3D-ESPNet with pyramidal refinement for volumetric brain tumor image segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Springer: Berlin/Heidelberg, Germany, 2019; pp. 245–253. [Google Scholar]
- Mehta, S.; Rastegari, M.; Caspi, A.; Shapiro, L.; Hajishirzi, H. Espnet: Efficient spatial pyramid of dilated convolutions for semantic segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 552–568. [Google Scholar]
- Wang, J.; Yu, Z.; Luan, Z.; Ren, J.; Zhao, Y.; Yu, G. RDAU-Net: Based on a residual convolutional neural network with DFP and CBAM for brain tumor segmentation. Front. Oncol. 2022, 12, 805263. [Google Scholar] [CrossRef]
- Chen, C.; Liu, X.; Ding, M.; Zheng, J.; Li, J. 3D dilated multi-fiber network for real-time brain tumor segmentation in MRI. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 184–192. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the 3rd International Workshop, DLMIA 2017 and 7th International Workshop, ML-CDS 2017, Québec City, QC, Canada, 14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 240–248. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 4th International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Luo, Z.; Jia, Z.; Yuan, Z.; Peng, J. HDC-Net: Hierarchical decoupled convolution network for brain tumor segmentation. IEEE Biomed. Health Inform. 2020, 25, 737–745. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, Proceedings of the 19th International Conference, Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Jiang, Y.; Zhang, Y.; Lin, X.; Dong, J.; Cheng, T.; Liang, J. SwinBTS: A method for 3D multimodal brain tumor segmentation using swin transformer. Brain Sci. 2022, 12, 797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, G.; Huang, H.; Yang, W.; Xu, X.; Liu, Y.; Lai, X. ME-Net: Multi-encoder net framework for brain tumor segmentation. Int. J. Imaging Syst. Technol. 2021, 31, 1834–1848. [Google Scholar] [CrossRef]
- Akbar, A.S.; Fatichah, C.; Suciati, N. Single level UNet3D with multipath residual attention block for brain tumor segmentation. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 3247–3258. [Google Scholar] [CrossRef]
- Liew, A.; Lee, C.C.; Lan, B.L.; Tan, M. CASPIANET++: A multidimensional channel-spatial asymmetric attention network with noisy student curriculum learning paradigm for brain tumor segmentation. Comput. Biol. Med. 2021, 136, 104690. [Google Scholar] [CrossRef]
- Brügger, R.; Baumgartner, C.F.; Konukoglu, E. A partially reversible U-Net for memory-efficient volumetric image segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 429–437. [Google Scholar]
- Zhang, D.; Huang, G.; Zhang, Q.; Han, J.; Han, J.; Wang, Y.; Yu, Y. Exploring task structure for brain tumor segmentation from multi-modality MR images. IEEE Trans. Image Process. 2020, 29, 9032–9043. [Google Scholar] [CrossRef]
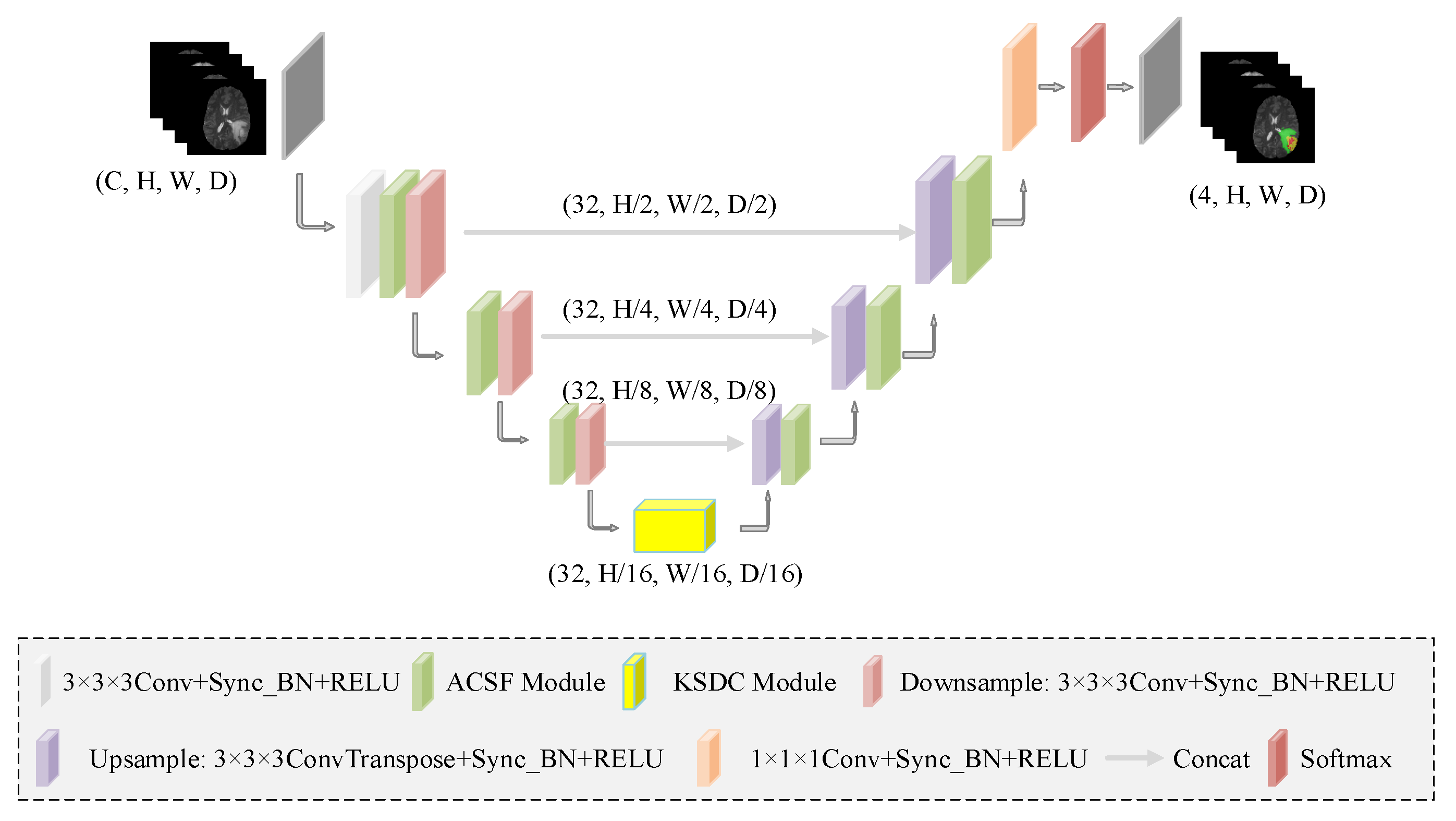
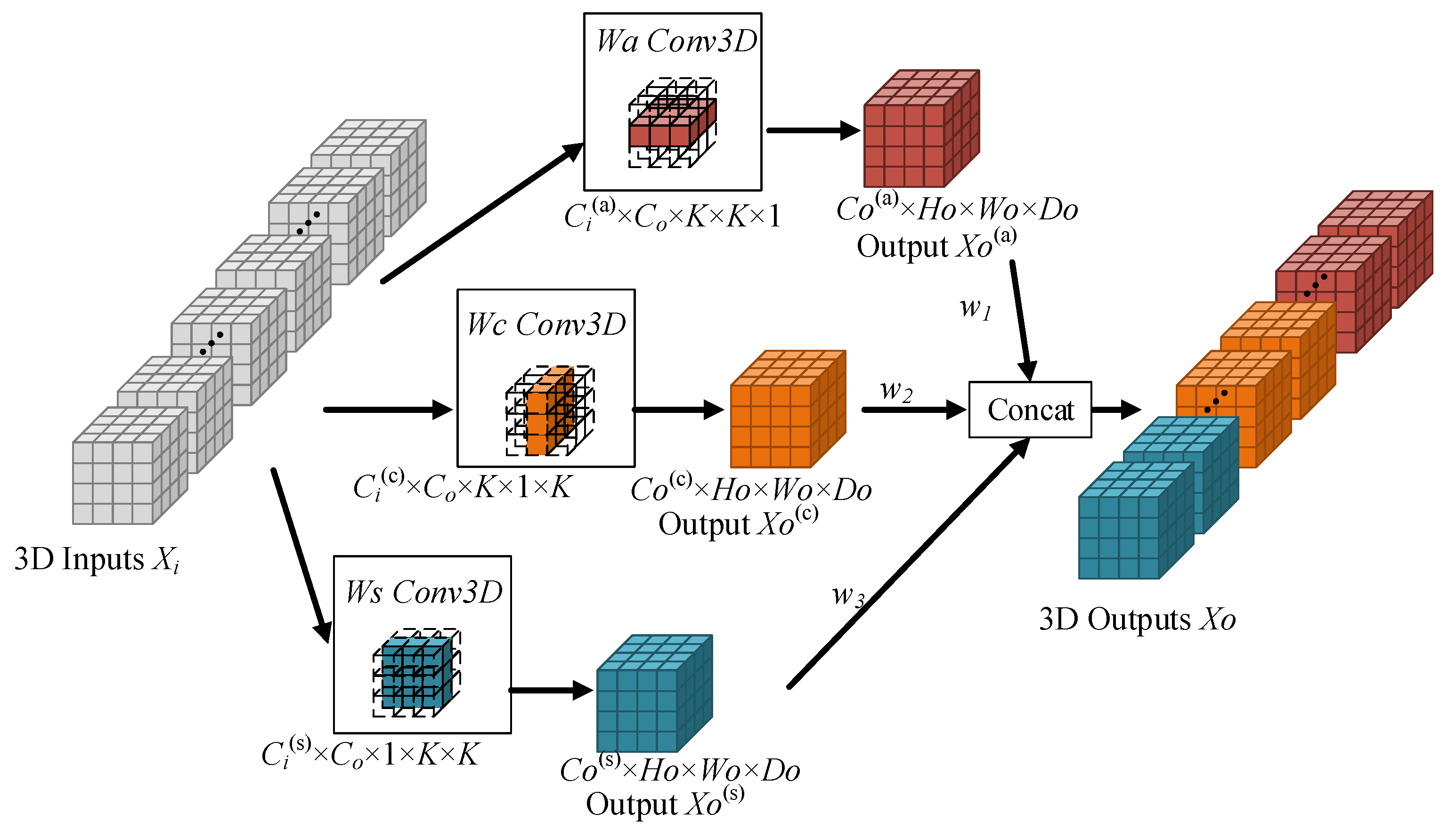
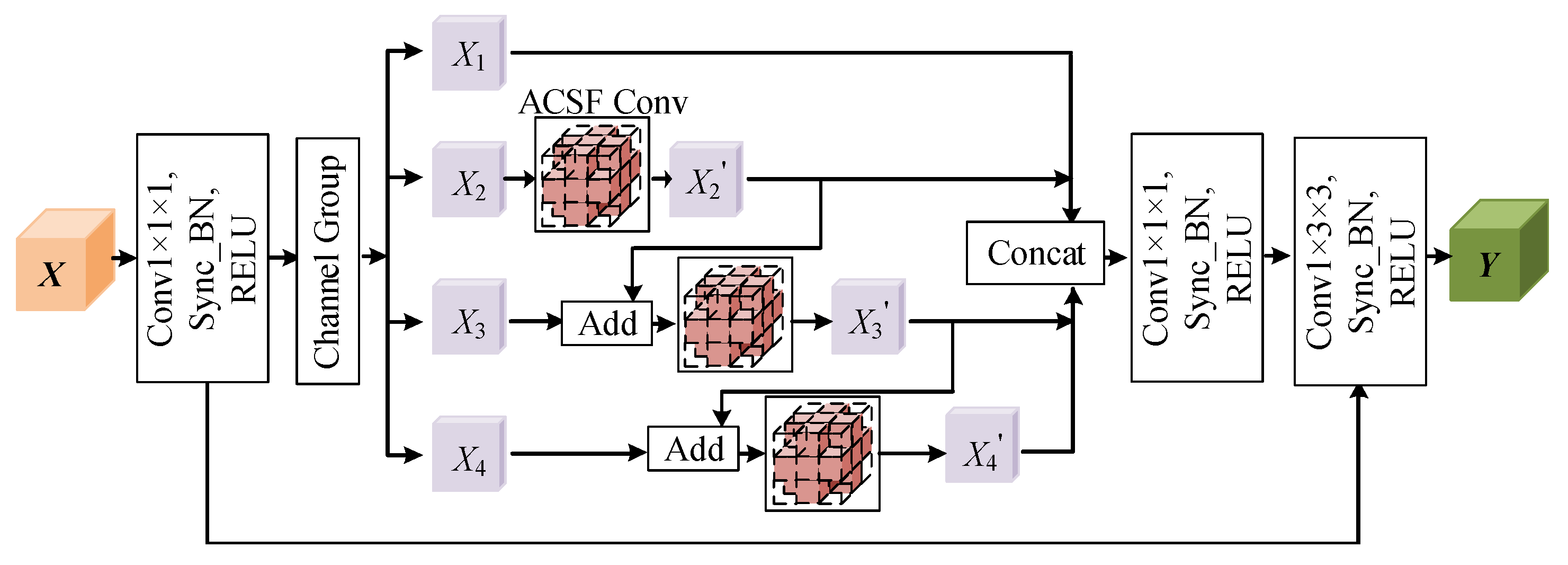

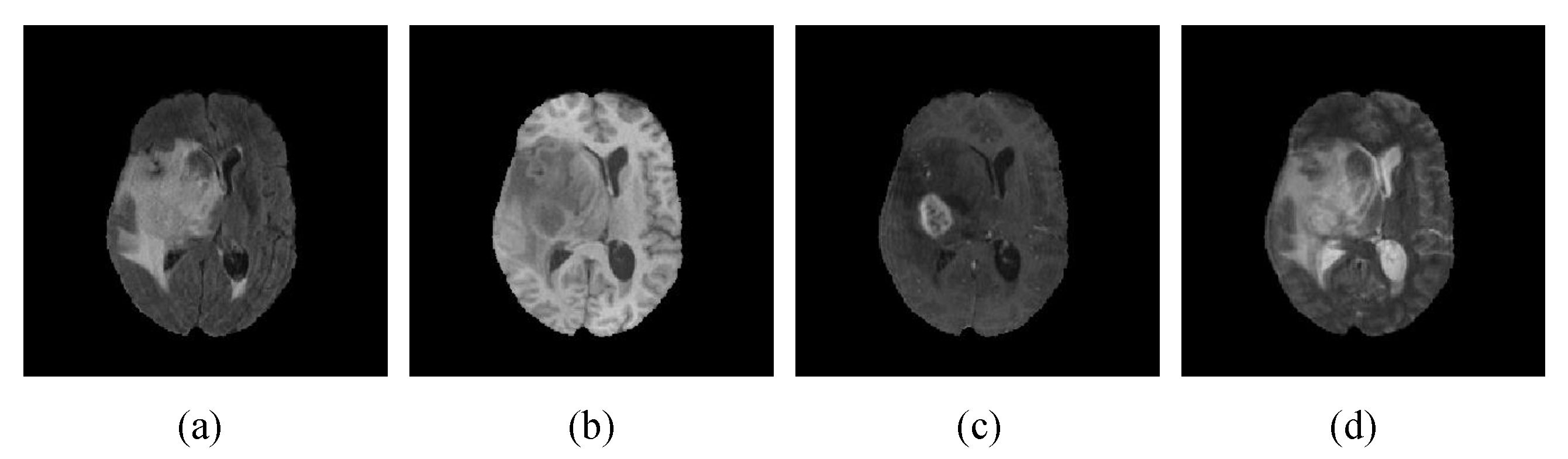

| Name | Details | Input | Output | |
|---|---|---|---|---|
| HDC_transform | Down_sampling | 4 × 128 × 128 × 128 | 32 × 64 × 64 × 64 | |
| Conv 3 × 3 × 3 | Conv 3 × 3 × 3 Block | 32 × 64 × 64 × 64 | 32 × 64 × 64 × 64 | |
| ACSF_module1 | Conv 1 × 1 × 1 Block, ACSF Conv × 3, Conv 1 × 1 × 1 Block, Conv 1 × 3 × 3 Block | 32 × 64 × 64 × 64 | 32 × 64 × 64 × 64 (x1) | |
| Encoding | Conv_down1 | Conv 3 × 3 × 3 Block (stride = 2) | 32 × 64 × 64 × 64 | 32 × 32 × 32 × 32 |
| ACSF_module2 | Conv 1 × 1 × 1 Block, ACSF Conv × 3, Conv 1 × 1 × 1 Block, Conv 1 × 3 × 3 Block | 32 × 32 × 32 × 32 | 32 × 32 × 32 × 32 (x2) | |
| Conv_down2 | Conv 3 × 3 × 3 Block (stride = 2) | 32 × 32 × 32 × 32 | 32 × 16 × 16 × 16 | |
| ACSF_module3 | Conv 1 × 1 × 1 Block, ACSF Conv × 3, Conv 1 × 1 × 1 Block, Conv 1 × 3 × 3 Block | 32 × 16 × 16 × 16 | 32 × 16 × 16 × 16 (x3) | |
| Conv_down3 | Conv 3 × 3 × 3 Block (stride = 2) | 32 × 16 × 16 × 16 | 32 × 8 × 8 × 8 | |
| KSDC_module | KSDC_module | 32 × 8 × 8 × 8 | 32 × 8 × 8 × 8 | |
| Conv_up1 | ConvTranspose 3 × 3 × 3 Block (stride = 2) | 32 × 8 × 8 × 8 | 32 × 16 × 16 × 16 (y1) | |
| Skip_connection1 | torch.cat (y1, x3) | 32 × 16 × 16 × 16 | 64 × 16 × 16 × 16 | |
| ACSF_module4 | Conv 1 × 1 × 1 Block, ACSF Conv × 3, Conv 1 × 1 × 1 Block, Conv 1 × 3 × 3 Block | 64 × 16 × 16 × 16 | 32 × 16 × 16 × 16 | |
| Conv_up2 | ConvTranspose 3 × 3 × 3 Block (stride = 2) | 32 × 16 × 16 × 16 | 32 × 32 × 32 × 32 (y2) | |
| Decoding | Skip_connection2 | torch.cat (y2, x2) | 32 × 32 × 32 × 32 | 64 × 32 × 32 × 32 |
| ACSF_module5 | Conv 1 × 1 × 1 Block, ACSF Conv × 3, Conv 1 × 1 × 1 Block, Conv 1 × 3 × 3 Block | 64 × 32 × 32 × 32 | 32 × 32 × 32 × 32 | |
| Conv_up3 | ConvTranspose 3 × 3 × 3 Block (stride = 2) | 32 × 32 × 32 × 32 | 32 × 64 × 64 × 64 (y3) | |
| Skip_connection1 | torch.cat (y3, x1) | 32 × 64 × 64 × 64 | 64 × 64 × 64 × 64 | |
| ACSF_module6 | Conv 1 × 1 × 1 Block, ACSF Conv × 3, Conv 1 × 1 × 1 Block, Conv 1 × 3 × 3 Block | 64 × 64 × 64 × 64 | 32 × 64 × 64 × 64 | |
| Upsample4 | Up_sampling | 32 × 64 × 64 × 64 | 32 × 128 × 128 × 128 | |
| Conv_output | Conv 1 × 1 × 1 | 32 × 128 × 128 × 128 | 4 × 128 × 128 × 128 | |
| Softmax | Softmax | 4 × 128 × 128 × 128 | 4 × 128 × 128 × 128 |
| Parameter | Value |
|---|---|
| Weight decay | |
| Initial learning rate | |
| Optimizer | Adam |
| Epoch | 900 |
| Batch size | 8 |
| Model | Params (M) | FLOPs (G) | Dice Coefficient (%) | Hausdorff95 (mm) | ||||
|---|---|---|---|---|---|---|---|---|
| ET | WT | TC | ET | WT | TC | |||
| HDC | 0.29 | 25.62 | 76.42 | 89.02 | 80.86 | 35.82 | 9.61 | 13.02 |
| HDC + ACSF | 0.32 | 28.08 | 77.73 | 89.46 | 82.20 | 30.21 | 6.21 | 12.63 |
| HDC + KSDC | 0.47 | 26.11 | 77.53 | 89.11 | 81.34 | 27.58 | 7.24 | 13.53 |
| HDC + ACSF + KSDC | 0.50 | 28.56 | 78.16 | 89.52 | 83.05 | 24.58 | 7.62 | 10.04 |
| Model | Params (M) | FLOPs (G) | Dice Coefficient (%) | Hausdorff95 (mm) | ||||
|---|---|---|---|---|---|---|---|---|
| ET | WT | TC | ET | WT | TC | |||
| 3D-ESPNet [29] | 3.36 | 76.51 | 69.0 | 87.10 | 78.60 | 31.29 | 7.10 | 14.61 |
| DMF-Net [32] | 3.88 | 27.04 | 76.41 | 90.08 | 81.50 | 35.17 | 7.17 | 12.17 |
| HDC-Net [37] | 0.29 | 25.62 | 76.42 | 89.02 | 80.86 | 35.82 | 9.61 | 13.02 |
| MVKS-Net (Ours) | 0.50 | 28.56 | 78.16 | 89.52 | 83.05 | 24.58 | 7.62 | 10.04 |
| Model | Params (M) | FLOPs (G) | Dice Coefficient (%) | Hausdorff95 (mm) | ||||
|---|---|---|---|---|---|---|---|---|
| ET | WT | TC | ET | WT | TC | |||
| 3D U-Net [38] | 16.21 | 1669.53 | 68.76 | 84.11 | 79.06 | 50.98 | 13.37 | 13.61 |
| V-Net [36] | - | - | 68.97 | 86.11 | 77.90 | 43.52 | 14.49 | 16.15 |
| Residual U-Net [39] | - | - | 71.63 | 83.46 | 76.47 | 37.42 | 12.34 | 13.11 |
| Attention U-Net [40] | - | - | 71.83 | 85.57 | 75.96 | 32.94 | 11.91 | 19.43 |
| SwinBTS [41] | - | - | 77.36 | 89.06 | 80.30 | 26.84 | 8.56 | 15.78 |
| ME-Net [42] | - | - | 70.0 | 88.0 | 74.0 | 38.6 | 6.95 | 30.18 |
| Akbar et al [43] | - | - | 72.91 | 88.57 | 80.19 | 31.97 | 10.26 | 13.58 |
| CASPIANET++ [44] | - | - | 77.37 | 89.26 | 81.56 | 27.13 | 7.22 | 9.45 |
| NoNew-Net [45] | 12.42 | 296.82 | 76.8 | 89.1 | 81.9 | 38.35 | 6.32 | 7.34 |
| MVKS-Net (Ours) | 0.50 | 28.56 | 78.16 | 89.52 | 83.05 | 24.58 | 7.62 | 10.04 |
| Model | Params (M) | FLOPs (G) | Dice Coefficient (%) | Hausdorff95 (mm) | ||||
|---|---|---|---|---|---|---|---|---|
| ET | WT | TC | ET | WT | TC | |||
| 3D U-Net [38] | 16.21 | 1669.53 | 75.96 | 88.53 | 71.77 | 6.04 | 17.10 | 11.62 |
| 3D-ESPNet [29] | 3.36 | 76.51 | 73.70 | 88.30 | 81.40 | 5.30 | 5.46 | 7.85 |
| DMF-Net [32] | 3.88 | 27.04 | 78.1 | 89.9 | 83.5 | 3.38 | 4.86 | 7.74 |
| HDC-Net [37] | 0.29 | 25.62 | 79.13 | 89.19 | 83.03 | 2.27 | 5.77 | 7.45 |
| Akbar et al. [43] | - | - | 77.71 | 89.59 | 79.77 | 3.90 | 9.13 | 8.67 |
| Zhang et al. [46] | - | - | 78.2 | 89.6 | 82.4 | 3.57 | 5.73 | 9.27 |
| MVKS-Net (Ours) | 0.50 | 28.56 | 79.88 | 90.00 | 83.39 | 2.31 | 3.95 | 7.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, X.; Zhao, Y.; Nyatega, C.O.; Li, Q. Brain Tumor Segmentation Network with Multi-View Ensemble Discrimination and Kernel-Sharing Dilated Convolution. Brain Sci. 2023, 13, 650. https://doi.org/10.3390/brainsci13040650
Guan X, Zhao Y, Nyatega CO, Li Q. Brain Tumor Segmentation Network with Multi-View Ensemble Discrimination and Kernel-Sharing Dilated Convolution. Brain Sciences. 2023; 13(4):650. https://doi.org/10.3390/brainsci13040650
Chicago/Turabian StyleGuan, Xin, Yushan Zhao, Charles Okanda Nyatega, and Qiang Li. 2023. "Brain Tumor Segmentation Network with Multi-View Ensemble Discrimination and Kernel-Sharing Dilated Convolution" Brain Sciences 13, no. 4: 650. https://doi.org/10.3390/brainsci13040650
APA StyleGuan, X., Zhao, Y., Nyatega, C. O., & Li, Q. (2023). Brain Tumor Segmentation Network with Multi-View Ensemble Discrimination and Kernel-Sharing Dilated Convolution. Brain Sciences, 13(4), 650. https://doi.org/10.3390/brainsci13040650






