Abstract
Brain tumours and Gliomas, in particular, are among the primary causes of cancer mortality worldwide. Glioma diagnosis and therapy have not significantly improved despite decades of efforts. Autocrine TGF-β signalling promotes glioma proliferation, invasion, epithelial-to-mesenchymal transition (EMT), and drug resistance. Non-coding RNAs such as miRNA, lncRNA, and circRNAs have emerged as critical transcriptional and post-transcriptional regulators of TGF-β pathway components in glioma. Here, we summarize the complex regulatory network among regulatory ncRNAs and TGF-β pathway during Glioma pathogenesis and discuss their role as potential therapeutic targets for Gliomas.
1. Introduction
1.1. Gliomas
Gliomas are a group of brain tumours clinically divided into four types from grade I to grade IV. Grade IV gliomas, known as Glioblastoma multiforme (GBM), are the most common form of adult brain cancer [1,2]. The etiology of GBMs is complex and involves mutation or overexpression of multiple genes, and they have high intra-tumour heterogeneity [3,4]. Based on the molecular characteristics of GBM, the World Health Organization (WHO) classifies it into three types: GBM isocitrate dehydrogenase (IDH) wild type, GBM IDH mutant, and GBM not otherwise specified (NOS) [5,6]. Mutations in the IDH group of genes represent the most critical genetic alterations in GBM, which plays a vital role in therapeutic responses [7]. GBMs likely originate from astrocytes; however, tracking the cell of origin in GBM is challenging due to their heterogeneity [8]. Diffuse intrinsic pontine glioma (DIPG) is a form of pediatric malignancy that primarily grows in the pons with a dismal prognosis [9]. DIPG shares resemblances with adult high-grade astrocytomas. However, this has been debated lately due to its distinct molecular alterations [9,10]. Glioma stem cells (GSCs) are glioma-initiating cells that form a small subpopulation of GBM tumour cells and express stemness markers, such as CD133 [8]. GSCs can differentiate into multiple tumour cell types, contributing to intratumour heterogeneity in GBM [4]. GSCs contribute to tumour initiation, therapeutic resistance, and recurrence [8].
Current therapeutic strategies for GBM include maximum surgical resection and radio- and chemotherapy with temozolomide (TMZ) [11,12]. TMZ is an oral alkylating agent that alkylates DNA bases; it causes mismatch during DNA replication, leading to futile rounds of DNA repair, DNA double-strand breaks, and apoptosis [13]. However, O6-methylguanine-DNA methyltransferase (MGMT) can resolve some TMZ-induced alterations and thus mediate the survival of GBM cells [14]. MGMT inhibitors are considered beneficial for improving the action of TMZ in GBM patients [15]. Localized application of pseudo-substrates or tumour-specific delivery of blocking peptides against MGMT increases TMZ efficiency [14]. Overall, TMZ treatment extends the survival of GBM patients from 12.1 to 14.6 months [11], while tumour recurrence in GBM patients is inevitable. Resistance to radiation and chemotherapy in gliomas is also due to various other adaptive mechanisms, such as enhanced DNA repair capacity, cytoprotective autophagy, deregulated signalling pathways, intratumoral heterogeneity, phenotypic plasticity, and hypoxia [16].
1.2. Transforming Growth Factor-β (TGF-β)
TGF-β is a pleiotropic cytokine that regulates cell proliferation, differentiation, tissue homeostasis, motility, invasion, extracellular matrix production, angiogenesis, epithelial to mesenchyme transition (EMT), chemoresistance, and immune response in various cancers, including GBM [17,18]. TGF-β also contributes to pathologies associated with virus and bacterial infections as an inflammatory cytokine [19]. TGF-β superfamily consists of a large spectrum of ligands, including TGF-β1, TGF-β2, TGF-β3, activin, nodal, bone morphogenetic protein (BMP), growth and differentiation factors (GDF), and anti-mullerian hormone (AMH) [20]. Seven types I and five types II transmembrane serine/threonine-protein kinase receptors exist for the TGF-β superfamily in the mammalian genome [21]. TGF-β ligand-mediated signalling is initiated by the binding of TGF-β to the type II TGF-β receptor (TGF-βR II), which gets phosphorylated, alters its conformation, and phosphorylates the type I TGF-β receptor (TGF-βR1). The signal through TGFβR1 is transduced downstream either through SMAD proteins—in the canonical TGF-β pathway or through other effectors like MAPK, ERK, and JUN kinase—in the non-canonical TGF-β pathway [20]. TGF-β target genes consist of evolutionarily conserved putative SMAD binding elements (SBEs) in their promoter regions. Nuclear translocation of the activated SMAD2/3 complex and binding of the translocated SMAD2/3 complex to the SBEs leads to the activation or repression of hundreds of TGF-β target genes [22]. Non-canonical or non-SMAD pathways include various branches of MAP kinase pathways, Rho-like GTPase signalling pathways, and phosphatidylinositol-3-kinase/AKT pathways. For example, in the ERK-mediated signal transduction, the activated TGFβRI phosphorylates and activates the ShcA protein, forming the SHCA/GRB22/SOS complex, followed by the sequential activation of c-RAF, MEK, ERK [23]. In normal cells and the early stages of cancer, TGF-β restrains cell proliferation whereas, in advanced stages of cancer, due to accumulation of mutations in the TGF-β pathway components or selective impairment of its tumour-suppressive function, it turns out to be oncogenic [24,25,26]. TGF-β levels are elevated in glioma and are associated with increased histologic grade [27,28]. TGF-β expression is also higher in the serum of GBM patients and correlates with poor survival [29]. TGF-β promotes TMZ resistance by activating genes, such as connective tissue growth factor (CTGF), ZEB1, and SNAIL1 [30,31,32]. TGF-β also promotes TMZ resistance in MGMT promoter hypomethylated GBM [33]. Several anti-TGF-β antibodies, inhibitors, and antisense oligonucleotides (ASOs) against TGF-β pathway components have been evaluated in the pre-clinical and clinical trials for GBM, with limited success [18,34,35,36]. Systemic inhibition of TGF-β may not be ideal as it has both pro and anti-tumour activities and might also hamper the normal physiological functions of the TGF-β pathway.
1.3. Non-Coding RNAs (ncRNAs)
The human genome may be categorized into protein-coding genes (PCGs) and non-protein-coding genes (NCGs). ncRNAs constitute >90% of the human genome. The non-coding category of the genome is highly heterogeneous, consisting of small non-coding RNAs < 200 bps in length and long ncRNAs (lncRNAs) > 200 bps in length.
1.3.1. miRNAs
miRNAs are small non-coding RNAs of 21–23 nucleotides in length that generally perform post-transcriptional gene silencing of their mRNA targets [37]. They regulate gene expression by primarily interacting with the miRNA response elements (MREs) present on the 3′ UTR of a transcript. However, interaction with 5′ UTR, coding sequences, and gene promoters are also observed [38]. They bind to the target gene and either degrade the transcript or, more often, limit the target gene’s translation [39]. The mRNA decay by miRNA occurs through the miRNA-induced silencing complex (miRISC) [40]. Argonaute and GW182 are the core components of the RISC complex. GW182 interacts with PABP and recruits the PAN2-PAN3 and CCR4-NOT deadenylase complex, causing deadenylation, decapping, and mRNA decay [41]. miRNAs also prevent protein synthesis by cap-dependent translational inhibition [42]. It interferes with the assembly or activity of the translation initiation complex, eIF4F, via eIF4E-T and DDX6 [42]. While these are the canonical miRISC-mediated silencing mechanisms, there are few non-canonical miRNA-mediated gene silencing. For example, complete sequence complementarity between miRNA and target mRNA results in direct AGO2-mediated target cleavage [43]. Also, the recruitment of Argonaute in the absence of GW182 inhibits translation without affecting mRNA stability [44]. The differential association of Argonaute protein with other proteins mainly decides the outcome of mRNA decay or translation inhibition [40]. Several miRNAs regulate multiple aspects of GBM pathogenesis and are potential diagnostic biomarkers and therapeutic targets for GBM [45].
1.3.2. LncRNAs
LncRNAs are transcripts longer than 200 bps with no ability to code for proteins [46,47,48,49,50,51,52,53]. More than 100,000 lncRNAs in the human genome are listed in the NONCODE and other lncRNA databases [50,54]. While most lncRNAs are generated from RNA polymerase II, which shares similarities with mRNAs, such as polyadenylation and 7-methylguanosine cap, some are generated from RNA pol I and RNA polymerase III [50]. LncRNAs have a wide range of functions to modulate gene expression, chromatin architecture, transcription, RNA processing, splicing, editing, localization, stability, and protein translation [50]. They modulate gene expression in cis and trans by interacting with DNA, RNAs, and proteins [55]. To regulate gene expression, lncRNAs modulate (i) recruitment of a regulatory/transcription factor/epigenetic protein to a gene locus; (ii) inhibit the binding of a transcription factor to gene promoter by acting as a decoy; (iii) by acting as scaffold for protein complexes to either positively or negatively regulate gene expression; and (iv) large number of lncRNAs localized in the cytoplasm function as competing endogenous RNAs (ceRNAs) for miRNAs and stabilize the mRNA target of those miRNAs. LncRNAs regulate multiple aspects of GBM pathogenesis, such as proliferation, invasion, metastasis, and drug resistance [56]. They have the potential to serve as potential diagnostic markers and therapeutic targets for GBM [57,58].
1.3.3. CircRNAs
Circular RNAs are covalently closed, single-stranded RNAs produced by a non-canonical back splicing of cellular non-coding RNAs and precursor messenger RNAs [54,55,56]. CircRNAs are generated in the nucleus, but most are primarily present in the cytoplasm. Their synthesis is regulated by specific cis-acting elements and trans-acting factors [59,60,61,62,63]. Many circRNAs act as non-coding RNAs and regulate gene expression by serving as decoys or competitors for microRNAs and proteins. In addition to sponging miRNAs and proteins, circRNAs also regulate the splicing of linear RNAs, regulate transcription, and form chromatin looping [60,64] A small percentage of circRNAs undergo cap-independent translation to encode functional peptides in response to specific cellular stresses [61]. CircRNAs regulate proliferation, angiogenesis, cancer cell migration and invasion, and apoptosis in cancer [62]. CircRNAs may act as diagnostic biomarkers and therapeutic targets in several cancer types, including GBM [62,63,65,66].
The miRNAs, lncRNAs, and circRNAs are aberrantly expressed in GBM tumour tissues and regulate GBM pathogenesis (Table 1, Table 2 and Table 3) [57,65,67]. They play critical regulatory functions in cancer by acting as oncogenes or tumour suppressors [68,69,70,71]. They have the potential for use as clinical biomarkers and therapeutic targets for cancers, including GBM [72,73,74,75,76,77].

Table 1.
TGF-β regulated oncogenic and tumour suppressor miRNAs in GBM.

Table 2.
TGF-β regulated oncogenic and tumour suppressor lncRNAs in GBM.

Table 3.
TGF-β regulated oncogenic and tumour suppressor circular RNAs in GBM.
Several miRNAs, lncRNAs, and circRNAs have recently been identified, which modulate the TGF-β pathway to promote or repress GBM (Table 1, Table 2 and Table 3). Since TGF-β plays a role in tumour suppression at early stages of cancer development, its complete inactivation for cancer treatment is not ideal [24,25,26]. NcRNAs regulated by the TGF-β pathway modulate numerous aspects of GBM pathogenesis. They may serve as attractive therapeutic targets downstream of TGF-β for countering the tumour-promoting effects of TGF-β. Here, we summarize the role of lncRNAs, miRNAs, and circRNAs in the TGF-β pathway in GBM pathogenesis (Figure 1, Figure 2 and Figure 3).
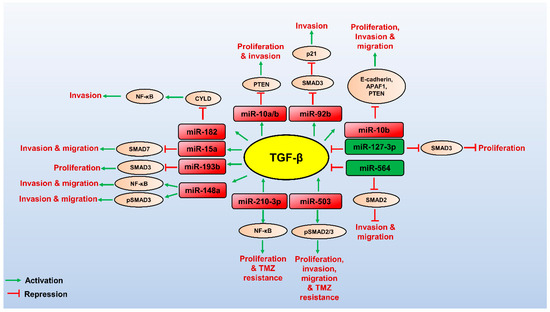
Figure 1.
Role miRNAs and their targets in regulating the TGF-β pathway in GBM. TGF-β (yellow circled) promotes the expression of oncogenic miRNAs (red boxed), which can control post-transcriptional gene expression of its targets (brown circled) to promote TGF-β-mediated GBM pathogenesis. Few tumour suppressor miRNAs (green boxed) target the TGF-β pathway’s components and downregulate the TGF-β signalling, thereby attenuating GBM progression. The red arrows indicate inhibitory function, and the green arrows indicate a stimulatory role.
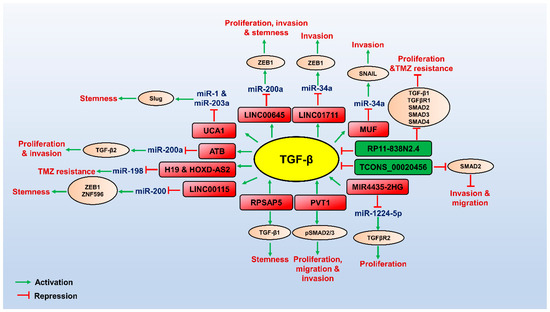
Figure 2.
Role of LncRNAs involved in TGF-β pathway in regulating GBM pathogenesis. TGF-β (yellow circled) promotes the expression of oncogenic lncRNAs (red boxed), which can control transcriptional/post-transcriptional gene expression of its targets (brown circled) by interacting with miRNAs or proteins to promote TGF-β-mediated GBM pathogenesis. Few oncogenic lncRNAs are not induced by TGF-β but promote the TGF-β signalling-mediated GBM pathogenesis. Tumour suppressor lncRNAs (green boxed) target the components of the TGF-β pathway and downregulate the TGF-β signalling, thereby attenuating GBM progression. The red arrows indicate inhibitory function, and the green arrows indicate a stimulatory role.
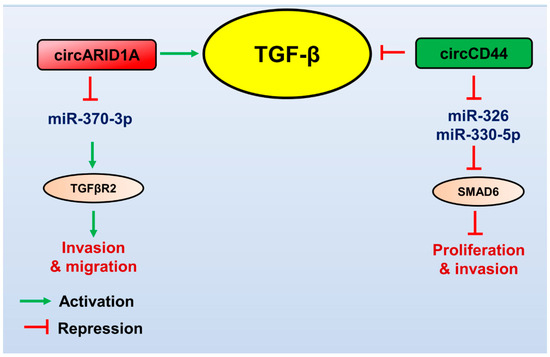
Figure 3.
Role of circular RNAs in regulating the TGF-β pathway in GBM. TGF-β (yellow circled) promotes the expression of oncogenic circRNA (red boxed), which controls the gene expression of its targets (brown circled) to promote TGF-β-mediated GBM pathogenesis. Tumour suppressor circRNA (green boxed) targets the TGF-β pathway’s components, downregulates the TGF-β signalling, and attenuates GBM progression. The red arrows indicate inhibitory function, and the green arrows indicate a stimulatory role.
2. miRNAs Involved in the TGF-β Pathway in GBM
2.1. Oncogenic miRNAs Involved in the TGF-β Pathway in Gliomas
2.1.1. miR-182
miR-182 is over-expressed in GBM tissues and cells, whereas CYLD, a negative regulator of the NFκB pathway, is downregulated [78]. The genomic location of miR-182, 7q32.1, is frequently amplified in gliomas [78]. Interestingly, miR-182 is induced by TGF-β in U373MG and LN229 cells through the SMAD signalling pathway [78]. miR-182 expression was also upregulated in Smad2/Smad4- overexpressing cells [78]. ChIP assays confirmed the binding of SMAD2/3 to the promoter of miR-182 [78]. These results suggest that TGF-β induced miR-182 expression in glioma cells through the SMAD signalling pathway. CYLD is a target of miR-182 [78]. Up-regulation of miR-182 in U373MG and LN229 cells decreased the expression of CYLD. Also, miR-182 interacts with and degrades CYLD. Further, overexpression of miR-182 increased, while inhibition of miR-182 reduced the luciferase activity of NF-κB reporter and expression of NF-κB target genes. miR-182 overexpression significantly induced the phosphorylation of IKKβ, while miR-182 inhibitor reversed this effect. Importantly, in vitro kinase assay showed that endogenous IKK kinase activity was prolonged in miR-182 –transduced cells. These results indicate that miR-182 suppresses CYLD and enhances and sustains NF-κB activity in GBM [78]. miR-182 up-regulation promotes anchorage-independent growth, colony formation, and invasion of GBM in vitro and in vivo [78]. At the same time, miR-182 inhibition reversed these effects [78]. Also, the tumour-promoting function of miR-182 overexpression was reversed with combined transfection of miR-182 mimics and IκBα dominant-negative mutant construct, indicating that miR-182 promotes GBM tumour through the NF-κB pathway [78]. Further, miR-182 suppression inhibits NF-κB activity and malignant properties of patient-derived glioma cells (PDGCs) [78]. The study also identified that TGF-β treatment in U373MG and LN229 cells significantly increased the NF-κB reporter activity, which was abolished upon miR-182 inhibition. This indicates that TGF-β-induced miR-182 is essential for sustained NF-κB activity in GBM [78]. Overexpression of pSMAD2/3, miR-182, and several NF-κB target genes was observed in clinical GBM samples, which conferred poor survival of the patients [78]. Also, the clinical samples showed a positive correlation between TGF-β, miR-182, and NF-κB target genes [78]. Upon induction by TGF-β, miR-182 promotes GBM pathogenesis by activating and sustaining NF-κB activity by downregulating CYLD [78].
2.1.2. miR-15a
Guo et al., using a microarray screen, observed that miR-15a-5p is upregulated in glioma tissues [79]. Bioinformatics analysis and luciferase reporter assay confirmed the direct interaction of miR-15a with SMAD7. Overexpression of miR-15a promoted migration and invasion of SHG139 cells [79]. Whereas inhibition of miR-15a displayed the opposite effect, indicating the tumour-promoting ability of miR-15a [79]. Anti-miRNA oligonucleotide (AMO)-mediated inhibition of miR-15a in SHG139 cells reduced migration and downregulated mesenchymal markers—Vimentin and N-cadherin. However, the combined knockdown of miR-15a and SMAD7 reversed these effects [79]. Further, miR-15a inhibition attenuated GBM tumour growth in vivo. Hence, this study indicates the oncogenic function of miR-15a in GBM by inhibiting SMAD7 activity [79].
2.1.3. miR-193b
miR-193b levels are upregulated in glioma cell lines and GBM tumour samples [80]. miR-193b depletion reduces the proliferation of U87 and U251 cells [80]. Cell cycle analysis upon miR-193b knockdown revealed G0/G1 arrest in U87 and U251 cells [80]. Bioinformatics analysis and luciferase reporter assays demonstrated that SMAD3 is the primary target of miR-193b [80]. Inhibition of miR-193b significantly increased the protein levels of SMAD3 in U87 and U251 cells [80]. Further, to know whether miR-193b regulates GBM proliferation through the TGF-β pathway, one of the primary targets of the TGF-β pathway, p21 levels were evaluated upon miR-193b inhibition. Inhibition of miR-193b and the subsequent up-regulation of SMAD3 in GBM cells displayed a significant accumulation of p21 [80]. Also, the down-regulation of miR-193b decreased the proliferation of U87 and U251 cells [80]. Hence, the study demonstrates that miR-193b is an oncogene that promotes cell growth by directly targeting SMAD3 and restricting the tumour-suppressive effects of SMAD3 through p21 down-regulation in GBM [80].
2.1.4. miR-210-3p
miR-210-3p is induced upon exposure to hypoxic conditions in GBM [81]. Treatment of the hypoxia-exposed cells with echinomycin, a transcriptional inhibitor of HIF-1α, significantly reduced hypoxia-induced miR-210 levels, indicating the transcriptional activation of miR-210-3p by HIF-1α in GBM [81]. HIF-1α promotes TGF-β expression, which was abrogated upon miR-210-3p inhibition. This indicates that miR-210-3p is an essential mediator in promoting HIF-1α-mediated TGF-β expression [81]. Exposure of U87-MG cells to hypoxic conditions and TGF-β individually promoted migration and invasion. However, this effect was abrogated upon miR-210-3p inhibition, indicating an oncogenic role of miR-210-3p in promoting hypoxia/TGF-β -mediated GBM pathogenesis [81]. Overexpression of miR-210-3p promotes resistance to TMZ in U87-MG cells. However, the chemoresistance induced by miR-210-3p overexpression was abrogated upon TGF-β knockdown [81]. Hypoxia-induced miR-210-3p also promotes the transcription activity of NF-κB in U87-MG cells [81]. This study establishes that HIF-1α-induced miR-210-3p promotes the TGF-β expression and TGF-β-mediated migration, invasion, and TMZ resistance in GBM [81]. However, further studies on the role of miR-210-3p on NF-κB-mediated GBM pathogenesis and the involvement of TGF-β in this context are yet to be studied.
2.1.5. miR-148a
Quaking (QKI), an essential negative regulator of the TGF-β signalling, is downregulated, and miR-148a is upregulated in GBM tissues and cell lines [82]. Low levels of QKI and overexpression of miR-148a confer poor prognosis in GBM patients [82]. Microribonucleoprotein (miRNP) IP and luciferase reporter assay confirmed the interaction between miR-148a and QKI [82]. Overexpression of miR-148a reduced the protein levels of QKI, while inhibition of miR-148a reduced this effect [82]. Moreover, SKP1, a member of the E3 ubiquitin ligase complex that degrades SMAD3, is also an essential target of miR-148a [82]. Consequently, miR-148a overexpression enhanced the SMAD luciferase reporter activity and phosphorylation of SMAD2/3 [82]. GSEA and TCGA database analysis revealed that miR-148a positively correlates with the gene signatures of TGF-β signalling [82]. Overexpression of miR-148a is associated with poor prognosis in GBM patients, indicating that miR-148a is an oncogenic miRNA [82]. High levels of miR-148a are positively correlated with p-SMAD3 expression and the DNA binding activity of NF-κB [82]. Also, the promoter of miR-148a contains multiple NF-κB binding domains. Consequently, NF-κB activation increased the expression of miR-148a in GBM cells [82]. Further, in vitro experiments in LN18 and U-138MG cells and in vivo experiments demonstrated that miR-148a promotes the invasion, migration, and aggressiveness of GBM [82]. These results illustrate that the hyperactive NF-κB signalling in GBM promotes the expression of miR-148a oncogene, which further promotes the aggressive phenotypes of GBM by downregulating the expression of QKI and SKP1 and activating the TGF-β signalling [82]. Thus, miR-148a establishes an essential link between NF-κB and TGF-β signalling in promoting GBM pathogenesis [82].
2.1.6. miR-10a/b
Liu et al. observed a significant positive correlation between the expression of miR-10a/b and TGF-β in glioma tissues [83]. miR-10a/10b is induced upon TGF-β treatment of U251 and SHG-44 cells [83]. miR-10a/10b overexpression significantly increased the migration of U251 cells [83]. Bioinformatics analysis and luciferase reporter assay revealed that PTEN, an important tumour suppressor gene, is a primary target of miR-10a/10b [83]. Also, overexpression of miR-10a/10b reduced the expression of PTEN, while depletion of miR-10a/10b displayed the reverse effect. These results indicate that miR-10a/10b, upon induction by TGF-β, promotes GBM pathogenesis by inhibiting PTEN [83].
2.1.7. miR-10b
miR-10b expression is upregulated upon TGF-β treatment in U87 and U251 cells through the SMAD signalling pathway [84]. Also, the induction of miR-10b upon TGF-β treatment in U251 cells was abrogated considerably upon treatment with TGF-β inhibitor SB431452 [84]. miR-10b mimics increased the proliferation in GBM [84]. The combination of anti-miR-10b and TGF-β treatment reversed the suppressive effects of miR-10b depletion [84]. This indicated that miR-10b promotes TGF-β1-induced glioma cell proliferation [84]. Similar to proliferation, the migration and invasion ability of GBM cells was significantly enhanced upon miR-10b mimics in combination with TGF-β treatment [84]. Bioinformatics analysis and luciferase reporter assay indicated that E-cadherin, APAF1, and PTEN are the primary targets of miR-10b [84]. TGF-β treatment and miR-10b mimics individually reduced E-cadherin, APAF1, and PTEN protein levels. At the same time, the suppression of miR-10b using anti-miR-10b enhanced their expression [84]. In vivo, the xenograft tumour model depicted that treatment with TGF-β1 or miR-10b agomir significantly promoted GBM tumour growth, whereas the miR-10b antagomir remarkably inhibited tumour growth, even in the presence of TGF-β1 [84]. The study demonstrates that TGF-β induces the expression of miR-10b, and it promotes TGF-β-mediated proliferation, migration, and invasion in GBM by suppressing E-cadherin, APAF1, and PTEN [84].
2.1.8. miR-92b
miR-92b levels are elevated, and SMAD3 expression is downregulated in GBM [85]. SMAD3 protein levels were downregulated upon miR-92b mimics and were upregulated by miR-92b inhibitors [85]. Subsequently, bioinformatics analysis and luciferase reporter assay demonstrated that SMAD3 is a direct target of miR-92b [85]. Also, the knockdown of miR-92b correlated with the reduced growth of U251 and SHG66 cells and restored G1 accumulation [85]. Knockdown of SMAD3 further decreased the TGF-β-mediated p21 induction [85]. Also, the high expression of miR-92b directly downregulates SMAD3 expression and inhibits the TGF-β/SMAD3/p21-mediated reduction in tumour cell growth [85]. Inhibition of miR-92b in vivo reduces tumour growth [85]. Hence, in this study, miR-92b promotes GBM growth by attenuating the inhibitory effects of TGF-β by targeting SMAD3 and thereby downregulating p21 [85].
2.1.9. miR-503
Analysis of the gene expression omnibus data of glioblastoma samples revealed that miR-503 is overexpressed in GBM tissue compared to normal tissue [86]. Also, TGF-β treatment significantly increased the expression of miR-503 in T98G cells through SMAD signalling [86]. miR-503 overexpression decreased the proliferation, migration, and colony formation ability and restricted apoptosis in GBM [86]. Dual luciferase reporter assay indicated that PDCD4 is the primary target of miR-503 [86]. Overexpression of miR-503 dramatically reduced the mRNA and protein levels of PDCD4 in GBM cells [86]. Further, the combination of miR-503 inhibition with varying doses of TMZ showed a synergistic decrease in proliferation and increased apoptosis of A172 and U251 cells, indicated by enhanced cleaved PARP [86]. These results suggest that miR-503 is an oncogenic miRNA induced by TGF-β in GBM [86]. Upon induction by TGF-β, miR-503 enhances the proliferation, invasion, migration, and drug resistance in GBM by targeting PDCD4 [86].
2.2. Tumour Suppressor miRNAs Involved in the TGF-β Pathway in Gliomas
2.2.1. miR-127-3p
RNAseq analysis of GBM and normal brain tissues indicated that miR-127 is down-regulated in GBM tissues compared to normal brain tissues [87]. miR-127-3p gene is located in chromosome 14 between two lincRNAs (ENSG00000214548 and ENSG00000258399). DNA methylation and histone acetylase inhibition resulted in the down-regulation of miR-127-3p in GBM tissues [87]. Additionally, overexpression of miR-127-3p in LN229 and T98G cells reduced the proliferation and caused cell cycle arrest compared to control cells. However, the overexpression of miR-127-3p did not significantly affect the apoptosis of glioma cells. In vivo nude mice model with miR-127-3p overexpressing LN229 cells displayed reduced colony formation and tumour volume compared to the control group. Hence, miR-127-3p functions as a tumour suppressor in GBM [87]. Bioinformatics analysis and luciferase reporter assays revealed that SKI, RGMA, ZWINT, SERPINB9, and SFRP1 are the primary targets of miR-127-3p. SKI is an essential negative regulator of the TGF-β pathway [87]. It binds to SMAD proteins, blocking the ability of the SMAD complexes to activate TGF-β signalling in GBM. Overexpression of miR-127-3p or knockdown of SKI promotes TGFBR1 expression, phosphorylation of SMAD3, and induces cell cycle arrest of LN229 cells [87]. Hence, miR-127-3p is an essential tumour suppressor miRNA that attenuates GBM proliferation by inhibiting the TGF-β signalling [87].
2.2.2. miR-564
miR-564 is downregulated in glioma cells and tumour tissues compared to normal human astrocytes and normal brain tissues, respectively [88]. miR-564 mimics decreased the proliferation and invasion of U87 cells [88]. Bioinformatics analysis and luciferase reporter assay indicated that TGF-β1 is the primary target of miR-564 [88]. miR-564 overexpression markedly decreased the mRNA and protein levels of TGF-β1 [88]. In addition, miR-564 also reduced the expression of SMAD4 protein and phosphorylated SMAD2 protein levels [88]. Moreover, protein levels of EGFR and MMP9 were also significantly reduced upon miR-564 overexpression in U87 cells [88]. EGFR and MMP9 are upregulated in GBM tissues compared to normal brain tissues [88]. Also, a significant negative correlation was observed between miR-564, EGFR, and MMP9 [88]. Cell proliferation and invasion assays indicated that the increase in proliferation and invasion by TGF-β treatment was attenuated by miR-564 overexpression [88]. Further, the U87-engrafted in vivo GBM tumour model indicated a reduction in tumour growth upon miR-564 overexpression [88]. Also, the mRNA and protein expression of TGF-β1 was lower in miR-564- treated tumours than in scramble-treated tumours [88]. Hence, miR-564 is a tumour suppressor miRNA, which restricts proliferation and invasion in GBM by targeting TGF-β1, and its downstream targets EGFR and MMP9 [88].
3. LncRNAs Involved in the TGF-β Pathway in GBM
3.1. Oncogenic lncRNAs Involved in the TGF-β Pathway in Gliomas
3.1.1. LncRNA-ATB
LncRNA-ATB levels are higher in glioma tissues and U251, A172 cell lines than in normal brain tissues [89]. Increased expression of lncRNA-ATB correlated with poor survival of GBM patients [89]. Further, loss of function studies depicted a reduced proliferation, migration, and invasion of U251 and A172 cells [89]. The study also indicated a negative correlation between the expression of lncRNA-ATB and miR-200a in GBM tissues. miR-200a is downregulated in GBM tissues and cell lines, and the knockdown of lncRNA-ATB significantly increased miR-200a expression in U251 and A172 cells [89]. Luciferase reporter assay and Ago2 pulldown assays validated that lncRNA-ATB and TGF-β2 are direct targets of miR-200a. Also, miR-200a inhibition results in up-regulation of TGF-β2 [89]. LncRNA-ATB knockdown mediated a reduction in cell proliferation, colony formation, and invasion of U251 and A172 cells, which is attenuated upon miR-200a inhibition. Additionally, the knockdown of lncRNA-ATB significantly reduced the levels of TGF-β2 expression, which was rescued by miR-200a inhibition. Glioma samples show a positive correlation between lncRNA-ATB and TGF-β2 and a negative correlation between miR-200a and TGF-β2. The reduction in mRNA and protein levels of TGF-β2 upon lncRNA-ATB depletion was further downregulated upon miR-200a overexpression. In contrast, TGF-β2 expression was rescued with the combination of lncRNA-ATB knockdown and miR-200a inhibition [89]. Studies in nude mice models upon lncRNA-ATB depletion demonstrated a reduction in tumour volume, tumour weight, and reduced proliferation index indicated by Ki67 staining, supporting the oncogenic role of lncRNA-ATB in GBM. These results suggest that lncRNA-ATB could competitively bind miR-200a to stabilize TGF-β2 and promote TGF-β2-mediated GBM pathogenesis [89].
Another study by Tang et al. reported that lncRNA-ATB is upregulated by TGF-β1 treatment in LN18 and U251 cells [90]. The up-regulation of lncRNA-ATB by TGF-β treatment was abrogated upon treatment with TGF-β inhibitor, SB505124, indicating the SMAD2/3 mediated regulation of lncRNA-ATB expression [90]. LncRNA-ATB overexpression combined with TGF-β1 treatment increases the invasion of U251 and LN18 cells. Also, lncRNA-ATB overexpression and TGF-β1 treatment promote the phosphorylation of p65, the nuclear translocation of p65, and the phosphorylation of p38. These results indicate the activation of the NF-κB and p38/MAPK pathways by TGF-β regulated lncRNA-ATB [90]. The increase in invasion in LN18 and U251 cells upon lncRNA-ATB overexpression and TGF-β treatment was significantly abolished upon treatment with NF-κB and p38/MAPK pathway inhibitors. These results suggest that the SMAD2/3 transcription factor induces lncRNA-ATB expression in GBM, and it promotes TGF-β-mediated GBM invasion through the NF-κB and p38/MAPK pathways.
3.1.2. LncRNA-UCA1
TGF-β significantly upregulates lncRNA-UCA1 expression in U87 and U251 cells [91]. Further, the knockdown of lncRNA-UCA1 attenuated the invasion and stemness of glioma cells induced by TGF-β [91]. Particularly, lncRNA-UCA1 knockdown downregulates SLUG, ALDH1, and NANOG, which are involved in TGF-β up-regulation. Luciferase reporter assay and Ago2 RIP suggest direct binding of lncRNA-UCA1, miR-1, and miR-203a. Moreover, miR-1 and miR-203a directly mediate SLUG down-regulation. The study also reported a positive correlation between lncRNA-UCA1 and SLUG expression in GBM tissues. LncRNA-UCA1 promotes SLUG expression by binding to and titrating miR-1 and miR-203a. Reduction in spheroid formation, SLUG expression, and ALDH1 activity upon lncRNA-UCA1 knockdown is partially rescued upon Slug overexpression. Hence, these results suggest that TGF-β induced lncRNA-UCA1 acts as a molecular sponge for miR-1 and miR-203a to promote SLUG expression and SLUG-mediated GBM cell stemness [91].
3.1.3. LINC00645
TCGA and GSEA data analysis by Li et al. indicated that LINC00645 is upregulated in GBM patients compared to normal brain tissues [92]. Expression of LINC00645 was high with increasing grades of glioma. Moreover, TGF-β induces LINC00645 expression in GBM [92]. Knockdown of LINC00645 attenuated the malignant behaviour of GBM by decreasing proliferation, invasion, migration, and EMT in T98G and U251 cells. Notably, LINC00645 knockdown reduces ZEB1 levels, an essential target of miR-205-3p [92]. miR-205-3p is downregulated in GBM tumour tissues and GBM cell lines compared to normal brain tissues [92]. Low expression of miR-205-3p indicates poor survival in GBM patients [92]. TGF-β treatment significantly downregulated miR-205-3p, and a negative correlation is observed between LINC00645 and miR-205-3p in GBM patients’ samples [92]. miR-205-3p overexpression significantly reduced the levels of LINC00645, while depletion of LINC00645 promoted miR-205-3p expression. Luciferase reporter assay and Ago2-RIP indicate a direct interaction between LINC00645 and miR-205-3p [92]. Epithelial marker, E-cadherin expression is downregulated, and expression of mesenchymal markers, vimentin, N-cadherin, SNAIL, and ZEB1 is upregulated upon TGF-β treatment in U251 and T98G cells. Whereas knockdown of LINC00645 in combination with TGF-β treatment increased E-cadherin levels and reduced the expression of mesenchymal markers. Further, TGF-β treatment in combination with miR-205-3p overexpression reversed these effects [92]. miR-205-3p is a tumour suppressor that directly targets and degrades ZEB1 [92]. TGF-β induced LINC00645 mediated GBM invasion, and migration is reversed upon miR-205-3p overexpression [92]. These results indicate that LINC00645 sponges miR-205-3p to stabilize ZEB1 and promote GBM pathogenesis [92]. In addition to its effect on GBM invasion, migration, and EMT, LINC00645 also induces stemness in GBM. Western blotting analysis upon LINC00645 depletion reduced the expression of stemness factors, BMI-1, OCT-4, SOX-2, and NANOG [92]. Sphere-forming assay indicated a decrease in sphere-forming ability on LINC00645 knockdown. In contrast, LINC00645 overexpression had the opposite effect [92]. Also, LINC00645 depletion partly decreased NESTIN expression and increased the GFAP expression in U251 cells [92]. Depletion of LINC00645 reduced tumour growth of the tumour xenograft model in vivo [92]. LINC00645/miR-205-3p/ZEB1 axis regulates invasion, migration, and EMT in GBM, and LINC00645 also promotes stemness in GSCs [92].
3.1.4. LINC00115
RNA-sequencing of TGF-β treated GSCs revealed up-regulation of LINC00115 [93]. Moreover, LINC00115 expression is higher in GBM tumour samples than in normal tissues and correlates with poor patient prognosis [93]. LINC00115 knockdown inhibits GSC proliferation and neurosphere formation in vitro and also inhibits tumour formation in the xenograft model [93]. LINC00115 physically associates with miR-200b and miR-200c [93]. Also, LINC00115 depletion reduced the expression of ZEB1 and ZNF596 and reduced invasion in GSCs [93]. Reporter assays indicate that ZEB1 and ZNF596 are targets of miR-200b and miR-200c. Down-regulation of ZEB1 and GBM invasion upon LINC00115 knockdown was reversed upon miR-200b overexpression, indicating that LINC00115 competitively binds to miR-200b to promote ZEB1-mediated GBM invasion [93]. LINC00115 and its target ZNG596 are co-expressed in clinical glioma samples. Concomitantly, exogenous expression of ZNF596 in LINC00115-depleted GSCs reversed the inhibition of cell proliferation caused by LINC00115 depletion, indicating that ZNF596 is the downstream effector of LINC00115-driven GBM tumourigenicity [93]. Also, LINC00115 binds to miR-200 to promote the expression of ZNF596 [93]. CRISPR-mediated knockout of ZNF596 indicated that EZH2 is a direct target of ZNF596. ZNF296 is a transcription factor promoting the expression of EZH2 [93]. LINC00115 depletion results in loss of EZH2 expression, which is reversed upon ZNF596 overexpression [93]. LINC00115 further activates STAT3 downstream of EZH2 through ZNF596, indicating that LINC00115 activates EZH2/STAT3 signalling through ZNF596, thereby promoting GSC self-renewal and tumourigenicity [93]. LINC00115 aids GSC’s self-renewal by acting as a ceRNA for transcription factors ZEB1 and ZNF596 by sponging miR-200 [93]. It also promotes GSC’s tumourigenicity through the ZNF596/EZH2/STAT3 signal axis [93].
3.1.5. H19 and HOXD-AS2
Nie et al. identified eight differentially regulated lncRNAs upon TGF-β treatment (H19, HOXD-AS2, LINC00635, LINC00277, RP11-196G11.2, LINC00152, MALAT1, and LOC100506207) in D54, P-GBM2 cells [33]. TGF-β induces LncRNAs H19 and HOXD-AS2 through SMAD signalling [33]. Further, RIP analysis of TGF-β treated cells depicted enhanced binding of H19 and HOXD-AS2 with K-homology (KH) splicing regulatory protein (KSRP) [33]. KSRP degrades follistatin-like 1 (FSTL1) and promotes the maturation of miR-198 in the nucleus [33]. TGF-β-induced H19 and HOXD-AS2 competitively bind to KSRP and thus prevent its nuclear translocation [33]. miR-198 is a tumour suppressor miRNA that promotes TMZ sensitivity in GBM by downregulating MGMT [33]. Overexpression of KSRP reversed the H19 and HOXD-AS2-mediated up-regulation of MGMT expression, which could reverse TGF-β-induced TMZ resistance [33]. H19 and HOXD-AS2 confer TMZ resistance by regulating miR-198 biogenesis by competing with KSRP [33].
3.1.6. MIR4435-2 Host Gene (MIR4435-2 HG)
Xu et al. reported the high expression of MIR4435-2HG in GBM tissue samples [98]. Loss-of-function studies of MIR4435-2HG in U251 and U87 cells decreased cell proliferation, colony formation, migration, and invasion [98]. In vivo, nude mice models also show reduced tumour volume and growth upon MIR4435-2HG depletion [98]. These results indicate an oncogenic role of MIR4435-2HG in GBM [98]. The starbase tool revealed that miR-1224-5p targets MIR4435-2HG [98]. Also, it was observed that miR-1224-5p is downregulated in LN229, U87-MG, and U251 cells compared to normal human astrocytes (NHA) [98]. Luciferase reporter assay confirmed the direct binding of MIR4435-2HG and miR-1224-5p [98]. Functional rescue experiments revealed that the increase in cell proliferation and colony formation induced by MIR4435-2HG overexpression was abrogated upon miR-1224-5p mimics, indicating that the MIR4435-2HG -miR-1224-5p axis promotes GBM pathogenesis [98]. Further, the starbase tool and luciferase reporter assay demonstrated that TGFBR2, a critical oncogene, is a direct target of miR-1224-5p [98]. Further, MIR4435-2HG overexpression promoted the expression of TGFBR2 at mRNA and protein levels [98]. Also, miR-1224-5p inhibition reduced proliferation and colony formation ability in GBM, which was rescued upon TGFBR2 overexpression [98]. In addition, TGFBR2 knockdown antagonized MIR4435-2HG overexpression-induced proliferation and colony formation in GBM, indicating that MIR4435-2HG promotes GBM proliferation by sponging miR-1224-5p and stabilizing TGFBR2 [98]. Hence, this study suggests an oncogenic role of lncRNA MIR4435-2HG in GBM by targeting the miR-1224-5p/TGFBR2 axis [98].
3.1.7. LncRNA RPSAP52
Wang et al. identified that lncRNA RPSAP52 is overexpressed in GBM tumour samples [97]. High expression of RPSAP52 is associated with poor survival in GBM patients [97]. Wang et al. also observed a positive correlation between RPSAP52 and TGF-β1expression in GBM samples [97]. Further, overexpression of RPSAP52 increased TGF-β1 protein expression, and knockdown of RPSAP52 exhibited the opposite effect [97]. Overexpression of RPSAP52 and TGF-β1 individually increased the percentage of CD133+ cells. Further, the overexpression of TGF-β1 rescued the reduction in the percentage of CD133+ cells observed upon RPSAP52 silencing [97].
3.1.8. LncRNA Plasmacytoma Variant Translocation-1 (PVT1)
Li et al. reported overexpression of lncRNA PVT1 and down-regulation of p53 in higher grades of glioma compared to normal brain cells [94]. Also, the expression of PVT1 increased with increasing grades of glioma [94]. Kaplan-Meier survival analysis revealed that high PVT1 levels are associated with poor survival in GBM patients. Clinical GBM samples show high expression of PVT1, TGF-β, and pSMAD2/3 levels and low p53 levels [94]. Further, the knockdown of p53 decreased PVT1 levels in U373 cells, while overexpression of p53 showed a reverse effect [94]. RIP assay revealed the direct interaction between PVT1 and p53 [94]. Bioinformatics analysis using lncATLAS provided evidence of the interaction of p53 with the PVT1 promoter. Also, the dual luciferase reporter assay indicated that p53 binds to the promoter of PVT1 to attenuate the expression of PVT1 [94]. Loss-of-function studies demonstrated that the knockdown of PVT1 reduced proliferation, viability, migration, and invasion, induced cell cycle arrest at S and G2/M phases, and promoted apoptosis in GBM [94]. However, the knockdown of p53 showed the opposite effects [94]. Expression of mesenchymal markers, N-cadherin, MMP-9, and MMP-2 was reduced, and E-cadherin was upregulated upon lncRNA PVT1 depletion. At the same time, the knockdown of p53 had a reverse effect. Furthermore, the study demonstrated that lncRNA PVT1 overexpression increased the transcription activity of the TGF-β, as depicted in the dual luciferase reporter assay, and increased the pSMAD2/3 levels [94]. Knockdown of lncRNA PVT1 decreased the transcription activity of TGF-β, while p53 knockdown displayed the opposite effects [94]. However, the combined knockdown of PVT1 and p53 counteracted the suppressive effects of p53 on TGF-β activity, indicating that p53 attenuated the TGF-β/SMAD pathway in GBM by targeting PVT1 [94]. In vivo nude mice model demonstrated that the knockdown of PVT1 and p53 individually suppressed the tumour growth [94]. In contrast, the combined knockdown of PVT1 and p53 counteracted the tumour-suppressive effects of p53 in GBM [94]. The levels of TGF-β and pSMAD2/3 were determined from in vivo nude tumour tissues transfected with shRNA against lncRNA PVT1 or p53 [94]. P53 knockdown increased PVT1 levels, TGF-β, and pSMAD2/3 levels [94]. At the same time, PVT1 knockdown displayed the opposite effects [94]. Also, the action of lncRNA PVT1 depletion on TGF-β activity was counteracted by p53 knockdown [94]. This study demonstrates that p53 potentially contributes to downregulating the oncogenic lncRNA PVT1, thereby suppressing the activation of TGF-β and TGF-β mediated GBM progression by modulating the lncRNA PVT1-TGF-β axis [94].
3.1.9. LncRNA-MUF
Using a genome-wide microarray screen, we identified that lncRNA-MUF is induced upon TGF-β treatment in T98G cells [95]. Also, lncRNA-MUF induction upon TGF-β treatment is observed across other GBM cell lines—LN229, U87-MG, and LN18. Moreover, levels of lncRNA-MUF are elevated in GBM tumour samples, and its expression is associated with poor survival and prognosis [95]. LncRNA-MUF induction by TGF-β is completely abolished upon treatment with TGFBR1 inhibitor SB505124 in glioma cells. In addition, the ChIP qPCR assay demonstrates the enrichment of SMAD2/3 antibody in the promoter of lncRNA-MUF upon TGF-β stimulation [95]. Loss-of-function assays using siRNA against lncRNA-MUF revealed that lncRNA-MUF promotes proliferation, migration, and invasion in GBM [95]. In addition, we show that loss of lncRNA-MUF sensitizes glioma cells to TMZ-induced cell death [95]. Knockdown of lncRNA-MUF downregulated its cis oncogene, CAPRIN2, and various trans genes from the TGF-β ontology group (VIMENTIN, CTGF, c-MYC, and SNAIL1) [95]. Western blotting analysis of mesenchymal markers revealed the down-regulation of N-cadherin, VIMENTIN, and SNAIL1 upon lncRNA-MUF knockdown in T98G and U87-MG cells [95]. Bioinformatics analysis and dual luciferase reporter assay demonstrated the direct interaction between lncRNA-MUF and miR-34a, and overexpression of miR-34a reduces lncRNA-MUF expression. miR-34a has a potential tumour-suppressor role in glioma by targeting several oncogenes, particularly SNAIL1, and it is downregulated in GBM tissues compared to normal tissues [95]. SNAIL1 is a crucial transcription factor that promotes tumour cell invasion and EMT and is upregulated in GBM. We observed a positive correlation between MUF and SNAIL1 expression in GBM tumour samples [95]. Down-regulation of Snail and reduction in invasion upon lncRNA-MUF knockdown was rescued upon miR-34a inhibition in T98G and U87-MG cells. These experiments indicate that TGF-β-induced lncRNA-MUF sponges miR-34a to promote SNAIL1-induced invasion in GBM [95]. Our study suggests that TGF-β induced lncRNA-MUF promotes GBM invasion through the miR-34a/Snail axis [95].
3.1.10. LINC01711
We have shown that TGF-β induces LINC01711 expression in glioma cells and that the levels of LINC01711 are elevated in GBM tumour samples, and its expression is associated with poor patients’ survival and prognosis [96]. Like lncRNA MUF, LINC01711 is also induced by the SMAD2/3 transcription factors downstream of TGF-β signalling. Down-regulation of LINC01711 reduces proliferation, migration, and invasion and induces apoptosis in GBM [96]. LINC01711 knockdown results in downregulating ZEB1, a crucial transcription factor that promotes tumour cell invasion and EMT [96]. LINC01711 also interacts with miR-34a, and ZEB1 is a target of miR-34a [96]. Reduction of ZEB1 expression due to LINC01711 knockdown was rescued upon miR-34a inhibition [96]. Further, the invasion assay revealed that miR-34a inhibition could reverse the reduction in invasion caused by LINC01711 knockdown in T98G and U87-MG cells [96]. We also observed that ZEB1 overexpression could partially reverse the LINC01711 knockdown-mediated reduction in invasion in GBM [96]. Further, we tested if LINC01711 could promote TMZ resistance in GBM. LINC01711 depletion significantly reduced proliferation and increased caspase 3/7 activity in T98G and LN229 cells, suggesting that LINC01711 promotes resistance to TMZ in GBM. Given the role of ZEB1 in TMZ resistance and that LINC01711 depletion results in ZEB1 inhibition, we evaluated if LINC01711 knockdown-mediated sensitization of GBM cells to TMZ-induced apoptosis is associated with a reduction in ZEB1 levels. ZEB1 protein levels were significantly downregulated during TMZ treatment in LINC01711-depleted cells compared to cells treated with TMZ alone [96]. We found that in addition to TMZ-mediated apoptosis, LINC01711 knockdown could induce cisplatin-mediated apoptosis in GBM. Hence, upon induction by TGF-β, LINC01711 promotes GBM proliferation, migration, invasion, and drug resistance by modulating the LINC01711/miR-34a/ZEBl signalling axis [96].
3.2. Tumour Suppressor lncRNAs Involved in the TGF-β Pathway in Gliomas
3.2.1. LncRNA TCONS_00020456
Tang et al., using a microarray screen, identified 1759 upregulated and 1932 downregulated lncRNAs in U251 cells [99]. Among these differentially expressed lncRNAs, they characterized the most downregulated lncRNA—TCONS_00020456 role in GBM pathogenesis [99]. The expression of TCONS_00020456 decreased with increasing glioma grades, and the low expression of TCONS_00020456 indicated poor survival of GBM patients [99]. Further, the siRNA-mediated knockdown of TCONS_00020456 significantly promoted the invasion and migration of U251 and U87 cells [99]. While overexpression of TCONS_00020456 significantly inhibited invasion and migration in GBM [99]. Bioinformatic analysis suggests that various mRNAs with oncogenic function negatively correlated with TCONS_00020456 expression [99]. Among them, SMAD2 and PKCα were the top hits [99]. Further, western blotting analysis revealed that the knockdown of TCONS_00020456 increased the expression of SMAD2, PKCα, N-cadherin, vimentin, and down-regulation of E-cadherin. Also, the phosphorylation of JNK and ERK was elevated upon TCONS_0002045 knockdown [99]. The overexpression of TCONS_0002045 reversed these effects [99]. These results indicate that TCONS_0002045 abrogates GBM invasion and migration by targeting SMAD2 and PKCα pathways [99]. In vivo, analysis of TCONS_0002045 in nude mice model revealed a decrease in tumour size and weight in the TCONS_0002045 overexpression group compared to the TCONS_002045 knockdown group [99]. In addition, the immunohistochemical staining of tumour tissues from the nude mice indicated increased expression of SMAD2 and PKCα in the TCONS_002045 knockdown group compared to the TCONS_0002045 overexpression group [99]. Computational analysis using the miRDB database revealed several miRNAs targeting TCONS_0002045, SMAD2, and PKCα [99]. Among these miRNAs, miR-1279 was identified as the common miRNA target between the three. LncRNA TCONS_0002045 abrogates GBM migration and invasion by targeting SMAD2 and PKCα [99]. However, the exact mechanism of down-regulation of SMAD2 and PKCα by TCONS_0002045 and the role of miR-1279 needs further investigation.
3.2.2. LncRNA RP11-838N2.4
RP11-838N2.4 expression is lower in TMZ-resistant cells (U87TR, U251TR) compared to the parental non-resistant cells (U87, U251) [100]. Moreover, RP11-838N2.4 down-regulation is associated with poor prognosis, a high risk of GBM relapse, and shorter postoperative survival times [100]. Overexpression of lncRNA RP11-838N2.4 enhances the cytotoxic effects of TMZ in vitro and in vivo [100]. Consequently, TMZ-resistant U251TR cells with high lncRNA RP11-838N2.4 displayed low levels of miR-10a [100]. The lncRNA acts as an endogenous sponge for EphA8 by competing with miR-10a and increasing the levels of EphA8, which promotes apoptosis in glioma cells [100]. Notably, lncRNA RP11-838N2.4 overexpression hindered the TGF-β pathway independent of miR-10a by reducing mRNA and protein levels of TGF-β1, TGFBR1, SMAD2, SMAD3, and SMAD4 levels [100]. LncRNA RP11-838N2.4 hinders GBM proliferation and promotes TMZ sensitivity and TMZ-mediated apoptosis by sponging miR-10a and stabilizing EphA8. In addition, it downregulates the TGF-β pathway by reducing the expression of the signalling pathway’s components. However, the exact molecular mechanism of lncRNA RP11-838N2.4-mediated down-regulation of the TGF-β pathway needs further investigation.
4. CircRNAs Involved in the TGF-β Pathway in GBM
4.1. Oncogenic circRNAs Involved in the TGF-β Pathway in Gliomas
CircARID1A
Li et al., using microarray analysis, identified differentially expressed circular RNAs in GBM tumour tissues [101]. CircARID1A was highly abundant in GBM samples and GBM cell lines and was also present in the blood exosomes of GBM patients [101]. shRNA-mediated knockdown of circARID1A attenuated the migration and invasion of GBM cells [101]. Also, MMP 2, MMP 9, and MMP 14 were downregulated upon circARID1A knockdown, indicating that circARID1A promotes GBM cell migration and invasion [101]. Bioinformatic analysis revealed that cicrARID1A interacts with miR-370-3p [101]. FISH experiments demonstrated the co-localization of circARID1A and miR-370-3p in the cytoplasm [101]. Dual luciferase reporter assay with the plasmid containing miR-370-3p binding sites of circARID1A, co-transfected with miR-370-3p mimics, significantly reduced the luciferase reporter activity [101]. These results suggest a direct interaction between cicrARID1A and miR-370-3p [101]. miR-370-3p expression is downregulated in GBM tissue compared to normal brain tissues [101]. Bioinformatics analysis and luciferase reporter assay showed that TGFBR2 is an essential target of miR-370-3p [101]. RNA pulldown assay with biotin-labelled miR-370-3p in U87 cells showed enrichment of TGFBR2 and circARID1A, indicating the possibility of circARID1A/miR-370-3p/TGFBR2 axis in GBM [101]. Further, silencing miR-370-3p increased the invasion of GBM cells [101]. Also, the knockdown of circARID1A attenuated TGFBR2 levels [101]. miR-370-3p inhibition promoted TGFBR2 protein levels, which was reversed upon combined knockdown of miR-370-3p and circARID1A [101]. GBM cell invasion and migration increased upon miR-370-3p inhibition, which was reversed upon combined knockdown of miR-370-3p and circARID1A [101]. Further, in vivo, the xenograft tumour model indicated reduced tumour growth and reduced TGFBR2 protein levels upon circARID1A knockdown. Thus, circARID1A promotes GBM invasion by sponging miR-370-3p to stabilize TGFBR2 [101].
Chen et al. performed circular RNA sequencing in LN229 and T98G GBM cells and identified several differentially expressed circular RNAs. KEGG and gene ontology analysis of the top-upregulated circular RNAs revealed that TGF-β is a vital pathway regulated by the top-candidate circular RNAs [102]. Further mechanistic studies are required to functionally characterize these candidate circular RNAs and their role in the TGF-β pathway in GBM [102].
4.2. Tumour Suppressor circRNAs Involved in the TGF-β Pathway in Gliomas
CircCD44
Leucine-rich repeat-containing 4 (LRRC4) is a tumour suppressor in GBM [103]. Feng et al. identified that LRCC4 promoted the generation of a circular RNA, circCD44, from the CD44 mRNA by inhibiting the interaction of CD44 pre-mRNA and SAM68 [103]. CircCD44 expression is downregulated in GBM tissues and cell lines [103]. Also, the overexpression of circCD44 attenuated the proliferation, colony formation, and invasion of GBM cells [103]. In vivo, the xenograft model also depicted a reduced tumour growth upon re-expression of circCD44 [103]. Bioinformatics analysis and reporter assays revealed that SMAD6 is an essential target of miR-326 and miR-330-5p [103]. Further mechanistic studies depicted that circCD44 sponges miR-326 and miR-330-5p to stabilize SMAD6 [103]. Thus, the LRRC4/SAM68/circCD44/miR-326/miR-330-5p/SMAD6 signalling axis is an essential regulator of GBM pathogenesis [103].
5. Discussion
The TGF-β signalling pathway is an attractive therapeutic target for GBM. However, the development of therapeutics targeting the TGF-β pathway has been hindered mainly by its critical regulatory roles in normal physiology and due to its ability to function as both tumour promoter and inhibitor in a context-dependent manner [104]. Hence, there is a need to specifically target tumour-promoting functions of TGF-β. Aberrant TGF-β signalling in GBM alters regulatory ncRNA expression and vice versa to promote GBM pathogenesis. NcRNAs can modulate the TGF-β pathway in GBM in the following ways (i) they act as downstream effectors of the TGF-β pathway, (ii) they can regulate components of the TGF-β pathway, and (iii) they can form a positive or negative feedback loop with the TGF-β pathway.
Many ncRNAs are in clinical trials for potential biomarkers and therapeutic targets for cancers and other diseases. LncRNA MFI2-AS1 is in clinical trials for use as a diagnostic biomarker for kidney cancer [105], and lncRNAs UCA1 and WRAP53 are in clinical trials for use as diagnostic biomarkers for hepatocellular carcinoma [106]. Results from safety trials of RNA-targeted therapies using ASO against lncRNAs are also promising [107]. Andes-1537, a short single-stranded phosphorothioate-deoxyoligonucleotide against antisense non-coding mitochondrial RNA (ASncmtRNA), was evaluated in phase I clinical trial by subcutaneous administration in patients with solid tumours. The results of this study displayed low toxicity of the oligonucleotide, with significant anti-tumour activity in pancreatic and cholangiocarcinoma patients [107]. A phase I trial for a liposomal mimic of miR-34a (MRX34) was carried out in patients with renal-cell carcinoma, hepatocellular carcinoma, melanoma, lung cancer, and gastrointestinal stromal tumours [108,109]. The MRX34 treatment in patients pretreated with dexamethasone displayed significant dose-dependent modulation of miR-34a target genes and manageable toxicity [109]. However, studies on ncRNAs as biomarkers and therapeutic agents in GBM are still in the pre-clinical testing stage. The major challenge associated with delivering RNA-targeted therapies to the brain is to cross the blood-brain barrier. ASO-mediated therapies against ncRNAs are best delivered when conjugated with nanoparticulate formulations [110]. ASO-loaded glucosylated-polyion complex micelles have shown promise to effectively deliver ASOs across the blood-brain barrier through the intravenous route [111].
TGF-β pathway promotes TMZ resistance in GBM [33]. Several TGF-β target genes, such as CTGF, ZEB1, and SNAIL1, are reported to promote TMZ resistance [31,32]. Many lncRNAs and miRNAs regulate TMZ-mediated cell death in GBM. For example, lncRNAs H19, HOXD-AS2, lncRNA-MUF, LINC01711, lncRNA RP11-838N2.4, miR-210-3p contribute to TMZ resistance [33,81,95,96,100]. Since TMZ is the preferred choice for glioma treatment, the potential of ASOs against these ncRNAs for promoting TMZ sensitivity should be tested in pre-clinical and clinical studies.
Furthermore, the non-canonical TGF-β downstream targets, such as NF-κB and PI3K/AKT/mTOR pathways, promote GBM pathogenesis. Specific lncRNAs and miRNAs establish a link between TGF-β and these non-canonical signalling pathways. For example, lncRNA-ATB, induced by TGF-β, promotes GBM invasion through the NF-κB and P38/MAPK pathways [90]. Also, TGF-β-induced miR-182 suppresses CYLD and promotes sustained activation of NF-κB in GBM [78]. Similarly, the hyperactive NF-κB signalling in GBM promotes the expression of miR-148a oncogene. miR-148a, upon induction, promotes the GBM pathogenesis by activating the TGF-β signalling by promoting the expression of pSMAD3 and downregulating the negative regulators (QKI and SKI) of the TGF-β signalling [82]. It needs to be tested if targeting such ncRNAs, which modulate crosstalk between multiple oncogenic pathways, can achieve better therapeutic efficacy for GBMs.
Most studies on the TGF-β modulating lncRNAs and circRNAs in GBM have focused on their ability to function as ceRNAs to sponge miRNAs. However, lncRNAs and circRNAs also function by interacting with proteins [49,59]. Further studies are needed to identify RNA binding proteins interacting with these lncRNAs and circRNAs to promote GBM pathogenesis and fully understand their potential as therapeutic targets. Dysregulation of several ncRNAs, such as: miR-129-2, lncRNAs AF086127, AF086217, AF086391, AF119852, AK021535, AK022370, AL050068, BC012548, and BC041658 occurs in DIPG [112]. However, studies are required to understand the relationship between ncRNAs and the TGF-β pathway in DIPG.
In summary, tumour-promoting ncRNAs involved in the TGF-β pathway have the potential to serve as attractive biomarkers and therapeutic targets for GBM.
Author Contributions
B.S. and V.S. designed and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by extramural grants from the Government of India (DST-SERB ECR/2017/001953 and DBT-RLS 102/IFD/SAN/3499/2016-17) and intramural funds from an OPERA grant from BITS Pilani to VS. BS was supported by an SRF from ICMR No. 2020-7940/GEN-BMS.
Conflicts of Interest
V.S. is a guest editor in the special issue in which this article is published.
References
- Stupp, R.; Brada, M.J.; van den Bent, M.; Tonn, J.-C.; Pentheroudakis, G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii93–iii101. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Raviram, R.; Raman, A.; Preissl, S.; Ning, J.; Wu, S.; Koga, T.; Zhang, K.; Brennan, C.W.; Zhu, C.; Luebeck, J.; et al. Integrated analysis of single-cell chromatin state and transcriptome identified common vulnerability despite glioblastoma heterogeneity. Proc. Natl. Acad. Sci. USA 2023, 120, e2210991120. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.Y.; Assem, M. Glioblastoma Genomics: A Very Complicated Story; Exon Publications: Brisbane, Australia, 2017; Chapter 1; pp. 3–25. [Google Scholar]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef]
- Damodharan, S.; Lara-Velazquez, M.; Williamsen, B.C.; Helgager, J.; Dey, M. Diffuse Intrinsic Pontine Glioma: Molecular Landscape, Evolving Treatment Strategies and Emerging Clinical Trials. J. Pers. Med. 2022, 12, 840. [Google Scholar] [CrossRef]
- Srikanthan, D.; Taccone, M.S.; Van Ommeren, R.; Ishida, J.; Krumholtz, S.L.; Rutka, J.T. Diffuse intrinsic pontine glioma: Current insights and future directions. Chin. Neurosurg. J. 2021, 7, 6. [Google Scholar] [CrossRef]
- Burster, T.; Traut, R.; Yermekkyzy, Z.; Mayer, K.; Westhoff, M.-A.; Bischof, J.; Knippschild, U. Critical View of Novel Treatment Strategies for Glioblastoma: Failure and Success of Resistance Mechanisms by Glioblastoma Cells. Front. Cell Dev. Biol. 2021, 9, 695325. [Google Scholar] [CrossRef]
- Aldape, K.; Brindle, K.; Chesler, L.; Chopra, R.; Gajjar, A.; Gilbert, M.R.; Gottardo, N.; Gutmann, D.H.; Hargrave, D.; Holland, E.C.; et al. Challenges to curing primary brain tumours. Nat. Rev. Clin. Oncol. 2019, 16, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, L.; Wei, Q.; Shao, A. O6-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front. Oncol. 2020, 9, 1547. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Somasundaram, K. Glioblastoma vs temozolomide: Can the red queen race be won? Cancer Biol Ther. 2019, 20, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Maksoud, S. The DNA Double-Strand Break Repair in Glioma: Molecular Players and Therapeutic Strategies. Mol. Neurobiol. 2022, 59, 5326–5365. [Google Scholar] [CrossRef]
- Kaminska, B.; Kocyk, M.; Kijewska, M. TGF Beta Signaling and Its Role in Glioma Pathogenesis. Adv. Exp. Med. Biol. 2013, 986, 171–187. [Google Scholar] [CrossRef]
- Han, J.; A Alvarez-Breckenridge, C.; Wang, Q.-E.; Yu, J. TGF-β signaling and its targeting for glioma treatment. Am. J. Cancer Res. 2015, 5, 945–955. [Google Scholar]
- Saghazadeh, A.; Rezaei, N. Central Inflammatory Cytokines in Tuberculous Meningitis: A Systematic Review and Meta-analysis. J. Interf. Cytokine Res. 2022, 42, 95–107. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Sedlarikova, L.; Hajek, R.; Sevcikova, S. TGF-β—An excellent servant but a bad master. J. Transl. Med. 2012, 10, 183. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef]
- David, C.J.; Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018, 19, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Pardoux, C.; Hall, M.C.; Lee, P.S.; Warburton, D.; Qing, J.; Smith, S.M.; Derynck, R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007, 26, 3957–3967. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Gomis, R.R. TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol. 2017, 9, a022277. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.H.; Thomas, T.Z.; Masumori, N.; Bhowmick, N.A.; Gorska, A.E.; Shyr, Y.; Kasper, S.; Case, T.; Roberts, R.L.; Shappell, S.B.; et al. The Loss of TGF-β Signaling Promotes Prostate Cancer Metastasis. Neoplasia 2003, 5, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Ling, L.; van Dam, H.; Zhou, F.; Zhang, L. TGF-β signaling in cancer metastasis. Acta Biochim. Biophys. Sin. 2018, 50, 121–132. [Google Scholar] [CrossRef]
- Frei, K.; Gramatzki, D.; Tritschler, I.; Schroeder, J.J.; Espinoza, L.; Rushing, E.J.; Weller, M. Transforming growth factor-β pathway activity in glioblastoma. Oncotarget 2015, 6, 5963–5977. [Google Scholar] [CrossRef]
- Roy, L.-O.; Poirier, M.-B.; Fortin, D. Transforming growth factor-beta and its implication in the malignancy of gliomas. Target. Oncol. 2015, 10, 1–14. [Google Scholar] [CrossRef]
- Urbanavičiūtė, R.; Zabitaitė, R.; Kriščiukaitis, A.; Deltuva, V.P.; Skiriutė, D. Serum protein triplet TGF-β1, TIMP-1, and YKL-40 serve as diagnostic and prognostic profile for astrocytoma. Sci. Rep. 2021, 11, 13100. [Google Scholar] [CrossRef]
- Zeng, H.; Yang, Z.; Xu, N.; Liu, B.; Fu, Z.; Lian, C.; Guo, H. Connective tissue growth factor promotes temozolomide resistance in glioblastoma through TGF-β1-dependent activation of Smad/ERK signaling. Cell Death Dis. 2017, 8, e2885. [Google Scholar] [CrossRef]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef]
- Liang, H.; Chen, G.; Li, J.; Yang, F. Snail expression contributes to temozolomide resistance in glioblastoma. Am. J. Transl. Res. 2019, 11, 4277–4289. [Google Scholar]
- Nie, E.; Jin, X.; Miao, F.; Yu, T.; Zhi, T.; Shi, Z.; Wang, Y.; Zhang, J.; Xie, M.; You, Y. TGF-β1 modulates temozolomide resistance in glioblastoma via altered microRNA processing and elevated MGMT. Neuro-Oncology 2020, 23, 435–446. [Google Scholar] [CrossRef]
- Bogdahn, U.; Hau, P.; Stockhammer, G.; Venkataramana, N.K.; Mahapatra, A.K.; Suri, A.A.; Balasubramaniam, A.; Nair, S.; Oliushine, V.; Parfenov, V.; et al. Targeted therapy for high-grade glioma with the TGF-β2 inhibitor trabedersen: Results of a randomized and controlled phase IIb study. Neuro-Oncology 2011, 13, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Rodon, J.; Carducci, M.A.; Sepulveda-Sánchez, J.M.; Azaro, A.; Calvo, E.; Seoane, J.; Braña, I.; Sicart, E.; Gueorguieva, I.; Cleverly, A.L.; et al. First-in-Human Dose Study of the Novel Transforming Growth Factor-β Receptor I Kinase Inhibitor LY2157299 Monohydrate in Patients with Advanced Cancer and Glioma. Clin. Cancer Res. 2015, 21, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Hau, P.; Jachimczak, P.; Bogdahn, U. Treatment of malignant gliomas with TGF-beta2 antisense oligonucleotides. Expert Rev. AnticancerTher. 2009, 9, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Niaz, S.; Hussain, M.U. Role of GW182 protein in the cell. Int. J. Biochem. Cell Biol. 2018, 101, 29–38. [Google Scholar] [CrossRef]
- Fukaya, T.; Iwakawa, H.-O.; Tomari, Y. MicroRNAs Block Assembly of eIF4F Translation Initiation Complex in Drosophila. Mol. Cell 2014, 56, 67–78. [Google Scholar] [CrossRef] [PubMed]
- de la Mata, M.; Gaidatzis, D.; Vitanescu, M.; Stadler, M.B.; Wentzel, C.; Scheiffele, P.; Filipowicz, W.; Grosshans, H. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015, 16, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Z.; Corey, D.R. The Requirement for GW182 Scaffolding Protein Depends on Whether Argonaute Is Mediating Translation, Transcription, or Splicing. Biochemistry 2018, 57, 5247–5256. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Medarova, Z.; Moore, A. Role of microRNAs in glioblastoma. Oncotarget 2021, 12, 1707–1723. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 2020, 89, 283–308. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Shree, B.; Mohapatra, S.; Swati; Basu, A.; Sharma, V. The Expanding Regulatory Mechanisms and Cellular Functions of Long Non-coding RNAs (lncRNAs) in Neuroinflammation. Mol. Neurobiol. 2021, 58, 2916–2939. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Shree, B.; Sharma, V. Linc‘ing’ RNA to DNA Repair. Proc. Indian Natn. Sci. Acad. 2018, 84, 521–529. [Google Scholar]
- Swati; Sharma, V. The interplay of cytokine signaling and non-coding RNAs in head and neck squamous cell carcinoma pathobiology. Mol. Biol. Rep. 2022, 49, 10825–10847. [Google Scholar] [CrossRef] [PubMed]
- Shree, B.; Das, K.; Sharma, V. Emerging role of transforming growth factor-β-regulated long non-coding RNAs in prostate cancer pathogenesis. Cancer Pathog. Ther. 2023, 1, 195–204. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2015, 44, D203–D208. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, O.; Tamizkar, K.H.; Sharifi, G.; Taheri, M.; Ghafouri-Fard, S. Emerging Role of Long Non-Coding RNAs in the Pathobiology of Glioblastoma. Front. Oncol. 2021, 10, 625884. [Google Scholar] [CrossRef]
- Wu, X.; Yang, L.; Wang, J.; Hao, Y.; Wang, C.; Lu, Z. The Involvement of Long Non-Coding RNAs in Glioma: From Early Detection to Immunotherapy. Front. Immunol. 2022, 13, 897754. [Google Scholar] [CrossRef]
- Stackhouse, C.T.; Gillespie, G.Y.; Willey, C.D. Exploring the Roles of lncRNAs in GBM Pathophysiology and Their Therapeutic Potential. Cells 2020, 9, 2369. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Liu, Y.; Su, H.; Zhang, J.; Liu, Y.; Feng, C.; Han, F. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol. 2020, 18, e3000582. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef]
- Rajappa, A.; Banerjee, S.; Sharma, V.; Khandelia, P. Circular RNAs: Emerging Role in Cancer Diagnostics and Therapeutics. Front. Mol. Biosci. 2020, 7, 577938. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Salami, R.; Salami, M.; Mafi, A.; Vakili, O.; Asemi, Z. Circular RNAs and glioblastoma multiforme: Focus on molecular mechanisms. Cell Commun. Signal. 2022, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Piao, H. Research Progress of circRNAs in Glioblastoma. Front. Cell Dev. Biol. 2021, 9, 791892. [Google Scholar] [CrossRef] [PubMed]
- Mafi, A.; Rahmati, A.; Babaei Aghdam, Z.; Salami, R.; Salami, M.; Vakili, O.; Aghadavod, E. Recent insights into the microRNA-dependent modulation of gliomas from pathogenesis to diagnosis and treatment. Cell. Mol. Biol. Lett. 2022, 27, 65. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Hu, F.; Peng, Y.; Fan, X.; Zhang, X.; Jin, Z. Circular RNAs: Implications of signaling pathways and bioinformatics in human cancer. Cancer Biol. Med. 2023, 20, 104–128. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Cheng, D.; Wang, J.; Dong, Z.; Li, X. Cancer-related circular RNA: Diverse biological functions. Cancer Cell Int. 2021, 21, 11. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef] [PubMed]
- Nandwani, A.; Rathore, S.; Datta, M. LncRNAs in cancer: Regulatory and therapeutic implications. Cancer Lett. 2021, 501, 162–171. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef]
- Papoutsoglou, P.; Moustakas, A. Long non-coding RNAs and TGF-β signaling in cancer. Cancer Sci. 2020, 111, 2672–2681. [Google Scholar] [CrossRef]
- Janakiraman, H.; House, R.P.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H.; Palanisamy, V. The Long (lncRNA) and Short (miRNA) of It: TGFβ-Mediated Control of RNA-Binding Proteins and Noncoding RNAs. Mol. Cancer Res. 2018, 16, 567–579. [Google Scholar] [CrossRef]
- Song, L.; Liu, L.; Wu, Z.; Li, Y.; Ying, Z.; Lin, C.; Wu, J.; Hu, B.; Cheng, S.Y.; Li, M.; et al. TGF-β induces miR-182 to sustain NF-κB activation in glioma subsets. J. Clin. Investig. 2012, 122, 3563–3578. [Google Scholar] [CrossRef]
- Guo, Y.; Gao, X.; An, S.; Li, X.; Pan, L.; Liu, H.; Liu, J.; Gao, J.; Zhao, Z.; Li, G.; et al. Deletion of miR-15a inhibited glioma development via targeting Smad7 and inhibiting EMT pathway. Aging 2021, 13, 24339–24348. [Google Scholar] [CrossRef]
- Zhong, Q.; Wang, T.; Lu, P.; Zhang, R.; Zou, J.; Yuan, S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma: miR-193b Promotes Cell Proliferation in Human Glioma. J. Neurosci. Res. 2014, 92, 619–626. [Google Scholar] [CrossRef]
- Liu, H.; Chen, C.; Zeng, J.; Zhao, Z.; Hu, Q. MicroRNA-210-3p is transcriptionally upregulated by hypoxia induction and thus promoting EMT and chemoresistance in glioma cells. PLoS ONE 2021, 16, e0253522. [Google Scholar] [CrossRef]
- Wang, H.; Pan, J.Q.; Luo, L.; Ning, X.J.; Ye, Z.P.; Yu, Z.; Li, W.S. NF-κB induces miR-148a to sustain TGF-β/Smad signaling activation in glioblastoma. Mol. Cancer 2015, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, J.; Lan, Q. TGF-β-induced miR10a/b expression promotes human glioma cell migration by targeting PTEN. Mol. Med. Rep. 2013, 8, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wei, F.; Xia, H.; Liu, H.; Dong, X.; Zhang, Y.; Luo, Q.; Liu, Y.; Li, Y. MicroRNA-10b mediates TGF-β1-regulated glioblastoma proliferation, migration and epithelial-mesenchymal transition. Int. J. Oncol. 2017, 50, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.B.; Cai, L.; Lin, S.J.; Lu, J.L.; Yao, Y.; Zhou, L.F. The miR-92b functions as a potential oncogene by targeting on Smad3 in glioblastomas. Brain Res. 2013, 1529, 16–25. [Google Scholar] [CrossRef]
- Guo, P.; Yu, Y.; Li, H.; Zhang, D.; Gong, A.; Li, S.; Liu, W.; Cheng, L.; Qiu, Y.; Yao, W.; et al. TGF-β1-induced miR-503 controls cell growth and apoptosis by targeting PDCD4 in glioblastoma cells. Sci. Rep. 2017, 7, 11569. [Google Scholar] [CrossRef]
- Jiang, H.; Jin, C.; Liu, J.; Hua, D.; Zhou, F.; Lou, X.; Zhao, N.; Lan, Q.; Huang, Q.; Yoon, J.-G.; et al. Next Generation Sequencing Analysis of miRNAs: MiR-127-3p Inhibits Glioblastoma Proliferation and Activates TGF-β Signaling by Targeting SKI. OMICS A J. Integr. Biol. 2014, 18, 196–206. [Google Scholar] [CrossRef]
- Jiang, C.; Shen, F.; Du, J.; Hu, Z.; Li, X.; Su, J.; Wang, X.; Huang, X. MicroRNA-564 is downregulated in glioblastoma and inhibited proliferation and invasion of glioblastoma cells by targeting TGF-β1. Oncotarget 2016, 7, 56200–56208. [Google Scholar] [CrossRef][Green Version]
- Ma, C.-C.; Xiong, Z.; Zhu, G.-N.; Wang, C.; Zong, G.; Wang, H.-L.; Bian, E.-B.; Zhao, B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J. Exp. Clin. Cancer Res. 2016, 35, 90. [Google Scholar] [CrossRef]
- Tang, F.; Wang, H.; Chen, E.; Bian, E.; Xu, Y.; Ji, X.; Yang, Z.; Hua, X.; Zhang, Y.; Zhao, B. LncRNA-ATB promotes TGF-β-induced glioma cells invasion through NF-κB and P38/MAPK pathway. J. Cell. Physiol. 2019, 234, 23302–23314. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; Zhong, Q.; Wu, J.; Tang, Z. Lnc RNA UCA 1 is necessary for TGF -β-induced epithelial–mesenchymal transition and stemness via acting as a ce RNA for Slug in glioma cells. FEBS Open Bio 2018, 8, 1855–1865. [Google Scholar] [CrossRef]
- Li, C.; Zheng, H.; Hou, W.; Bao, H.; Xiong, J.; Che, W.; Gu, Y.; Sun, H.; Liang, P. Long non-coding RNA linc00645 promotes TGF-β-induced epithelial–mesenchymal transition by regulating miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 2019, 10, 717. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yu, B.; Li, Y.; Zhang, W.; Alvarez, A.A.; Hu, B.; Cheng, S.Y.; Feng, H. TGF-β-activated lncRNA LINC00115 is a critical regulator of glioma stem-like cell tumorigenicity. EMBO Rep. 2019, 20, e48170. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.; Xia, P.; Wang, L.; Lu, Z. Targeting long non-coding RNA PVT1/TGF-β/Smad by p53 prevents glioma progression. Cancer Biol. Ther. 2022, 23, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Shree, B.; Tripathi, S.; Sharma, V. Transforming Growth Factor-Beta-Regulated LncRNA-MUF Promotes Invasion by Modulating the miR-34a Snail1 Axis in Glioblastoma Multiforme. Front. Oncol. 2022, 11, 788755. [Google Scholar] [CrossRef] [PubMed]
- Shree, B.; Sengar, S.; Tripathi, S.; Sharma, V. LINC01711 promotes transforming growth factor-beta (TGF-β) induced invasion in glioblastoma multiforme (GBM) by acting as a competing endogenous RNA for miR-34a and promoting ZEB1 expression. Neurosci. Lett. 2023, 792, 136937. [Google Scholar] [CrossRef]
- Wang, S.; Guo, X.; Lv, W.; Li, Y.; Zhang, L.; Dong, C.; Zhang, J.; Cheng, G. LncRNA RPSAP52 Upregulates TGF-β1 to Increase Cancer Cell Stemness and Predict Postoperative Survival in Glioblastoma. Cancer Manag. Res. 2020, 12, 2541–2547. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, B.; Yang, Y.; Li, Z.; Zhao, P.; Wu, W.; Zhang, H.; Mao, J. LncRNA MIR4435-2HG potentiates the proliferation and invasion of glioblastoma cells via modulating miR-1224-5p/TGFBR2 axis. J. Cell. Mol. Med. 2020, 24, 6362–6372. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, Y.; Zhang, L.; Wang, J.; Wang, W.; Han, X.; Mu, C.; Gao, D. Identification of novel LncRNA targeting Smad2/PKCα signal pathway to negatively regulate malignant progression of glioblastoma. J. Cell. Physiol. 2020, 235, 3835–3848. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, N.; Liu, B.; Huang, Y.; Zeng, H.; Yang, Z.; He, Z.; Guo, H. Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget 2016, 7, 43835–43851. [Google Scholar] [CrossRef]
- Li, B.; Chen, J.; Wu, Y.; Luo, H.; Ke, Y. Decrease of circARID1A retards glioblastoma invasion by modulating miR-370-3p/ TGFBR2 pathway. Int. J. Biol. Sci. 2022, 18, 5123–5135. [Google Scholar] [CrossRef]
- Chen, Z.; Su, S.; Yang, M.; Wang, F.; Chen, M. Profiling and Bioinformatics Analyses of Differential Circular RNA Expression in Glioblastoma Multiforme Cells under Hypoxia. J. Mol. Neurosci. 2022, 72, 2451–2463. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Ren, X.; Fu, H.; Li, D.; Chen, X.; Zu, X.; Liu, Q.; Wu, M. LRRC4 mediates the formation of circular RNA CD44 to inhibit GBM cell proliferation. Mol. Ther.-Nucleic Acids 2021, 26, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.C.; Zhang, Y.Y.; Li, J.S.; Chan, M.K.; Chen, J.; Tang, Y.; Zhou, Y.; Zhang, D.; Leung, K.T.; To, K.F.; et al. LncRNA-Dependent Mechanisms of Transforming Growth Factor-β: From Tissue Fibrosis to Cancer Progression. ncRNA 2022, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Song, J.; Zhang, W.; Ran, L. Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer. Dig. Dis. Sci. 2020, 65, 3538–3550. [Google Scholar] [CrossRef]
- Abdelmoety, A.A.; Elhassafy, M.Y.; Said, R.S.O.; Elsheaita, A.; Mahmoud, M.M. The role of UCA1 and WRAP53 in diagnosis of hepatocellular carcinoma: A single-center case-control study. Clin. Exp. Hepatol. 2023, 9, 129–137. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2016, 35, 180–188. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Bennett, C.F.; Krainer, A.R.; Cleveland, D.W. Antisense Oligonucleotide Therapies for Neurodegenerative Diseases. Annu. Rev. Neurosci. 2019, 42, 385–406. [Google Scholar] [CrossRef]
- Min, H.S.; Kim, H.J.; Naito, M.; Ogura, S.; Toh, K.; Hayashi, K.; Kim, B.S.; Fukushima, S.; Anraku, Y.; Miyata, K.; et al. Systemic Brain Delivery of Antisense Oligonucleotides across the Blood-Brain Barrier with a Glucose-Coated Polymeric Nanocarrier. Angew. Chem. Int. Ed. Engl. 2020, 59, 8173–8180. [Google Scholar] [CrossRef]
- Chen, S.; Deng, X.; Sheng, H.; Rong, Y.; Zheng, Y.; Zhang, Y.; Lin, J. Noncoding RNAs in pediatric brain tumors: Molecular functions and pathological implications. Mol. Ther. Nucleic Acids 2021, 26, 417–431. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).