PD-L1/miR-155 Interplay in Pediatric High-Grade Glioma
Abstract
:1. Introduction
2. Methods
2.1. Subjects
2.2. Tumor Samples Collection
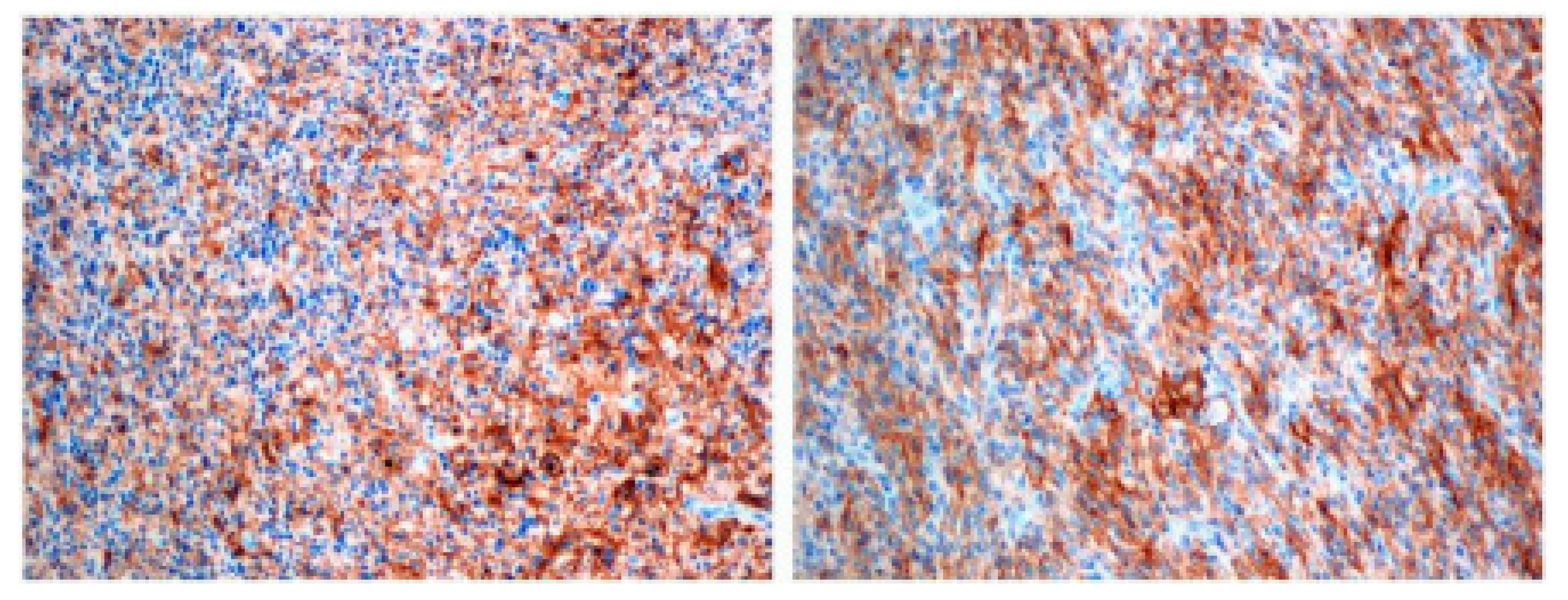
2.3. Evaluation of Immunohistochemistry Findings
2.4. Detection of miR-155 Expression in Tumor Samples
2.5. Statistical Analysis
3. Results
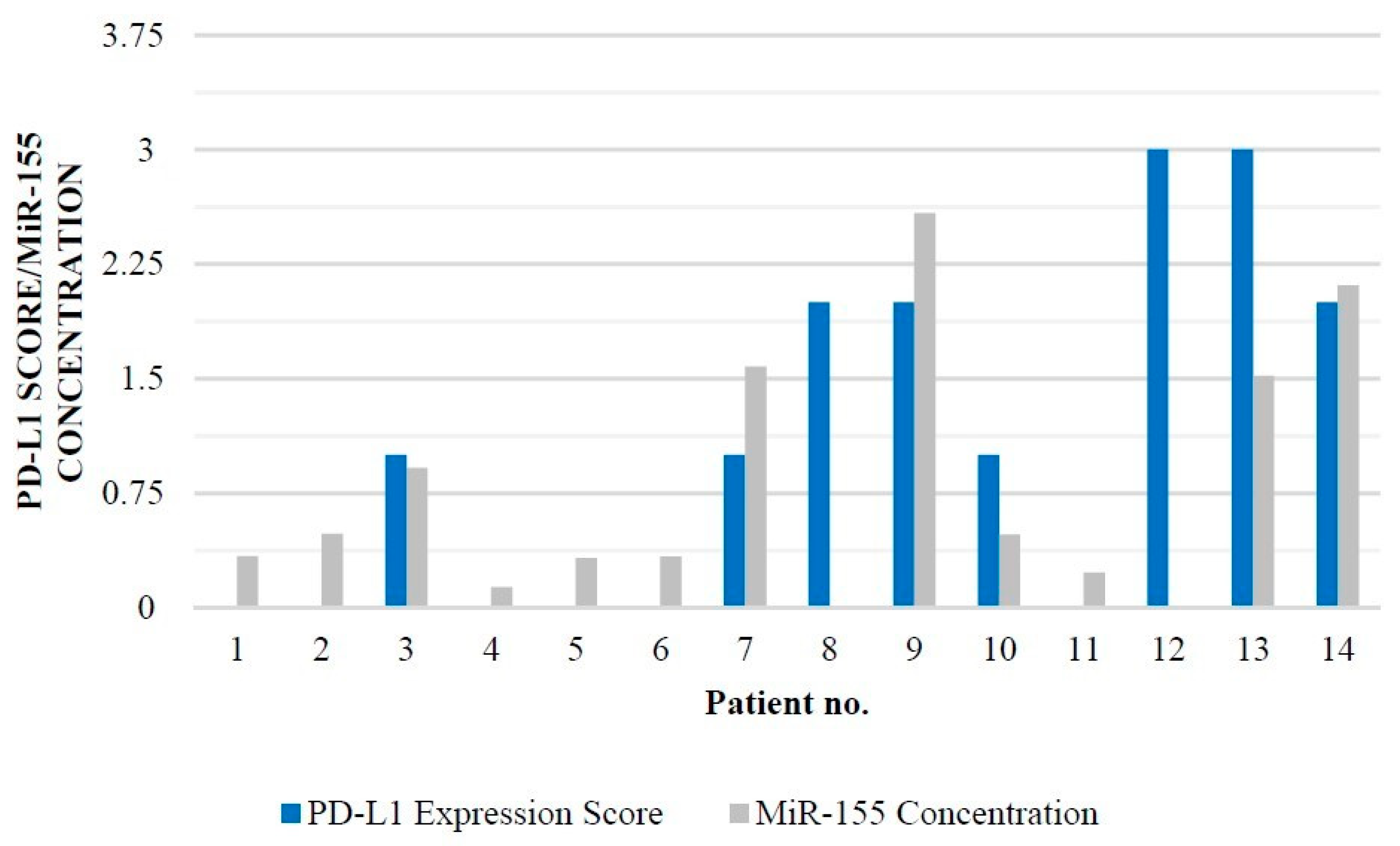
PD-L1 and miR-155 Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Das, K.K.; Kumar, R. Pediatric Glioblastoma. In Glioblastoma [Internet]; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017; Chapter 15. Available online: https://www.ncbi.nlm.nih.gov/books/NBK469983/ (accessed on 23 February 2022). [CrossRef]
- Jones, C.; Karajannis, M.A.; Jones, D.T.W.; Kieran, M.W.; Monje, M.; Baker, S.J.; Becher, O.J.; Cho, Y.-J.; Gupta, N.; Hawkins, C.; et al. Pediatric high-grade glioma: Biologically and clinically in need of new thinking. Neuro Oncol. 2016, 19, 153–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanan, M.I.; Eisenstat, D.D. Management of high-grade gliomas in the pediatric patient: Past, present, and future. Neuro-Oncol. Pract. 2014, 1, 145–157. [Google Scholar] [CrossRef]
- Lam, S.; Lin, Y.; Zinn, P.; Su, J.; Pan, I.W. Patient and treatment factors associated with survival among pediatric glioblastoma patients: A Surveillance, Epidemiology, and End Results study. J. Clin. Neurosci. 2018, 47, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, M.; Grochowski, C.; Litak, J.; Osuchowska, I.; Maciejewski, R.; Kamieniak, P. Recent Trends of microRNA Significance in Pediatric Population Glioblastoma and Current Knowledge of Micro RNA Function in Glioblastoma Multiforme. Int. J. Mol. Sci. 2020, 21, 3046. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbato, S.; Solaini, G.; Fabbri, M. MicroRNAs in Oncogenesis and Tumor Suppression. Int. Rev. Cell Mol. Biol. 2017, 333, 229–268. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, T.; Huang, C.; Hu, T.; Li, J. The pivotal role of microRNA-155 in the control of cancer. J. Cell. Physiol. 2014, 229, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Srivastava, N.; Yadav, A.; Ateeq, B. Targeting AGTR1/NF-κB/CXCR4 axis by miR-155 attenuates oncogenesis in glioblastoma. Neoplasia 2020, 22, 497–510. [Google Scholar] [CrossRef]
- Litak, J.; Grajkowska, W.; Szumiło, J.; Krukow, P.; Maciejewski, R.; Roliński, J.; Grochowski, C. PD-L1 Expression Correlated with p53 Expression in Pediatric Glioblastoma Multiforme. Brain Sci. 2021, 11, 262. [Google Scholar] [CrossRef]
- Litak, J.; Mazurek, M.; Grochowski, C.; Kamieniak, P.; Roliński, J. PD-L1/PD-1 Axis in Glioblastoma Multiforme. Int. J. Mol. Sci. 2019, 20, 5347. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhou, S.; Yang, F.; Qi, X.; Wang, X.; Guan, X.; Shen, C.; Duma, N.; Vera Aguilera, J.; Chintakuntlawar, A.; et al. Treatment-Related Adverse Events of PD-1 and PD-L1 Inhibitors in Clinical Trials: A Systematic Review and Meta-analysis. JAMA Oncol. 2019, 5, 1008–1019. [Google Scholar] [CrossRef]
- Martin, A.M.; Bell, W.R.; Yuan, M.; Harris, L.; Poore, B.; Arnold, A.; Engle, E.L.; Asnaghi, L.; Lim, M.; Raabe, E.H.; et al. PD-L1 Expres- sion in Pediatric Low-Grade Gliomas Is Independent of BRAF V600E Mutational Status. J. Neuropathol. Exp. Neurol. 2019, 79, 74–85. [Google Scholar] [CrossRef]
- Majzner, R.G.; Martinez, D.; Pawel, B.; Santi, M.; Sorensen, P.; Mackall, C.; Maris, J.M. Assessment of PD-L1 expression and tumor associated immune cells in pediatric cancer tissues. J. Clin. Oncol. 2016, 34, 11542. [Google Scholar] [CrossRef]
- Bernstock, J.D.; Vicario, N.; Rong, L.; Valdes, P.A.; Choi, B.D.; Chen, J.A.; DiToro, D.; Osorio, D.S.; Kachurak, K.; Gessler, F.; et al. A novel in situ multiplex immunofluorescence panel for the assessment of tumor immunopathology and response to virotherapy in pediatric glioblastoma reveals a role for checkpoint protein inhibition. Oncoimmunology 2019, 8, e1678921. [Google Scholar] [CrossRef]
- Blumenthal, D.T.; Yalon, M.; Vainer, G.W.; Lossos, A.; Yust, S.; Tzach, L.; Cagnano, E.; Limon, D.; Bokstein, F. Pembrolizumab: First experience with recurrent primary central nervous system (CNS) tumors. J. Neurooncol. 2016, 129, 453–460. [Google Scholar] [CrossRef]
- Lombardi, G.; Barresi, V.; Indraccolo, S.; Simbolo, M.; Fassan, M.; Mandruzzato, S.; Simonelli, M.; Caccese, M.; Pizzi, M.; Fassina, A.; et al. Pembrolizumab Activity in Recurrent High-Grade Gliomas with Partial or Complete Loss of Mismatch Repair Protein Expression: A Monocentric, Observational and Prospective Pilot Study. Cancers 2020, 12, 2283. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018, 37, 33–44. [Google Scholar] [CrossRef]
- Michaille, J.J.; Awad, H.; Fortman, E.C.; Efanov, A.A.; Tili, E. miR-155 expression in antitumor immunity: The higher the better. Genes Chromosomes Cancer 2019, 58, 208–218. [Google Scholar] [CrossRef]
- Bondada, M.S.; Yao, Y.; Nair, V. Multifunctional miR-155 Pathway in Avian Oncogenic Virus-Induced Neoplastic Diseases. Noncoding RNA 2019, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Zou, R.; Zhou, R.; Gong, C.; Wang, Z.; Cai, T.; Tan, C.; Fang, J. miR-155 Regulates Glioma Cells Invasion and Chemosensitivity by p38 Isforms In Vitro. J. Cell. Biochem. 2015, 116, 1213–1221. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Yu, T.; Nie, E.; Hu, Q.; Wu, W.; Zhi, T.; Jiang, K.; Wang, X.; Lu, X.; et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro-Oncol. 2017, 19, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Litak, J.; Grochowski, C.; Litak, J.; Osuchowska, I.; Gosik, K.; Radzikowska, E.; Kamieniak, P.; Rolinski, J. TLR- 4 Signaling vs. Immune Checkpoints, miRNAs Molecules, Cancer Stem Cells, and Wingless-Signaling Interplay in Glioblastoma Multiforme-Future Perspectives. Int. J. Mol. Sci. 2020, 21, 3114. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Chen, Z.; Zhao, H. MicroRNA-155-3p promotes glioma progression and temozolomide resistance by targeting Six1. J. Cell. Mol. Med. 2020, 24, 5363–5374. [Google Scholar] [CrossRef] [Green Version]
- Schliesser, M.G.; Claus, R.; Hielscher, T.; Grimm, C.; Weichenhan, D.; Blaes, J.; Wiestler, B.; Hau, P.; Schramm, J.; Sahm, F.; et al. Prognostic relevance of miRNA-155 methylation in anaplastic glioma. Oncotarget 2016, 7, 82028–82045. [Google Scholar] [CrossRef] [Green Version]
- Banelli, B.; Forlani, A.; Allemanni, G.; Morabito, A.; Pistillo, M.P.; Romani, M. MicroRNA in Glioblastoma: An Overview. Int. J. Genomics 2017, 2017, 7639084. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Tang, F. The potency of lncRNA MALAT1/miR-155/CTLA4 axis in altering Th1/Th2 balance of asthma. Biosci Rep. 2020, 40, BSR20190397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atwa, S.M.; Handoussa, H.; Hosny, K.M.; Odenthal, M.; El Tayebi, H.M. Pivotal role of long non-coding ribonucleic acid-X-inactive specific transcript in regulating immune checkpoint programmed death ligand 1 through a shared pathway between miR-194-5p and miR-155-5p in hepatocellular carcinoma. World J. Hepatol. 2020, 12, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Chen, Z.; Chen, Y.; Wang, X.; Tang, N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019, 8, 7161–7173. [Google Scholar] [CrossRef] [Green Version]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef]
- Zhang, J.; Dang, F.; Ren, J.; Wei, W. Biochemical aspects of PD-L1 regulation in cancer immunotherapy. Trends Biochem. Sci. 2018, 43, 1014–1032. [Google Scholar] [CrossRef] [PubMed]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 2013, 532, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhang, H.; Zhang, B.; Zhang, E.; Wu, Y. Tumor Necrosis Factor-alpha (TNF-α) Enhances miR-155-Mediated Endothelial Senescence by Targeting Sirtuin1 (SIRT1). Med. Sci. Monit. 2019, 25, 8820–8835. [Google Scholar] [CrossRef]
- Mazurek, M.; Litak, J.; Kamieniak, P.; Osuchowska, I.; Maciejewski, R.; Roliński, J.; Grajkowska, W.; Grochowski, C. Micro RNA Molecules as Modulators of Treatment Resistance, Immune Checkpoints Controllers and Sensitive Biomarkers in Glioblastoma Multiforme. Int. J. Mol Sci. 2020, 21, 1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiffer, D.; Annovazzi, L.; Casalone, C.; Corona, C.; Mellai, M. Glioblastoma: Microenvironment and Niche Concept. Cancers 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakrzewska, M.; Gruszka, R.; Stawiski, K.; Fendler, W.; Kordacka, J.; Grajkowska, W.; Daszkiewicz, P.; Liberski, P.P.; Zakrzewski, K. Expression-based decision tree model reveals distinct microRNA expression pattern in pediatric neuronal and mixed neuronal-glial tumors. BMC Cancer 2019, 19, 544. [Google Scholar] [CrossRef]
- Wu, W.; Yu, T.; Wu, Y.; Tian, W.; Zhang, J.; Wang, Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J. Exp. Clin. Cancer Res. 2019, 38, 133. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, M.; Guo, Y.; Zhong, Y.; He, Z.; Xu, Y.; Zou, J. Immune response in glioma’s microenvironment. Innov. Surg. Sci. 2020, 5, 115–125. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, J.; Jiang, J.; He, Y.; Zhang, W.; Mo, X.; Kang, X.; Xu, Q.; Wang, B.; Huang, Y. Remodeling tumor immune microenvironment (TIME) for glioma therapy using multi-targeting liposomal codelivery. J. Immunother. Cancer 2020, 8, e000207. [Google Scholar] [CrossRef]
- Venneti, S.; Santi, M.; Felicella, M.M.; Yarilin, D.; Phillips, J.J.; Sullivan, L.M.; Martinez, D.; Perry, A.; Lewis, P.W.; Thompson, C.B.; et al. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014, 128, 743–753. [Google Scholar] [CrossRef] [Green Version]

| n (%) | |
|---|---|
| Male vs. Female | 10 vs. 4 (71% vs. 29%) |
| Mean age 9.64 years old (SD = 6.68) | |
| Surgery | 14 (100%) |
| GTR | 10 (71%) |
| PR | 4 (29%) |
| Adjuvant treatment | 14 (100%) |
| n (%) | |
|---|---|
| Single lesion | 14 (100%) |
| Supratentorial | 11 (78%) |
| Infratentorial | 3 (22%) |
| Right | 3 (22%) |
| Left | 9 (64%) |
| Midline | 2 (14%) |
| Age | Sex | Tumor | PD-L1 Expression Score | MiR-155 |
|---|---|---|---|---|
| Localization | Amol/Mikrol | |||
| 14 | Male | Left temporal lobe | 0 | 0.337 |
| 11 | Male | Left frontal lobe | 0 | 0.483 |
| 13 | Female | Left temporal lobe | 1 | 0.916 |
| 12 | Female | Left occipital lobe | 0 | 0.133 |
| 6 | Male | Brainstem | 0 | 0.324 |
| 3 | Female | Left temporal | 0 | 0.333 |
| 10 | Male | Right parietal | 1 | 1.579 |
| 11 | Male | Right temporal | 2 | 1.609 |
| 13 | Male | Right temporal | 2 | 2.584 |
| 5 | Male | Left frontal | 1 | 0.479 |
| 12 | Male | Left cerebellum | 0 | 0.228 |
| 11 | Male | Left temporal | 3 | 1.609 |
| 6 | Female | Left frontal | 3 | 1.518 |
| 8 | Male | Brainstem | 2 | 2.112 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Litak, J.; Grajkowska, W.; Bogucki, J.; Kowalczyk, P.; Petniak, A.; Podkowiński, A.; Szumiło, J.; Kocki, J.; Roliński, J.; Rahnama-Hezavah, M.; et al. PD-L1/miR-155 Interplay in Pediatric High-Grade Glioma. Brain Sci. 2022, 12, 324. https://doi.org/10.3390/brainsci12030324
Litak J, Grajkowska W, Bogucki J, Kowalczyk P, Petniak A, Podkowiński A, Szumiło J, Kocki J, Roliński J, Rahnama-Hezavah M, et al. PD-L1/miR-155 Interplay in Pediatric High-Grade Glioma. Brain Sciences. 2022; 12(3):324. https://doi.org/10.3390/brainsci12030324
Chicago/Turabian StyleLitak, Jakub, Wiesława Grajkowska, Jacek Bogucki, Paweł Kowalczyk, Alicja Petniak, Arkadiusz Podkowiński, Justyna Szumiło, Janusz Kocki, Jacek Roliński, Mansur Rahnama-Hezavah, and et al. 2022. "PD-L1/miR-155 Interplay in Pediatric High-Grade Glioma" Brain Sciences 12, no. 3: 324. https://doi.org/10.3390/brainsci12030324
APA StyleLitak, J., Grajkowska, W., Bogucki, J., Kowalczyk, P., Petniak, A., Podkowiński, A., Szumiło, J., Kocki, J., Roliński, J., Rahnama-Hezavah, M., Roszkowski, M., & Grochowski, C. (2022). PD-L1/miR-155 Interplay in Pediatric High-Grade Glioma. Brain Sciences, 12(3), 324. https://doi.org/10.3390/brainsci12030324






