Applications of Machine Learning to Diagnosis of Parkinson’s Disease
Abstract
:1. Introduction
2. Methods
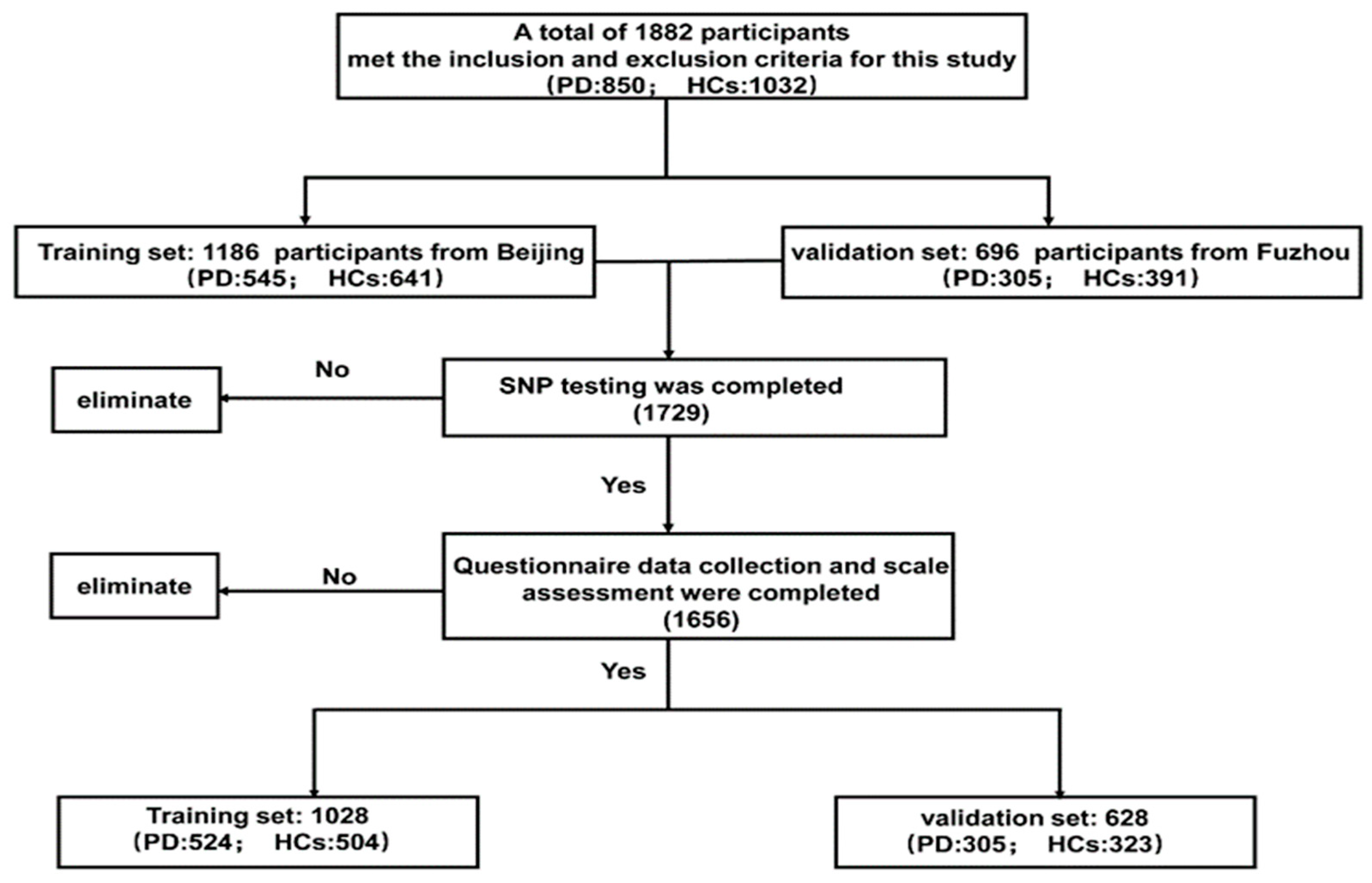
2.1. Study Design and Data Source
2.2. Candidate Variables
2.3. Genotype Analysis and Classification
2.4. Machine Learning Algorithm
2.5. Statistical Analysis
3. Results
3.1. Clinical and Demographic Characteristics
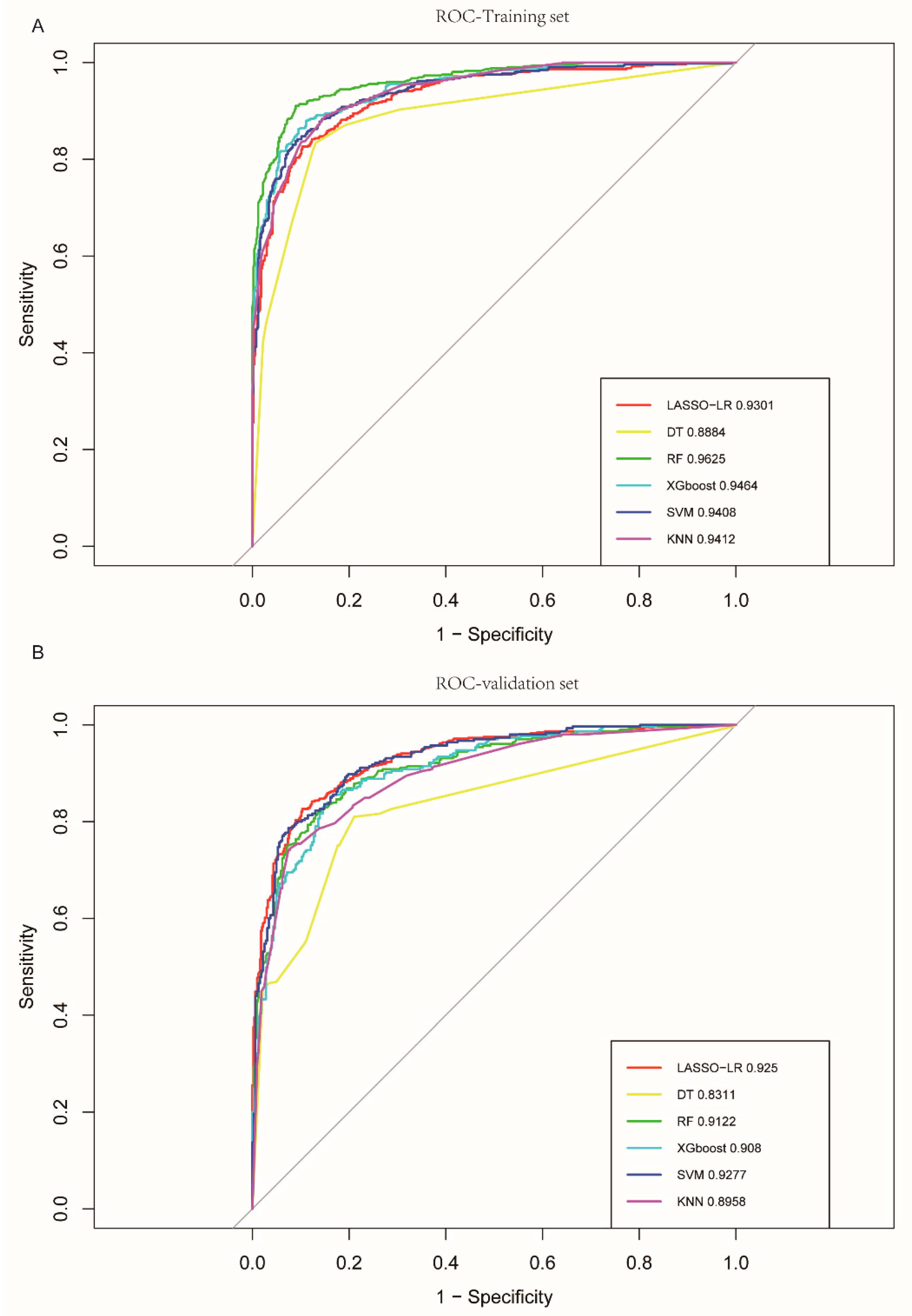
3.2. Comparison of Algorithms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- De Lau, L.M.L.; Breteler, M.M.B. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef]
- Ascherio, A.; Schwarzschild, M.A. The epidemiology of Parkinson’s disease: Risk factors and prevention. Lancet Neurol. 2016, 15, 1257–1272. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-X.; Chen, L. Economic Burden Analysis of Parkinson’s Disease Patients in China. Park. Dis. 2017, 2017, 8762939. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.J.; Diener, M.D.; Kaltenboeck, A.; Birnbaum, H.G.; Siderowf, A.D. An economic model of Parkinson’s disease: Implications for slowing progression in the United States. Mov. Disord. Off. J. Mov. Disord. Soc. 2013, 28, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Wilczyński, J.; Ścipniak, M.; Ścipniak, K.; Margiel, K.; Wilczyński, I.; Zieliński, R.; Sobolewski, P. Assessment of Risk Factors for Falls among Patients with Parkinson’s Disease. BioMed Res. Int. 2021, 2021, 5531331. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, J.M.; Lees, A.J. Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain J. Neurol. 1991, 114 Pt 5, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- Rocca, W.A.; McDonnell, S.K.; Strain, K.J.; Bower, J.H.; Ahlskog, J.E.; Elbaz, A.; Schaid, D.J.; Maraganore, D.M. Familial aggregation of Parkinson’s disease: The Mayo Clinic family study. Ann. Neurol. 2004, 56, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Pezzoli, G.; Cereda, E. Exposure to pesticides or solvents and risk of Parkinson disease. Neurology 2013, 80, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Heinzel, S.; Berg, D.; Gasser, T.; Chen, H.; Yao, C.; Postuma, R.B.; The MDS Task Force on the Definition of Parkinson’s Disease. Update of the MDS research criteria for prodromal Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2019, 34, 1464–1470. [Google Scholar] [CrossRef]
- Noyce, A.J.; Bestwick, J.P.; Silveira-Moriyama, L.; Hawkes, C.H.; Giovannoni, G.; Lees, A.J.; Schrag, A. Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann. Neurol. 2012, 72, 893–901. [Google Scholar] [CrossRef]
- Hu, G.; Bidel, S.; Jousilahti, P.; Antikainen, R.; Tuomilehto, J. Coffee and tea consumption and the risk of Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2007, 22, 2242–2248. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Guo, X.; Park, Y.; Huang, X.; Sinha, R.; Freedman, N.D.; Hollenbeck, A.R.; Blair, A.; Chen, H. Caffeine Intake, Smoking, and Risk of Parkinson Disease in Men and Women. Am. J. Epidemiol. 2012, 175, 1200–1207. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Han, D.; Cheng, Q.; Zhang, P.; Zhao, C.; Min, J.; Wang, F. Association of Levels of Physical Activity With Risk of Parkinson Disease: A Systematic Review and Meta-analysis. JAMA Netw. Open 2018, 1, e182421. [Google Scholar] [CrossRef] [PubMed]
- Foo, J.N.; Chew, E.G.Y.; Chung, S.J.; Peng, R.; Blauwendraat, C.; Nalls, M.A.; Mok, K.Y.; Satake, W.; Toda, T.; Chao, Y.; et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk between Asians and Europeans: A Genome-Wide Association Study. JAMA Neurol. 2020, 77, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.S.; Goto, C.; et al. Application of Artificial Intelligence for Preoperative Diagnostic and Prognostic Prediction in Epithelial Ovarian Cancer Based on Blood Biomarkers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Angraal, S.; Mortazavi, B.J.; Gupta, A.; Khera, R.; Ahmad, T.; Desai, N.R.; Jacoby, D.L.; Masoudi, F.A.; Spertus, J.A.; Krumholz, H.M. Machine Learning Prediction of Mortality and Hospitalization in Heart Failure with Preserved Ejection Fraction. JACC Heart Fail. 2020, 8, 12–21. [Google Scholar] [CrossRef]
- Wang, H.-H.; Wang, Y.-H.; Liang, C.-W.; Li, Y.-C. Assessment of Deep Learning Using Nonimaging Information and Sequential Medical Records to Develop a Prediction Model for Nonmelanoma Skin Cancer. JAMA Dermatol. 2019, 155, 1277–1283. [Google Scholar] [CrossRef]
- Ali, L.; Zhu, C.; Zhang, Z.; Liu, Y. Automated Detection of Parkinson’s Disease Based on Multiple Types of Sustained Phonations Using Linear Discriminant Analysis and Genetically Optimized Neural Network. IEEE J. Transl. Eng. Health Med. 2019, 7, 2000410. [Google Scholar] [CrossRef]
- Pereira, C.R.; Pereira, D.R.; Rosa, G.H.; Albuquerque, V.H.; Weber, S.A.; Hook, C.; Papa, J.P. Handwritten dynamics assessment through convolutional neural networks: An application to Parkinson’s disease identification. Artif. Intell. Med. 2018, 87, 67–77. [Google Scholar] [CrossRef]
- Caramia, C.; Torricelli, D.; Schmid, M.; Munoz-Gonzalez, A.; Gonzalez-Vargas, J.; Grandas, F.; Pons, J.L. IMU-Based Classification of Parkinson’s Disease From Gait: A Sensitivity Analysis on Sensor Location and Feature Selection. IEEE J. Biomed. Health Inform. 2018, 22, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Mining imaging and clinical data with machine learning approaches for the diagnosis and early detection of Parkinson’s disease. NPJ Park. Dis. 2022, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Maass, F.; Michalke, B.; Willkommen, D.; Leha, A.; Schulte, C.; Tönges, L.; Mollenhauer, B.; Trenkwalder, C.; Rückamp, D.; Börger, M.; et al. Elemental fingerprint: Reassessment of a cerebrospinal fluid biomarker for Parkinson’s disease. Neurobiol. Dis. 2020, 134, 104677. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Tong, J.; Wang, F. Mining genetic and transcriptomic data using machine learning approaches in Parkinson’s disease. NPJ Park. Dis. 2020, 6, 24. [Google Scholar] [CrossRef]
- Kang, J.-H.; Irwin, D.J.; Chen-Plotkin, A.S.; Siderowf, A.; Caspell, C.; Coffey, C.S.; Waligórska, T.; Taylor, P.; Pan, S.; Frasier, M.; et al. Association of Cerebrospinal Fluid β-Amyloid 1-42, T-tau, P-tau181, and α-Synuclein Levels with Clinical Features of Drug-Naive Patients with Early Parkinson Disease. JAMA Neurol. 2013, 70, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Silveira-Moriyama, L.; Carvalho, M.d.J.; Katzenschlager, R.; Petrie, A.; Ranvaud, R.; Barbosa, E.R.; Lees, A.J. The use of smell identification tests in the diagnosis of Parkinson’s disease in Brazil. Mov. Disord. Off. J. Mov. Disord. Soc. 2008, 23, 2328–2334. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Prasad, S.; Saboo, Y.; Kaushick, R.; Saini, J.; Pal, P.K.; Ingalhalikar, M. Predictive markers for Parkinson’s disease using deep neural nets on neuromelanin sensitive MRI. NeuroImage Clin. 2019, 22, 101748. [Google Scholar] [CrossRef]
- Armañanzas, R.; Bielza, C.; Chaudhuri, K.R.; Martinez-Martin, P.; Larrañaga, P. Unveiling relevant non-motor Parkinson’s disease severity symptoms using a machine learning approach. Artif. Intell. Med. 2013, 58, 195–202. [Google Scholar] [CrossRef]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2015, 30, 1591–1601. [Google Scholar] [CrossRef]
- Belvisi, D.; Pellicciari, R.; Fabbrini, A.; Costanzo, M.; Pietracupa, S.; De Lucia, M.; Modugno, N.; Magrinelli, F.; Dallocchio, C.; Ercoli, T.; et al. Risk factors of Parkinson disease: Sim-ultaneous assessment, interactions, and etiologic subtypes. Neurology 2020, 95, e2500–e2508. [Google Scholar] [CrossRef]
- Belvisi, D.; Pellicciari, R.; Fabbrini, G.; Tinazzi, M.; Berardelli, A.; Defazio, G. Modifiable risk and protective factors in disease development, progression and clinical subtypes of Parkinson’s disease: What do prospective studies suggest? Neurobiol. Dis. 2020, 134, 104671. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Cai, Y.; Zheng, Z.; Tang, B.-S.; Xu, Y.; Wang, T.; Ma, J.; Chen, S.-D.; Langston, J.W.; Tanner, C.M.; et al. Penetrance of LRRK2 G2385R and R1628P is modified by common PD-associated genetic variants. Park. Relat. Disord. 2012, 18, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Liu, Y.; Mi, Y.; Zhao, J.; Liu, D.; Tian, Q. Alpha-synuclein (SNCA) polymorphisms and susceptibility to Parkinson’s disease: A meta-analysis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2015, 168, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.-L.; Mao, X.-Y.; Li, H.-H.; Zhang, J.-H.; Li, N.-N.; Burgunder, J.-M.; Peng, R.; Tan, E.-K. Association of GWAS loci with PD in China. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2011, 156, 334–339. [Google Scholar] [CrossRef] [PubMed]
- International Parkinson Disease Genomics Consortium. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet 2011, 377, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, L.; Lu, Z.-J.; Sun, X.-Y.; Li, J.-Y.; Peng, R. Association of three candidate genetic variants in RAB7L1/NUCKS1, MCCC1 and STK39 with sporadic Parkinson’s disease in Han Chinese. J. Neural Transm. 2016, 123, 425–430. [Google Scholar] [CrossRef]
- Zhao, Y.; Qin, L.; Pan, H.; Liu, Z.; Jiang, L.; He, Y.; Zeng, Q.; Zhou, X.; Zhou, X.; Zhou, Y.; et al. The role of genetics in Parkinson’s disease: A large cohort study in Chinese mainland population. Brain J. Neurol. 2020, 143, 2220–2234. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, R.; Roy, S.D.; Mandal, P.K.; Ghosh, S. High-Accuracy Detection of Early Parkinson’s Disease through Multimodal Features and Machine Learning. Int. J. Med. Inform. 2016, 90, 13–21. [Google Scholar] [CrossRef]
- Shu, Z.; Pang, P.; Wu, X.; Cui, S.; Xu, Y.; Zhang, M. An Integrative Nomogram for Identifying Early-Stage Parkinson’s Disease Using Non-motor Symptoms and White Matter-Based Radiomics Biomarkers from Whole-Brain MRI. Front. Aging Neurosci. 2020, 12, 548616. [Google Scholar] [CrossRef]
- Karabayir, I.; Butler, L.; Goldman, S.M.; Kamaleswaran, R.; Gunturkun, F.; Davis, R.L.; Ross, G.W.; Petrovitch, H.; Masaki, K.; Tanner, C.M.; et al. Predicting Parkinson’s Disease and Its Pathology via Simple Clinical Variables. J. Park. Dis. 2022, 12, 341–351. [Google Scholar] [CrossRef]
- Lin, S.; Gao, C.; Li, H.; Huang, P.; Ling, Y.; Chen, Z.; Ren, K.; Chen, S. Wearable sensor-based gait analysis to discriminate early Parkinson’s disease from essential tremor. J. Neurol. 2023, 270, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- Govindu, A.; Palwe, S. Early detection of Parkinson’s disease using machine learning. Procedia Comput. Sci. 2023, 218, 249–261. [Google Scholar] [CrossRef]
- He, S.; Huang, L.; Shao, C.; Nie, T.; Xia, L.; Cui, B.; Lu, F.; Zhu, L.; Chen, B.; Yang, Q. Several miRNAs derived from serum extracellular vesicles are potential biomarkers for early diagnosis and progression of Parkinson’s disease. Transl. Neurodegener. 2021, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Gopar-Cuevas, Y.; Duarte-Jurado, A.P.; Diaz-Perez, R.N.; Saucedo-Cardenas, O.; Loera-Arias, M.J.; Montes-De-Oca-Luna, R.; Rodriguez-Rocha, H.; Garcia-Garcia, A. Pursuing Multiple Biomarkers for Early Idiopathic Parkinson’s Disease Diagnosis. Mol. Neurobiol. 2021, 58, 5517–5532. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.; Lehéricy, S.; Chiu, S.Y.; Strafella, A.P.; Stoessl, A.J.; Vaillancourt, D.E. Emerging Neuroimaging Biomarkers across Disease Stage in Parkinson Disease. JAMA Neurol. 2021, 78, 1262–1272. [Google Scholar] [CrossRef]
- Li, X.; Fan, X.; Yang, H.; Liu, Y. Review of Metabolomics-Based Biomarker Research for Parkinson’s Disease. Mol. Neurobiol. 2022, 59, 1041–1057. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, H. Premotor Diagnosis of Parkinson’s Disease. Neurosci. Bull. 2017, 33, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.G.; Postuma, R. Premotor and nonmotor features of Parkinson’s disease. Curr. Opin. Neurol. 2014, 27, 434–441. [Google Scholar] [CrossRef]
- Fullard, M.E.; Morley, J.F.; Duda, J.E. Olfactory Dysfunction as an Early Biomarker in Parkinson’s Disease. Neurosci. Bull. 2017, 33, 515–525. [Google Scholar] [CrossRef]
- Haehner, A.; Boesveldt, S.; Berendse, H.; Mackay-Sim, A.; Fleischmann, J.; Silburn, P.; Johnston, A.; Mellick, G.; Herting, B.; Reichmann, H.; et al. Prevalence of smell loss in Parkinson’s disease—A multicenter study. Park. Relat. Disord. 2009, 15, 490–494. [Google Scholar] [CrossRef]
- Ponsen, M.M.; Stoffers, D.; Booij, J.; van Eck-Smit, B.L.F.; Wolters, E.C.; Berendse, H.W. Idiopathic hyposmia as a preclinical sign of Parkinson’s disease. Ann. Neurol. 2004, 56, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Ghebremedhin, E.; Rüb, U.; Bratzke, H.; Del Tredici, K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004, 318, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.D.; Petrovitch, H.; White, L.R.; Masaki, K.H.; Tanner, C.M.; Curb, J.D.; Grandinetti, A.; Blanchette, P.L.; Popper, J.S.; Ross, G.W. Frequency of bowel movements and the future risk of Parkinson’s disease. Neurology 2001, 57, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Cai, Y.; Hou, Y.; Ou, R.; Jiang, Z.; Shang, H. Excessive daytime sleepiness in Parkinson’s disease: A systematic review and meta-analysis. Park. Relat. Disord. 2021, 85, 133–140. [Google Scholar] [CrossRef]
- Abbott, R.D.; Ross, G.W.; White, L.R.; Tanner, C.M.; Masaki, K.H.; Nelson, J.S.; Curb, J.D.; Petrovitch, H. Excessive daytime sleepiness and subsequent development of Parkinson disease. Neurology 2005, 65, 1442–1446. [Google Scholar] [CrossRef] [PubMed]
- Williams-Gray, C.H.; Mason, S.L.; Evans, J.R.; Foltynie, T.; Brayne, C.; Robbins, T.W.; Barker, R.A. The CamPaIGN study of Parkinson’s disease: 10-year outlook in an incident population-based cohort. J. Neurol. Neurosurg. Psychiatry 2013, 84, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Schuurman, A.G.; Akker, M.v.D.; Ensinck, K.T.; Metsemakers, J.F.; Knottnerus, J.A.; Leentjens, A.F.; Buntinx, F. Increased risk of Parkinson’s disease after depression: A retrospective cohort study. Neurology 2002, 58, 1501–1504. [Google Scholar] [CrossRef] [PubMed]
- Leentjens, A.F.; Akker, M.V.D.; Metsemakers, J.F.; Lousberg, R.; Verhey, F.R. Higher incidence of depression preceding the onset of Parkinson’s disease: A register study. Mov. Disord. Off. J. Mov. Disord. Soc. 2003, 18, 414–418. [Google Scholar] [CrossRef]
- Howell, M.J.; Schenck, C.H. Rapid Eye Movement Sleep Behavior Disorder and Neurodegenerative Disease. JAMA Neurol. 2015, 72, 707–712. [Google Scholar] [CrossRef]
- Postuma, R.B.; Iranzo, A.; Hu, M.; Högl, B.; Boeve, B.F.; Manni, R.; Oertel, W.H.; Arnulf, I.; Ferini-Strambi, L.; Puligheddu, M.; et al. Risk and predictors of dementia and parkinsonism in idiopathic REM sleep behaviour disorder: A multicentre study. Brain J. Neurol. 2019, 142, 744–759. [Google Scholar] [CrossRef]

| Training Set (n = 1028) | p | Validation Set (n = 628) | p | |||
|---|---|---|---|---|---|---|
| PD | HC | PD | HC | |||
| (n = 524) | (n = 504) | (n = 305) | (n = 323) | |||
| Demographics | ||||||
| Age, year | 66 (61, 71.25) | 67 (63, 70) | 0.222 | 69 (61, 75) | 68 (65, 73) | 0.159 |
| Male | 257 (49) | 227 (45) | 0.221 | 173 (56.7) | 162 (50.2) | 0.117 |
| Education, year | 9 (5, 11.12) | 9 (9, 12) | <0.001 | 8 (5, 11) | 9 (6, 11) | 0.001 |
| Family History of PD | 44 (8.4) | 12 (2.4) | <0.001 | 27 (8.9) | 3 (0.9) | <0.001 |
| Lifestyle behaviors | ||||||
| Smoking | 124 (23.7) | 188 (37.3) | <0.001 | 82 (26.9) | 121 (37.5) | 0.006 |
| Alcohol | 175 (33.4) | 209 (41.5) | 0.009 | 62 (20.3) | 145 (44.9) | <0.001 |
| Tea | 203 (38.7) | 323 (64.1) | <0.001 | 57 (18.7) | 213 (65.9) | <0.001 |
| Coffee | 66 (12.6) | 93 (18.5) | 0.012 | 8 (2.6) | 32 (9.9) | <0.001 |
| Environmental exposure | ||||||
| Pesticides | 157 (30) | 110 (21.8) | 0.004 | 91 (29.8) | 80 (24.8) | 0.181 |
| Organic solvent | 13 (2.5) | 18 (3.6) | 0.401 | 5 (1.6) | 10 (3.1) | 0.351 |
| Heavy metal | 12 (2.3) | 18 (3.6) | 0.301 | 4 (1.3) | 6 (1.9) | 0.753 |
| Head injury | 21 (4) | 12 (2.4) | 0.193 | 2 (0.7) | 6 (1.9) | 0.288 |
| General anesthesia | 105 (20) | 117 (23.2) | 0.245 | 44 (14.4) | 51 (15.8) | 0.715 |
| Non-motor symptoms | ||||||
| pRBD | 172 (32.8) | 28 (5.6) | <0.001 | 75 (24.6) | 12 (3.7) | <0.001 |
| Olfactory dysfunction | 319 (60.9) | 63 (12.5) | <0.001 | 217 (71.1) | 38 (11.8) | <0.001 |
| Constipation | 347 (66.2) | 73 (14.5) | <0.001 | 169 (55.4) | 54 (16.7) | <0.001 |
| Global cognitive deficit | 273 (52.1) | 97 (19.2) | <0.001 | 185 (60.7) | 103 (31.9) | <0.001 |
| Depression | 242 (46.2) | 63 (12.5) | <0.001 | 44 (14.4) | 17 (5.3) | <0.001 |
| Daytime somnolence | 216 (41.2) | 37 (7.3) | <0.001 | 161 (52.8) | 47 (14.6) | <0.001 |
| Gene mutation | ||||||
| MMRN1 rs6532194 | 208 (39.7) | 143 (28.4) | <0.001 | 106 (34.8) | 105 (32.5) | 0.609 |
| RAB7L1 rs823144 | 192 (36.6) | 144 (28.6) | 0.007 | 127 (41.6) | 94 (29.1) | 0.001 |
| SNCA rs356182 | 405 (77.3) | 326 (64.7) | <0.001 | 248 (81.3) | 182 (56.3) | <0.001 |
| LRRK2 rs34778348 | 65 (12.4) | 40 (7.9) | 0.024 | 23 (7.5) | 23 (7.1) | 0.961 |
| SNCA rs356219 | 208 (39.7) | 134 (26.6) | <0.001 | 109 (35.7) | 98 (30.3) | 0.176 |
| MCCC1 rs12637471 | 352 (67.2) | 294 (58.3) | 0.004 | 218 (71.5) | 182 (56.3) | <0.001 |
| GBA rs421016 | 7 (1.3) | 0 (0) | 0.015 | 5 (1.6) | 0 (0) | 0.027 |
| LASSO-LR | DT | RF | XGboost | SVM | KNN | |
| Training set | ||||||
| AUC | 0.930 (95% CI: 0.915–0.945) | 0.888 (95% CI: 0.868–0.909) | 0.963 (95% CI: 0.952 −0.973) | 0.946 (95% CI: 0.946–0.959) | 0.941 (95% CI: 0.927–0.955) | 0.941 (95% CI: 0.928–0.954) |
| Accuracy | 0.861 (95% CI: 0.840–0.881) | 0.851 (95% CI: 0.829–0.873) | 0.911 (95% CI: 0.892–0.928) | 0.884 (95% CI: 0.865–0.904) | 0.874 (95% CI: 0.854–0.896) | 0.870 (95% CI: 0.849–0.890) |
| Sensitivity (Recall) | 0.826 (95% CI: 0.789–0.860) | 0.834 (95% CI: 0.803–0.866) | 0.910 (95% CI: 0.884–0.936) | 0.880 (95% CI: 0.850–0.909) | 0.838 (95% CI: 0.805–0.872) | 0.891 (95% CI: 0.863–0.919) |
| Specificity | 0.897 (95% CI: 0.870–0.924) | 0.869 (95% CI: 0.840–0.898) | 0.911 (95% CI: 0.884–0.936) | 0.889 (95% CI: 0.860–0.916) | 0.911 (95% CI: 0.885–0.936) | 0.847 (95% CI: 0.817–0.880) |
| Precision | 0.893 (95% CI: 0.866–0.919) | 0.869 (95% CI: 0.839–0.897) | 0.914 (95% CI: 0.890–0.937) | 0.892 (95% CI: 0.867–0.918) | 0.907 (95% CI: 0.882–0.933) | 0.859 (95% CI: 0.831–0.888) |
| F1 score | 0.858 (95% CI: 0.834–0.879) | 0.851 (95% CI: 0.828–0.875) | 0.912 (95% CI: 0.894–0.930) | 0.886 (95% CI: 0.866–0.905) | 0.871 (95% CI: 0.849–0.895) | 0.875 (95% CI: 0.854–0.896) |
| Validation set | ||||||
| AUC | 0.925 (95% CI: 0.905–0.945) | 0.831 (95% CI: 0.80–0.862) | 0.912 (95% CI: 0.890–0.934) | 0.908 (95% CI: 0.886–0.931) | 0.928 (95% CI: 0.908–0.947) | 0.896 (95% CI: 0.871–0.921) |
| Accuracy | 0.842 (95% CI: 0.812–0.869) | 0.787 (95% CI: 0.753–0.820) | 0.834 (95% CI: 0.804–0.863) | 0.819 (95% CI: 0.788–0.849) | 0.844 (95% CI: 0.814–0.871) | 0.811 (95% CI: 0.779–0.841) |
| Sensitivity | 0.810 (95% CI: 0.764–0.853) | 0.751 (95% CI: 0.701–0.80) | 0.866 (95% CI: 0.826–0.902) | 0.889 (95% CI: 0.851–0.922) | 0.826 (95% CI: 0.786–0.866) | 0.830 (95% CI: 0.779–0.841) |
| Specificity | 0.873 (95% CI: 0.834–0.909) | 0.820 (95% CI: 0.777–0.862) | 0.805 (95% CI: 0.764–0.848) | 0.752 (95% CI: 0.707–0.799) | 0.861 (95% CI: 0.820–0.898) | 0.793 (95% CI: 0.747–0.835) |
| Precision | 0.858 (95% CI: 0.816–0.90) | 0.798 (95% CI: 0.751–0.845) | 0.807 (95% CI: 0.765–0.851) | 0.772 (95% CI: 0.727–0.819) | 0.849 (95% CI: 0.807–0.891) | 0.791 (95% CI: 0.746–0.838) |
| F1 score | 0.833 (95% CI: 0.799–0.864) | 0.774 (95% CI: 0.731–0.811) | 0.835 (95% CI: 0.802–0.867) | 0.826 (95% CI: 0.794–0.858) | 0.837 (95% CI: 0.803–0.868) | 0.810 (95% CI: 0.774–0.842) |
| Reference | Modeling Approach | Features Selection | Objective | Source of Data | No. of Subjects | Evaluation |
|---|---|---|---|---|---|---|
| R. Prashanth et al. [38] | Naïve Bayes, SVM, Boosted Trees, RF | NMS, CSF, dopaminergic imaging markers | Classification of PD from HC | PPMI | 401 PD + 183 HC | Best performing: SVM with Accuracy = 96.40%, Sensitivity = 97.03%, Specificity = 95.01%, AUC = 98.88%. |
| Shu et al. [39] | Nomogram | MRI, clinical characteristics, NMS | Identification of early-stage PD | PPMI | 168 PD + 168HC + atypical PD 58 | AUC of training, testing and verification sets were 0.937, 0.922, and 0.836, respectively; the specificity were 83.8, 88.2, and 91.38%, respectively; and the sensitivity were 84.6, 82.4, and 70.69%. |
| Karabayir et al. [40] | Light Gradient Boosting | Questionnaires; simple non-invasive clinical tests | To predict a future diagnosis of PD | HAAS | 292 subjects | Individuals who developed PD within 3 years: AUC = 0.82, (95% CI 0.76–0.89), 5 years: AUC = 0.77 (95% CI 0.71–0.84). |
| Lin et al. [41] | SVM RF | Gathering gait and postural transition data using wearable sensors. | To diferentiate early-stage PD from ET | Ruijin Hospital, Shanghai Jiao Tong University School of Medicine | 84 early-stage PD and 80 ET subjects | Best performing: weighted average ensemble classifcation models with accuracy = 84%, sensitivity = 85.9%, Specifcity = 82.1%, AUC = 0.912. |
| Govindu et al. [42] | SVM, RF, KNN, Logistic Regression models | MDVP audio data | Early detection of PD | PPMI | 23PD + 8 HC | Best performing: RF with accuracy = 91.83%, sensitivity = 0.95. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, H.; Li, X.-Y.; Xu, F.; Zhu, J.; Li, X.; Song, Y.; Wang, X.; Wang, Z.; Wang, C. Applications of Machine Learning to Diagnosis of Parkinson’s Disease. Brain Sci. 2023, 13, 1546. https://doi.org/10.3390/brainsci13111546
Lai H, Li X-Y, Xu F, Zhu J, Li X, Song Y, Wang X, Wang Z, Wang C. Applications of Machine Learning to Diagnosis of Parkinson’s Disease. Brain Sciences. 2023; 13(11):1546. https://doi.org/10.3390/brainsci13111546
Chicago/Turabian StyleLai, Hong, Xu-Ying Li, Fanxi Xu, Junge Zhu, Xian Li, Yang Song, Xianlin Wang, Zhanjun Wang, and Chaodong Wang. 2023. "Applications of Machine Learning to Diagnosis of Parkinson’s Disease" Brain Sciences 13, no. 11: 1546. https://doi.org/10.3390/brainsci13111546
APA StyleLai, H., Li, X.-Y., Xu, F., Zhu, J., Li, X., Song, Y., Wang, X., Wang, Z., & Wang, C. (2023). Applications of Machine Learning to Diagnosis of Parkinson’s Disease. Brain Sciences, 13(11), 1546. https://doi.org/10.3390/brainsci13111546






