Multi-Slice Generation sMRI and fMRI for Autism Spectrum Disorder Diagnosis Using 3D-CNN and Vision Transformers
Abstract
:1. Introduction
- Extracting 1, 10, and 50 slices along each brain plane (axial, sagittal, and coronal) to generate sequences of 2D images from raw 3D sMRI scans.
- Extracting 10, 30, and 50 slices along all brain planes (axial, sagittal, and coronal) to generate sequences of 3D images from raw 4D fMRI scans.
- Extracting all slices or giving some exceptions to the beginnings and the ends along all brain planes (axial, sagittal, and coronal) to generate sequences of 3D images from 3D sMRI and 4D fMRI scans.
- The study explores the diagnostic capability of the proposed 3D-CNN model to automatically leverage ASD biomarkers from 3D sMRI and fMRI generated data, and the classifiability of ASD subjects versus typical control (TC) subjects.
- Finally, this research contributes to the progress of the ASD field by evaluating the proposed methods with state-of-the-art models that have utilized either transfer learning or 3D-CNN models. These models are considered benchmarks for comparison with our models, as they also made use of the same dataset that we used in our study.
2. Related Works
3. Methods
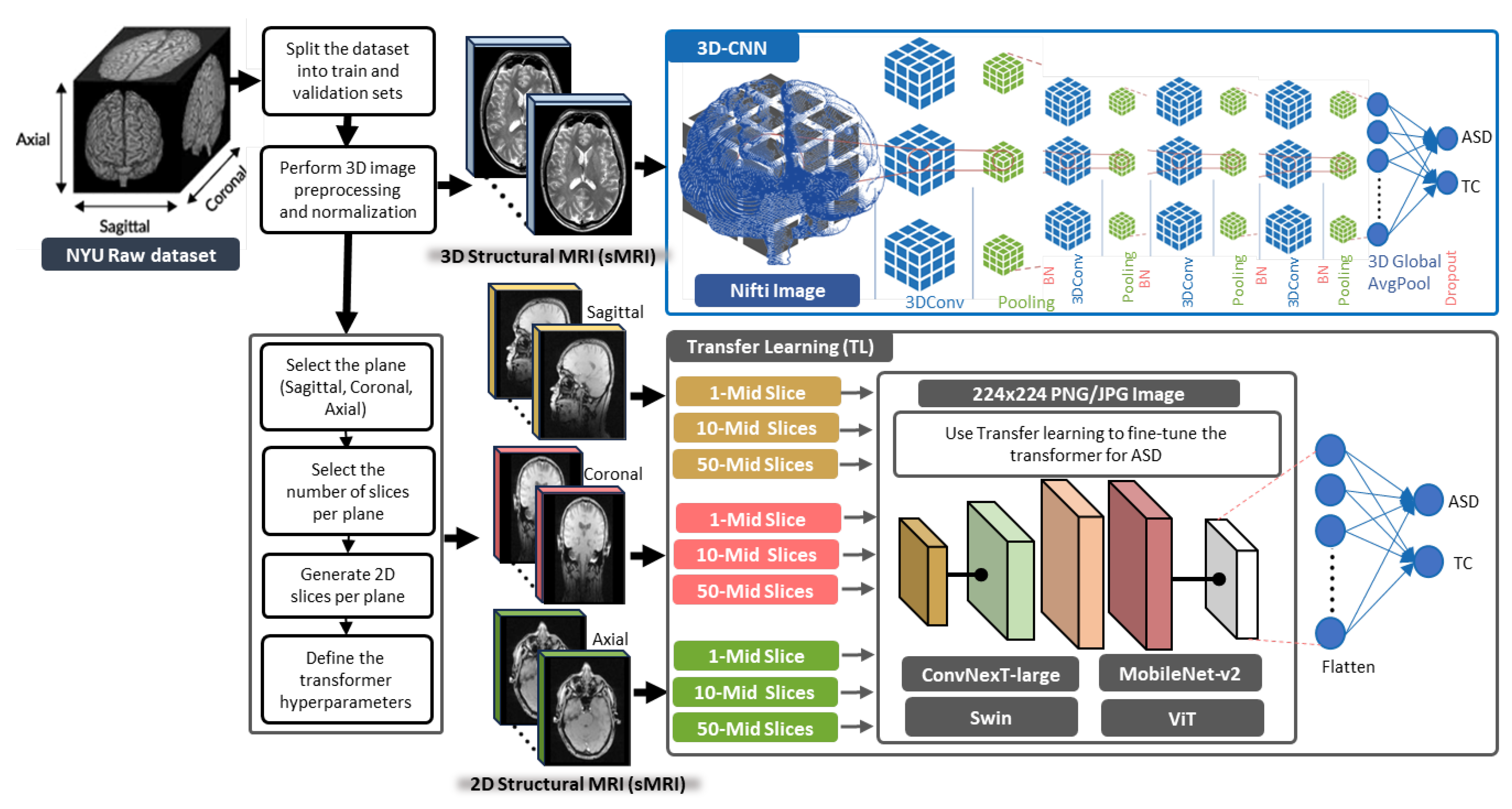
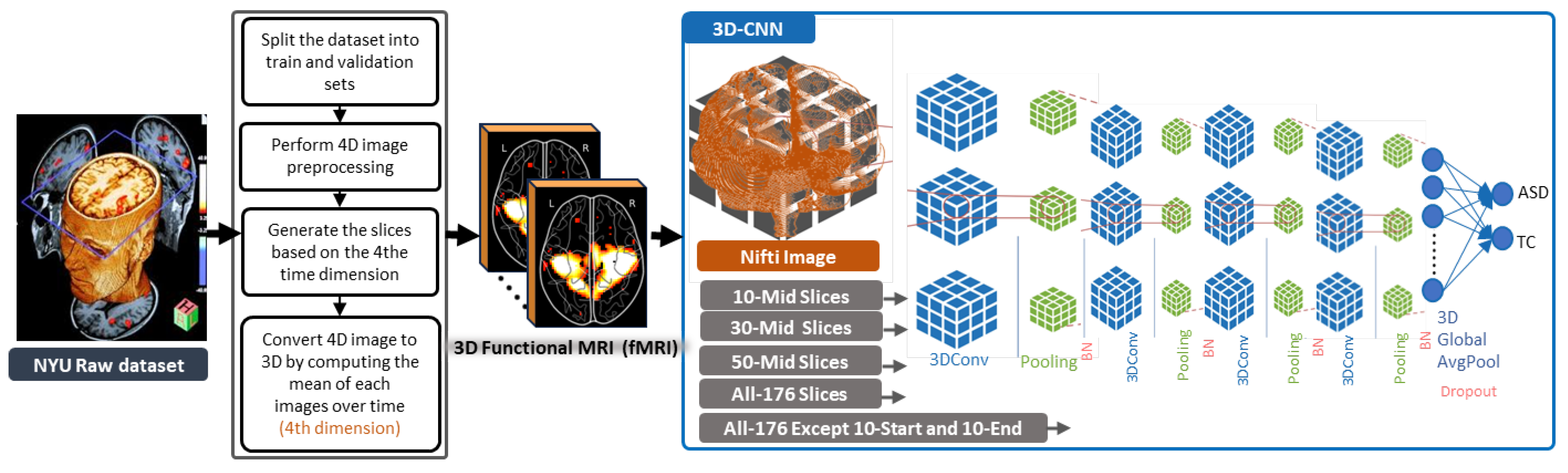
3.1. General Framework of the Proposed Methods
3.2. 3D-CNN Architecture for ASD Diagnosis
3.3. TL Vision Transformers for ASD Diagnosis
4. Experimental Setup
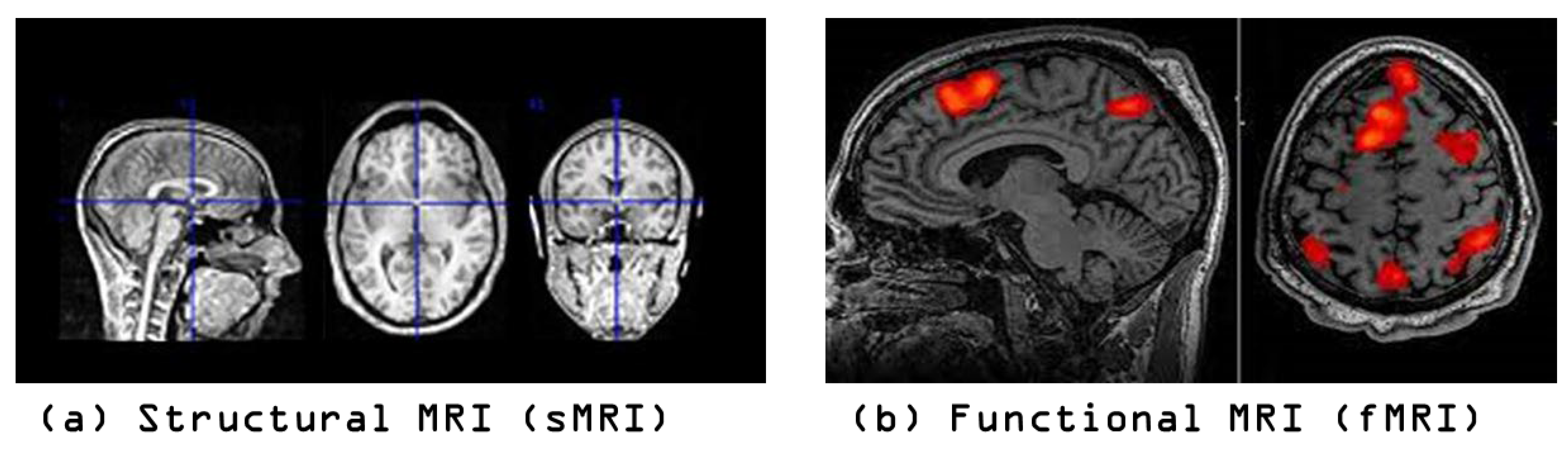
4.1. Datasets
- The ABIDE dataset includes a heterogeneous mix of subjects and imaging modalities with a variety of scanning methods, and some datasets of lower quality, which could impact the outcomes of our study.
- Our decision to use data exclusively from a single site enhanced our findings, and we plan to scale our future research to include additional sites that maintain the same level of scanning and data quality as NYU.
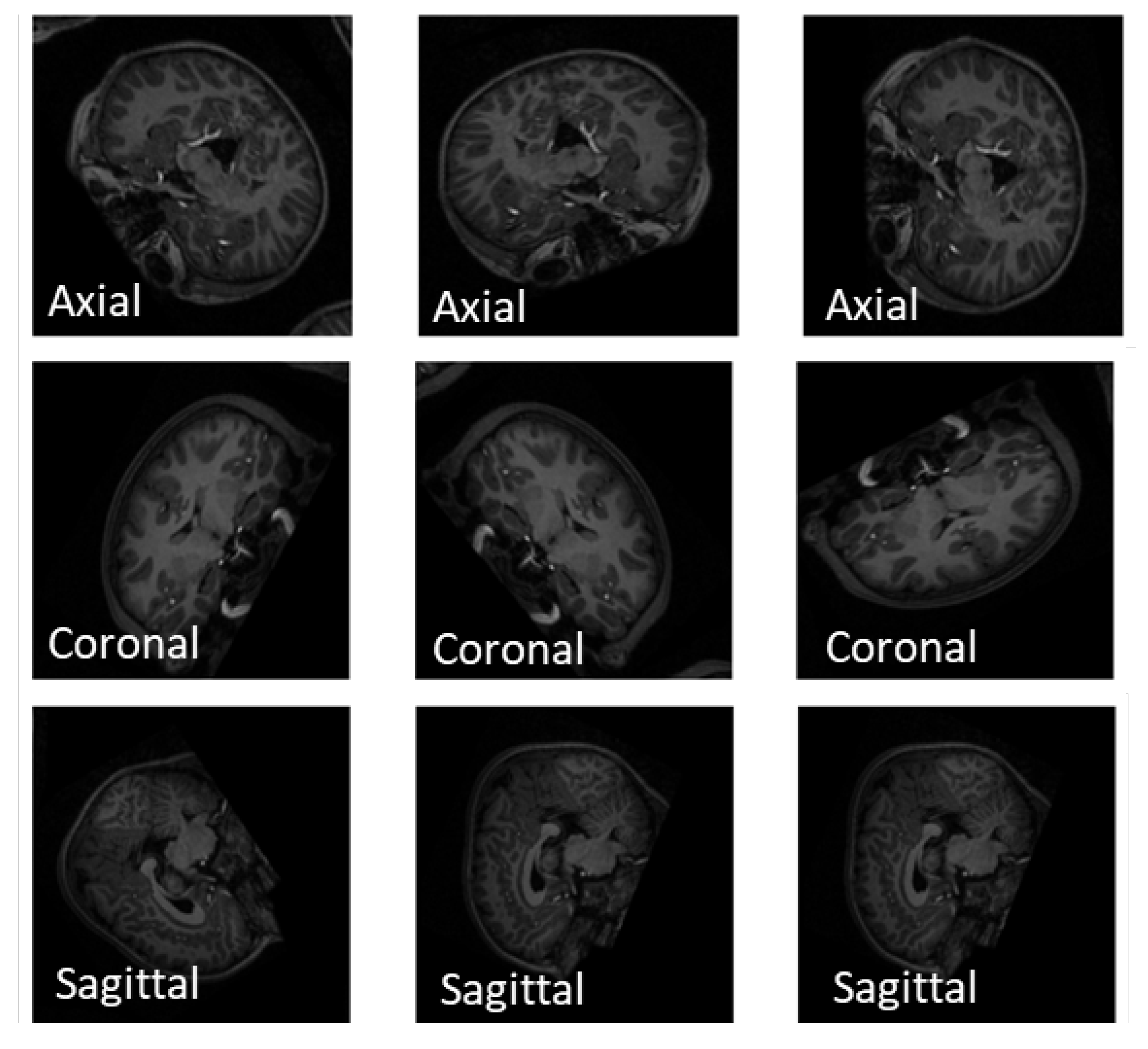
4.2. Slice Extraction from sMRI Modalities
4.3. Slice Extraction from fMRI Modalities
4.4. Baselines
4.5. Evaluation
4.6. Resources and Tools
5. Experimental Results
5.1. ASD Classification Results from sMRI Modalities
5.2. ASD Classification Results from fMRI Modalities
5.3. Comparison with the Baselines for ASD Diagnosis
6. Conclusions and Future Works
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, S.; Summers, R.M. Machine learning and radiology. Med. Image Anal. 2012, 16, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Moridian, P.; Ghassemi, N.; Jafari, M.; Salloum-Asfar, S.; Sadeghi, D.; Khodatars, M.; Shoeibi, A.; Khosravi, A.; Ling, S.H.; Subasi, A.; et al. Automatic autism spectrum disorder detection using artificial intelligence methods with MRI neuroimaging: A review. Front. Mol. Neurosci. 2022, 15, 999605. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, Y.-K.; Xue, S.-W.; Luo, H.; Lu, X.-D.; Zhang, L.-H. Identifying Boys with Autism Spectrum Disorder Based on Whole-Brain Resting-State Interregional Functional Connections Using a Boruta-Based Support Vector Machine Approach. Front. Neurosci. 2022, 16, 761942. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yao, D.; Ma, L.; Liu, M. Multi-site clustering and nested feature extraction for identifying autism spectrum disorder with resting-state fMRI. Med. Image Anal. 2021, 75, 102279. [Google Scholar] [CrossRef]
- Sharif, H.; Khan, R.A. A Novel Machine Learning Based Framework for Detection of Autism Spectrum Disorder (ASD). Appl. Artif. Intell. 2021, 36, 2004655. [Google Scholar] [CrossRef]
- Reiter, M.A.; Jahedi, A.; Fredo, A.R.J.; Fishman, I.; Bailey, B.; Müller, R.-A. Performance of machine learning classification models of autism using resting-state fMRI is contingent on sample heterogeneity. Neural Comput. Appl. 2020, 33, 3299–3310. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Usman, O.L.; Muniyandi, R.C.; Sahran, S.; Mohamed, S.; Razak, R.A. A Review of Machine Learning Methods of Feature Selection and Classification for Autism Spectrum Disorder. Brain Sci. 2020, 10, 949. [Google Scholar] [CrossRef]
- Kunda, M.; Zhou, S.; Gong, G.; Lu, H. Improving Multi-Site Autism Classification via Site-Dependence Minimization and Second-Order Functional Connectivity. IEEE Trans. Med. Imaging 2022, 42, 55–65. [Google Scholar] [CrossRef]
- Chaitra, N.; Vijaya, P.; Deshpande, G. Diagnostic prediction of autism spectrum disorder using complex network measures in a machine learning framework. Biomed. Signal Process. Control 2020, 62, 102099. [Google Scholar] [CrossRef]
- Liu, M.; Li, B.; Hu, D. Autism Spectrum Disorder Studies Using fMRI Data and Machine Learning: A Review. Front. Neurosci. 2021, 15, 697870. [Google Scholar] [CrossRef]
- Kazeminejad, A.; Sotero, R.C. Topological Properties of Resting-State fMRI Functional Networks Improve Machine Learning-Based Autism Classification. Front. Neurosci. 2019, 12, 1018. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Zhang, Y.; Liu, Y.; Liao, H. Single Volume Image Generator and Deep Learning-Based ASD Classification. IEEE J. Biomed. Health Inform. 2020, 24, 3044–3054. [Google Scholar] [CrossRef] [PubMed]
- Shahamat, H.; Abadeh, M.S. Brain MRI analysis using a deep learning based evolutionary approach. Neural Netw. 2020, 126, 218–234. [Google Scholar] [CrossRef]
- Gao, J.; Chen, M.; Li, Y.; Gao, Y.; Li, Y.; Cai, S.; Wang, J. Multisite Autism Spectrum Disorder Classification Using Convolutional Neural Network Classifier and Individual Morphological Brain Networks. Front. Neurosci. 2021, 14, 629630. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, F.; Han, T.; Gong, X.; Duan, F. Detection of Autism Spectrum Disorder using fMRI Functional Connectivity with Feature Selection and Deep Learning. Cogn. Comput. 2022, 15, 1106–1117. [Google Scholar] [CrossRef]
- Yin, W.; Mostafa, S.; Wu, F.-X. Diagnosis of Autism Spectrum Disorder Based on Functional Brain Networks with Deep Learning. J. Comput. Biol. 2021, 28, 146–165. [Google Scholar] [CrossRef]
- Yang, M.; Cao, M.; Chen, Y.; Chen, Y.; Fan, G.; Li, C.; Wang, J.; Liu, T. Large-Scale Brain Functional Network Integration for Discrimination of Autism Using a 3-D Deep Learning Model. Front. Hum. Neurosci. 2021, 15, 687288. [Google Scholar] [CrossRef]
- Thomas, R.M.; Gallo, S.; Cerliani, L.; Zhutovsky, P.; El-Gazzar, A.; van Wingen, G. Classifying Autism Spectrum Disorder Using the Temporal Statistics of Resting-State Functional MRI Data With 3D Convolutional Neural Networks. Front. Psychiatry 2020, 11, 440. [Google Scholar] [CrossRef]
- Subah, F.Z.; Deb, K.; Dhar, P.K.; Koshiba, T. A Deep Learning Approach to Predict Autism Spectrum Disorder Using Multisite Resting-State fMRI. Appl. Sci. 2021, 11, 3636. [Google Scholar] [CrossRef]
- Shao, L.; Fu, C.; You, Y.; Fu, D. Classification of ASD based on fMRI data with deep learning. Cogn. Neurodyn. 2021, 15, 961–974. [Google Scholar] [CrossRef]
- Khodatars, M.; Shoeibi, A.; Sadeghi, D.; Ghaasemi, N.; Jafari, M.; Moridian, P.; Khadem, A.; Alizadehsani, R.; Zare, A.; Kong, Y.; et al. Deep learning for neuroimaging-based diagnosis and rehabilitation of Autism Spectrum Disorder: A review. Comput. Biol. Med. 2021, 139, 104949. [Google Scholar] [CrossRef] [PubMed]
- Kashef, R. ECNN: Enhanced convolutional neural network for efficient diagnosis of autism spectrum disorder. Cogn. Syst. Res. 2021, 71, 41–49. [Google Scholar] [CrossRef]
- Eslami, T.; Saeed, F. Auto-ASD-Network: A Technique Based on Deep Learning and Support Vector Machines for Diagnosing Autism Spectrum Disorder Using FMRI Data. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Niagara Falls, NY, USA, 7–10 September 2019; pp. 646–651. [Google Scholar] [CrossRef]
- Cao, M.; Yang, M.; Qin, C.; Zhu, X.; Chen, Y.; Wang, J.; Liu, T. Using DeepGCN to identify the autism spectrum disorder from multi-site resting-state data. Biomed. Signal Process. Control 2021, 70, 103015. [Google Scholar] [CrossRef]
- Almuqhim, F.; Saeed, F. ASD-SAENet: A Sparse Autoencoder, and Deep-Neural Network Model for Detecting Autism Spectrum Disorder (ASD) Using fMRI Data. Front. Comput. Neurosci. 2021, 15, 654315. [Google Scholar] [CrossRef]
- Ahammed, S.; Niu, S.; Ahmed, R.; Dong, J.; Gao, X.; Chen, Y. DarkASDNet: Classification of ASD on Functional MRI Using Deep Neural Network. Front. Neurosci. 2021, 15, 635657. [Google Scholar] [CrossRef] [PubMed]
- Sewani, H.; Kashef, R. An Autoencoder-Based Deep Learning Classifier for Efficient Diagnosis of Autism. Children 2020, 7, 182. [Google Scholar] [CrossRef]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A Hybrid Learning Approach for Detection of Autism Spectrum Disorder Using fMRI Data. Front. Neurosci. 2019, 13, 70. [Google Scholar] [CrossRef]
- Devika, K.; Mahapatra, D.; Subramanian, R.; Oruganti, V.R.M. Outlier-Based Autism Detection Using Longitudinal Structural MRI. IEEE Access 2022, 10, 27794–27808. [Google Scholar] [CrossRef]
- Yang, C.; Wang, P.; Tan, J.; Liu, Q.; Li, X. Autism spectrum disorder diagnosis using graph attention network based on spatial-constrained sparse functional brain networks. Comput. Biol. Med. 2021, 139, 104963. [Google Scholar] [CrossRef]
- Wang, Z.; Peng, D.; Shang, Y.; Gao, J. Autistic Spectrum Disorder Detection and Structural Biomarker Identification Using Self-Attention Model and Individual-Level Morphological Covariance Brain Networks. Front. Neurosci. 2021, 15, 756868. [Google Scholar] [CrossRef]
- Niu, K.; Guo, J.; Pan, Y.; Gao, X.; Peng, X.; Li, N.; Li, H. Multichannel Deep Attention Neural Networks for the Classification of Autism Spectrum Disorder Using Neuroimaging and Personal Characteristic Data. Complexity 2020, 2020, 1357853. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, L.; Li, J.; Yu, J.Y.A.X.; Yu, X. Attentional Connectivity-based Prediction of Autism Using Heterogeneous rs-fMRI Data from CC200 Atlas. Exp. Neurobiol. 2020, 29, 27–37. [Google Scholar] [CrossRef]
- Hu, J.; Cao, L.; Li, T.; Dong, S.; Li, P. GAT-LI: A graph attention network based learning and interpreting method for functional brain network classification. BMC Bioinform. 2021, 22, 379. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Vasilev, I.; Slater, D.; Spacagna, G.; Roelants, P.; Zocca, V. Python Deep Learning, 2nd ed.; 2019; preprints. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. arXiv 2016, arXiv:1603.05027. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Tang, M.; Kumar, P.; Chen, H.; Shrivastava, A. Deep Multimodal Learning for the Diagnosis of Autism Spectrum Disorder. J. Imaging 2020, 6, 47. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2018, arXiv:1801.04381. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv 2021, arXiv:2103.14030. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Akter, T.; Ali, M.H.; Khan, I.; Satu, S.; Uddin, J.; Alyami, S.A.; Ali, S.; Azad, A.; Moni, M.A. Improved Transfer-Learning-Based Facial Recognition Framework to Detect Autistic Children at an Early Stage. Brain Sci. 2021, 11, 734. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Perkowski, M. Deep Learning Approach for Screening Autism Spectrum Disorder in Children with Facial Images and Analysis of Ethnoracial Factors in Model Development and Application. Brain Sci. 2021, 11, 1446. [Google Scholar] [CrossRef]
- Tampu, I.E.; Eklund, A.; Haj-Hosseini, N. Inflation of test accuracy due to data leakage in deep learning-based classification of OCT images. Sci. Data 2022, 9, 580. [Google Scholar] [CrossRef]
- Reinhold, J.C.; Dewey, B.E.; Carass, A.; Prince, J.L. Evaluating the Impact of Intensity Normalization on MR Image Synthesis. Proc. SPIE Int. Soc. Opt. Eng. 2019, 10949, 890–898. [Google Scholar] [CrossRef]
- Zhang, H.; Goodfellow, I.; Metaxas, D.; Odena, A. Self-Attention Generative Adversarial Networks. arXiv 2018, arXiv:1805.08318. [Google Scholar] [CrossRef]
- Mehta, S.; Rastegari, M. MobileViT: Light-weight, General-purpose, and Mobile-friendly Vision Transformer. arXiv 2021, arXiv:2110.02178. [Google Scholar] [CrossRef]
- Jun, E.; Jeong, S.; Heo, D.-W.; Suk, H.-I. Medical Transformer: Universal Brain Encoder for 3D MRI Analysis. arXiv 2021, arXiv:2104.13633. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2023, 3, 59–78. [Google Scholar] [CrossRef]
- Jun, L.; Junyu, C.; Yucheng, T.; Ce, W.; Bennett, A.L.; Zhou, S.K. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. arXiv 2022, arXiv:2206.01136. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Wu, F.-X.; Hayrat, R.; Liu, J. AIMAFE: Autism spectrum disorder identification with multi-atlas deep feature representation and ensemble learning. J. Neurosci. Methods 2020, 343, 108840. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lin, L.; Sun, S.; Wu, S. A review of the application of three-dimensional convolutional neural networks for the diagnosis of Alzheimer’s disease using neuroimaging. Rev. Neurosci. 2023, 34, 649–670. [Google Scholar] [CrossRef] [PubMed]



| Site | ASD | TC | ||||
|---|---|---|---|---|---|---|
| AGE AVG | GENDER | TOTAL COUNT | AGE AVG | GENDER | TOTAL COUNT | |
| NYU (sMRI) | 14.51 | 11 F, 68 M | 79 | 15.8 | 26 F, 79 M | 105 |
| NYU (fMRI) | 14.76 | 10 F, 64 M | 74 | 15.75 | 26 F, 72 M | 98 |
| Site | 3D sMRI | Brain View | No Slices/Plane | Train Set (80%) Case No | Test Set (20%) Case No | ||
|---|---|---|---|---|---|---|---|
| 0952 to 1018 | 1036 to 1122 | 1019 to 1035 | 1123 to 1159 | ||||
| ASD | TC | ASD | TC | ||||
| NYU | ASD 79, TC 105 | Axial | 1 | 64 | 84 | 15 | 21 |
| NYU | ASD 79, TC 105 | Coronal | 1 | 64 | 84 | 15 | 21 |
| NYU | ASD 79, TC 105 | Sagittal | 1 | 64 | 84 | 15 | 21 |
| NYU | ASD 79, TC 105 | Axial | 10 | 640 | 840 | 150 | 210 |
| NYU | ASD 79, TC 105 | Coronal | 10 | 640 | 840 | 150 | 210 |
| NYU | ASD 79, TC 105 | Sagittal | 10 | 640 | 840 | 150 | 210 |
| NYU | ASD 79, TC 105 | Axial | 50 | 3200 | 4200 | 750 | 1050 |
| NYU | ASD 79, TC 105 | Coronal | 50 | 3200 | 4200 | 750 | 1050 |
| NYU | ASD 79, TC 105 | Sagittal | 50 | 3200 | 4200 | 750 | 1050 |
| Method | Brain Plane | No Slices | Loss | Acc | F1 | No Slices | Loss | Acc | F1 | No Slices | Loss | Acc | F1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ViT | Axial | 1 slice | 0.68 | 0.58 | 0.36 | 10 slices | 0.69 | 0.63 | 0.51 | 50 slices | 0.67 | 0.61 | 0.54 |
| Coronal | 1 slice | 0.68 | 0.58 | 0.36 | 10 slices | 0.69 | 0.61 | 0.44 | 50 slices | 0.68 | 0.59 | 0.43 | |
| Sagittal | 1 slice | 0.67 | 0.58 | 0.36 | 10 slices | 0.68 | 0.60 | 0.46 | 50 slices | 0.68 | 0.58 | 0.36 | |
| Swin | Axial | 1 slice | 0.67 | 0.58 | 0.36 | 10 slices | 0.67 | 0.58 | 0.36 | 50 slices | 0.67 | 0.58 | 0.36 |
| Coronal | 1 slice | 0.67 | 0.58 | 0.36 | 10 slices | 0.68 | 0.58 | 0.36 | 50 slices | 0.67 | 0.58 | 0.36 | |
| Sagittal | 1 slice | 0.67 | 0.58 | 0.36 | 10 slices | 0.67 | 0.58 | 0.36 | 50 slices | 0.67 | 0.58 | 0.36 | |
| MobileNet | Axial | 1 slice | 0.70 | 0.72 | 0.70 | 10 slices | 0.67 | 0.68 | 0.62 | 50 slices | 0.66 | 0.64 | 0.59 |
| Coronal | 1 slice | 0.71 | 0.66 | 0.58 | 10 slices | 0.67 | 0.69 | 0.64 | 50 slices | 0.65 | 0.65 | 0.59 | |
| Sagittal | 1 slice | 0.67 | 0.61 | 0.59 | 10 slices | 0.68 | 0.60 | 0.45 | 50 slices | 0.66 | 0.64 | 0.58 | |
| ConvNeXt | Axial | 1 slice | 0.63 | 0.77 | 0.76 | 10 slices | 0.64 | 0.71 | 0.68 | 50 slices | 0.67 | 0.65 | 0.58 |
| Coronal | 1 slice | 0.64 | 0.69 | 0.66 | 10 slices | 0.64 | 0.67 | 0.62 | 50 slices | 0.64 | 0.68 | 0.61 | |
| Sagittal | 1 slice | 0.66 | 0.69 | 0.68 | 10 slices | 0.76 | 0.63 | 0.53 | 50 slices | 0.68 | 0.66 | 0.61 | |
| 3D-CNN | 3D (Axial, Coronal, Sagittal) | All slices | 0.73 | 0.66 | 0.57 | ||||||||
| Method | Brain Plane | No Slices | Loss | Accuracy | F1 |
|---|---|---|---|---|---|
| 3D-CNN | 3D (Axial, Coronal, Sagittal) | 10-Mid Slices | 0.8793 | 0.7581 | 0.6809 |
| 3D (Axial, Coronal, Sagittal) | 30-Mid Slices | 1.4709 | 0.7903 | 0.6667 | |
| 3D (Axial, Coronal, Sagittal) | 50-Mid Slices | 0.4044 | 0.8710 | 0.8261 | |
| 3D (Axial, Coronal, Sagittal) | All-176 Slices | 1.2591 | 0.7581 | 0.6512 | |
| 3D (Axial, Coronal, Sagittal) | All-176 Except 10-Start & 10-End | 0.5081 | 0.8387 | 0.7727 |
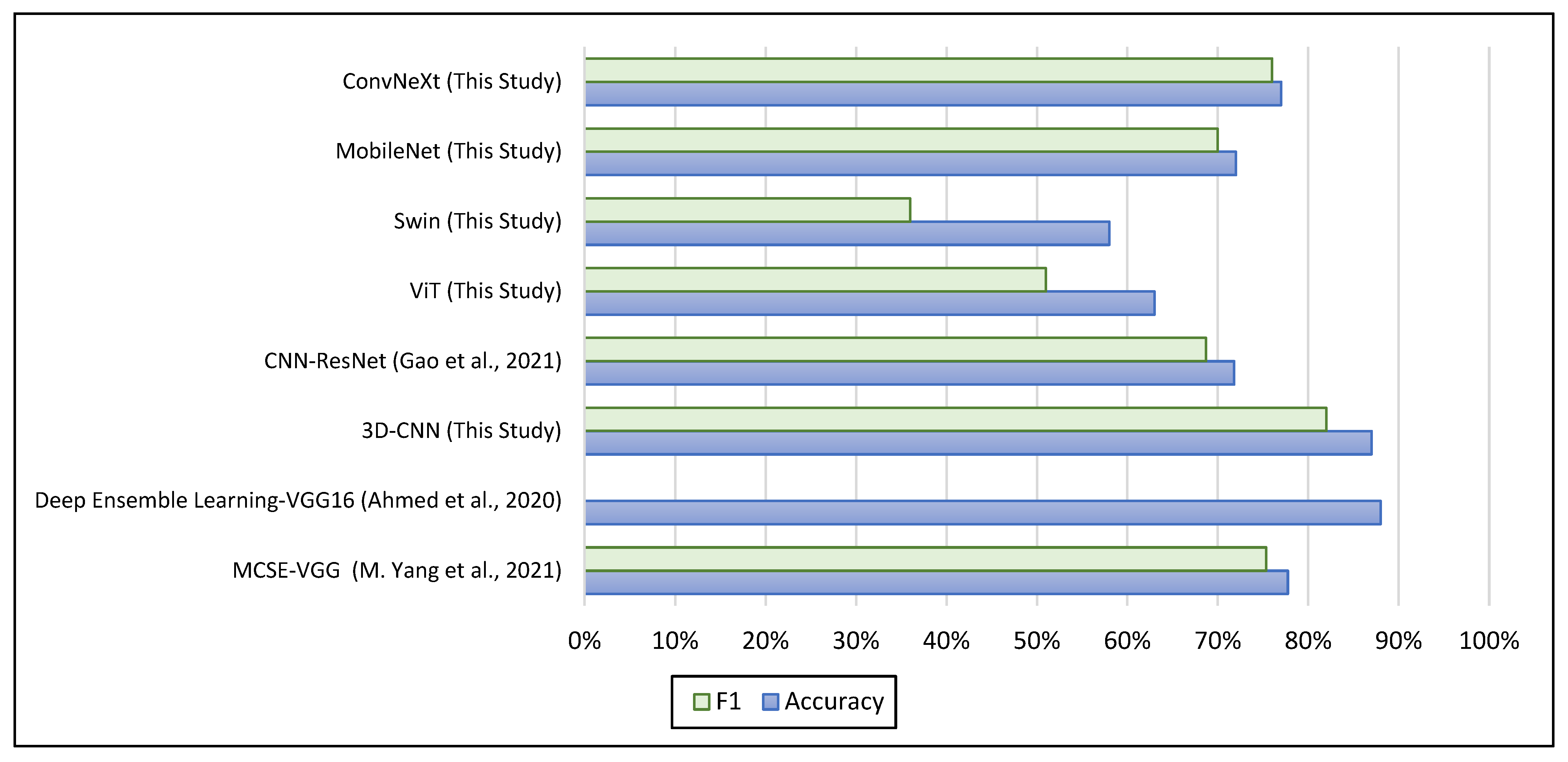
| Study | Site | Input | No Subjects | Accuracy | F1 |
|---|---|---|---|---|---|
| MCSE—VGG [17] | NYU | fMRI—Brain Functional Networks | ASD 79; TC 105 | 77.74% | 75.33% |
| Deep Ensemble Learning—VGG16 [12] | NYU | fMRI—Glass Brain | ASD 79; TC 105 | 88% | — |
| 3D-CNN (This Study) | NYU | fMRI—Mean Image | ASD 74; TC 98 | 87% | 82% |
| Study | Site | Input | No Subjects | Accuracy | F1 |
|---|---|---|---|---|---|
| CNN—ResNet [14] | ABIDE-I | sMRI—Covariance Brain Networks | ASD 518; TC 567 | 71.81% | 68.68% |
| ViT (This Study) | NYU | sMRI—2D Slices | ASD 79; TC 105 | 63% | 51% |
| Swin (This Study) | NYU | sMRI—2D Slices | ASD 79; TC 105 | 58% | 36% |
| MobileNet (This Study) | NYU | sMRI—2D Slices | ASD 79; TC 105 | 72% | 70% |
| ConvNeXt (This Study) | NYU | sMRI—2D Slices | ASD 79; TC 105 | 77% | 76% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharthi, A.G.; Alzahrani, S.M. Multi-Slice Generation sMRI and fMRI for Autism Spectrum Disorder Diagnosis Using 3D-CNN and Vision Transformers. Brain Sci. 2023, 13, 1578. https://doi.org/10.3390/brainsci13111578
Alharthi AG, Alzahrani SM. Multi-Slice Generation sMRI and fMRI for Autism Spectrum Disorder Diagnosis Using 3D-CNN and Vision Transformers. Brain Sciences. 2023; 13(11):1578. https://doi.org/10.3390/brainsci13111578
Chicago/Turabian StyleAlharthi, Asrar G., and Salha M. Alzahrani. 2023. "Multi-Slice Generation sMRI and fMRI for Autism Spectrum Disorder Diagnosis Using 3D-CNN and Vision Transformers" Brain Sciences 13, no. 11: 1578. https://doi.org/10.3390/brainsci13111578
APA StyleAlharthi, A. G., & Alzahrani, S. M. (2023). Multi-Slice Generation sMRI and fMRI for Autism Spectrum Disorder Diagnosis Using 3D-CNN and Vision Transformers. Brain Sciences, 13(11), 1578. https://doi.org/10.3390/brainsci13111578






