A Macaque Brain Extraction Model Based on U-Net Combined with Residual Structure
Abstract
:1. Introduction
2. Materials and Methods
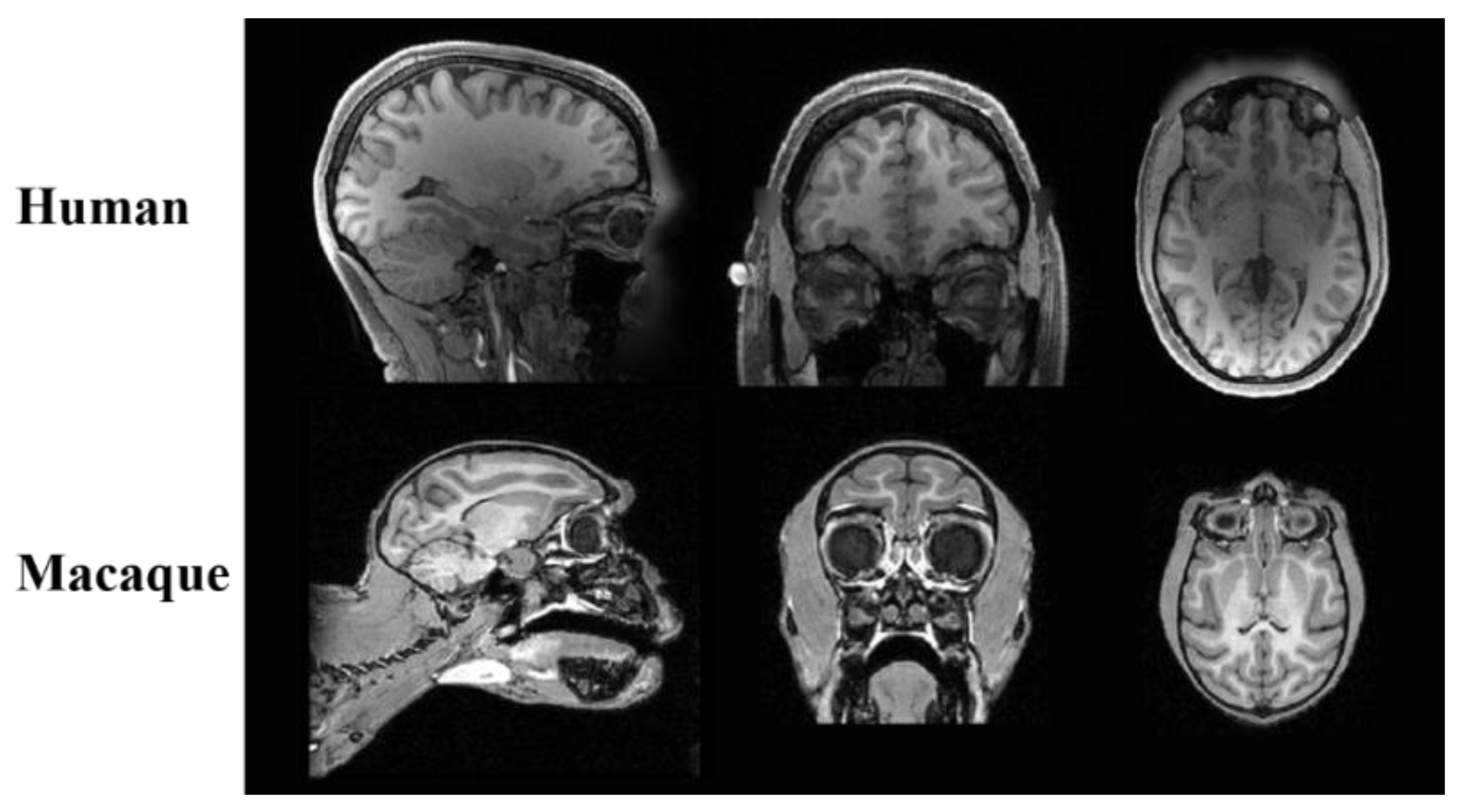
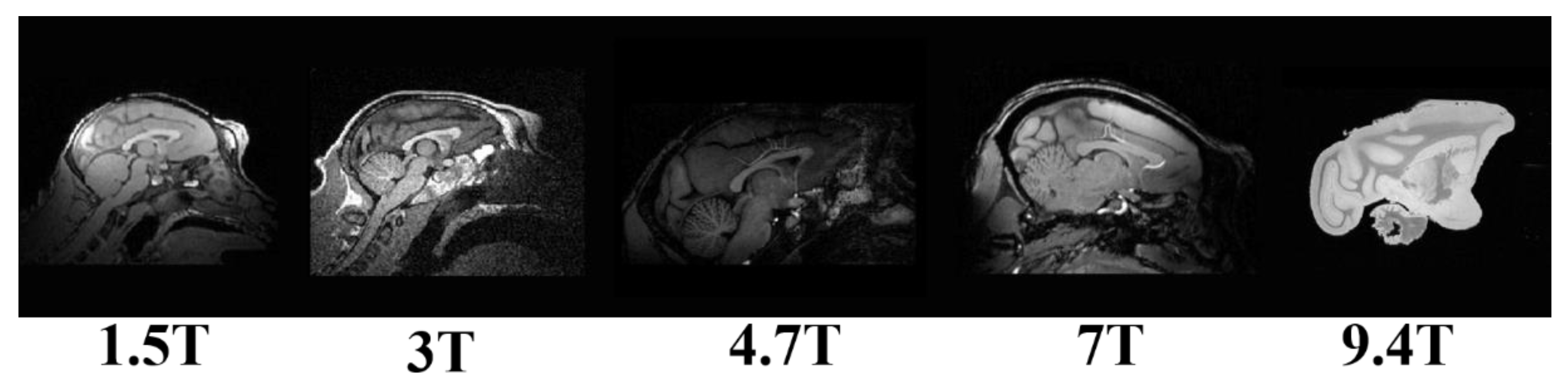
2.1. Dataset
2.2. Resampling
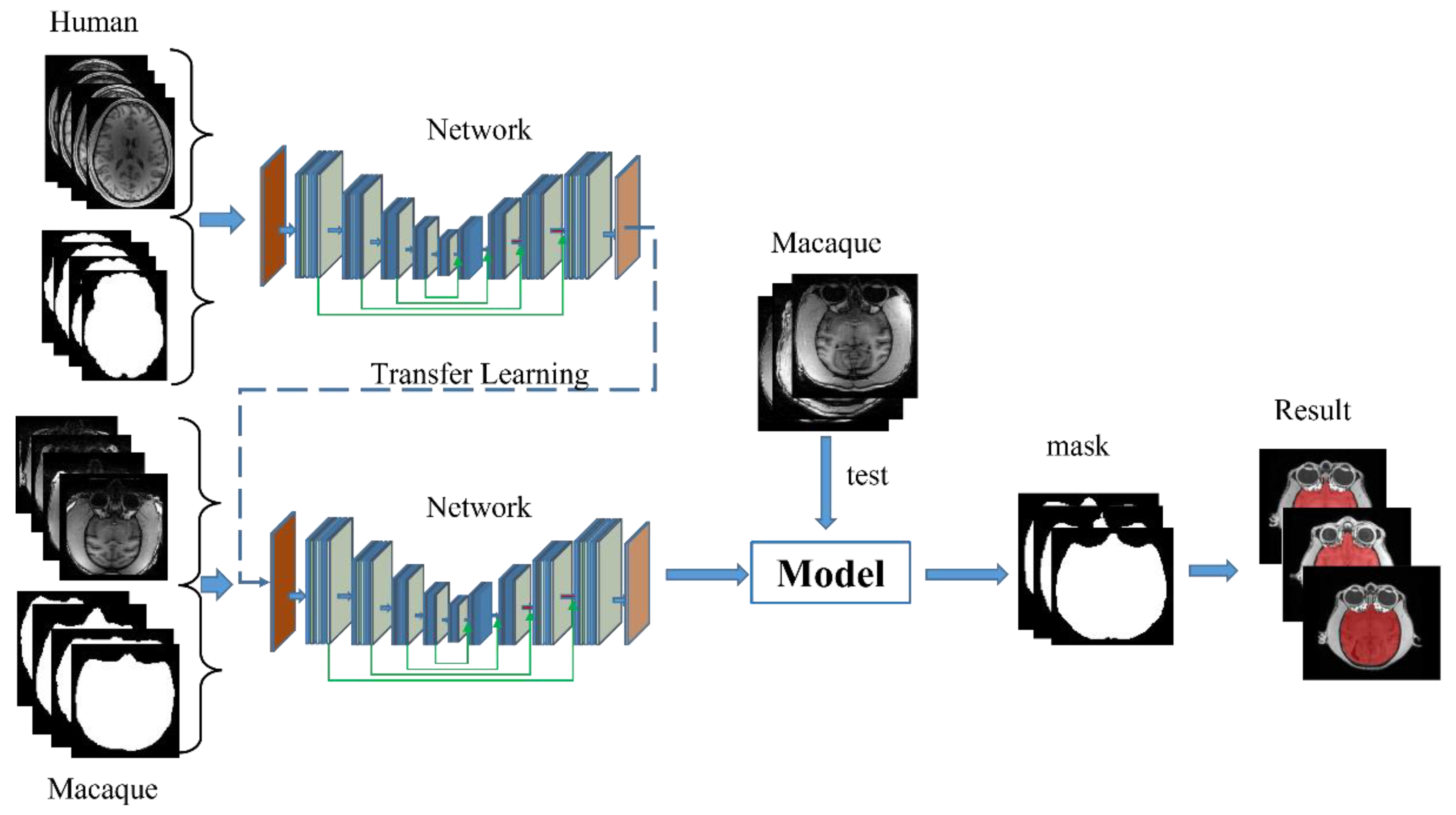
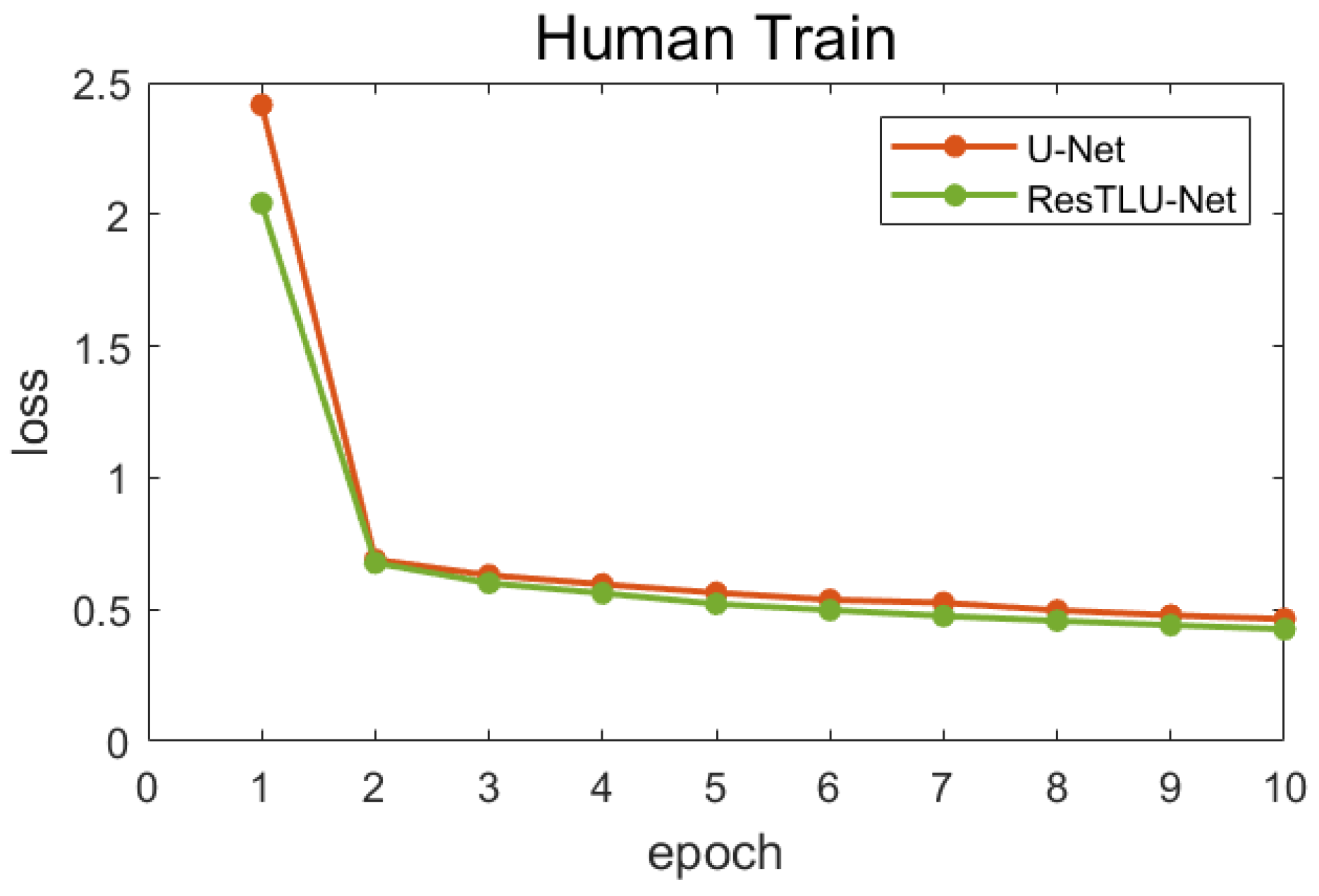
- Dataset I is trained for 10 epochs to generate a pretraining model. After that, the best performing pretraining model is selected through the validation dataset for subsequent training of macaque images.
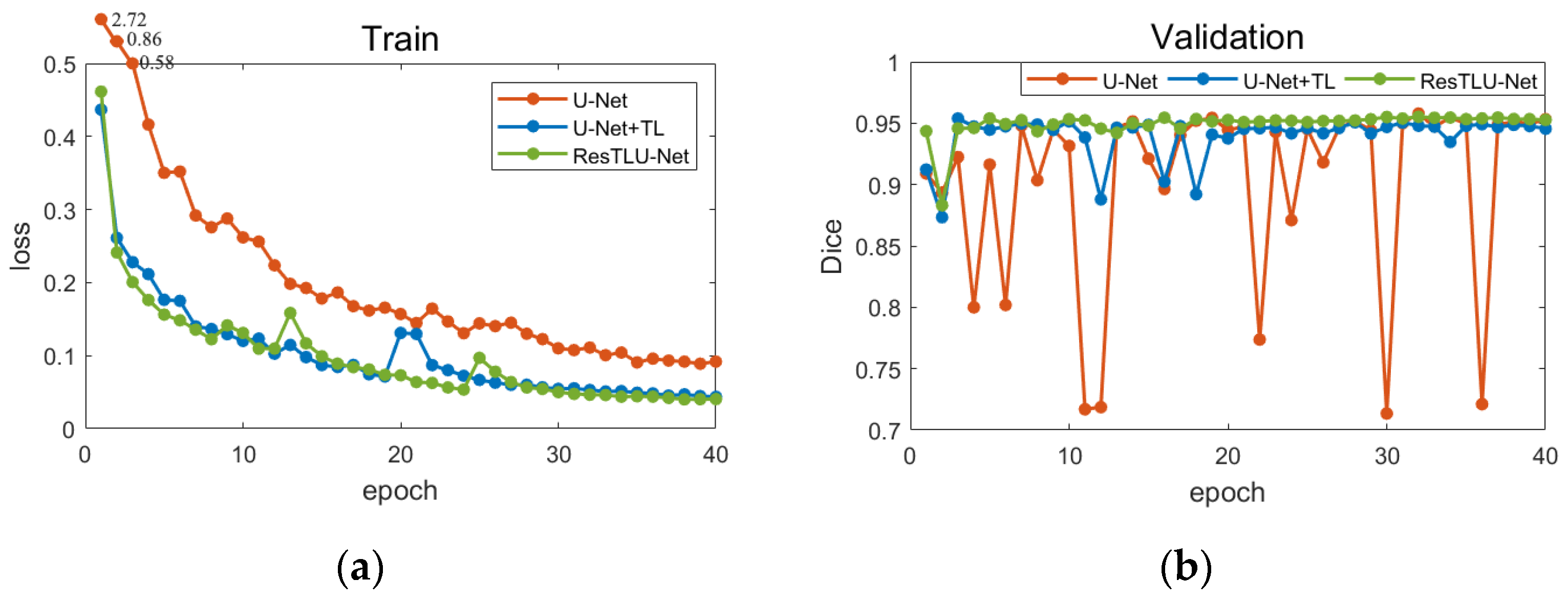
- The model will be trained on Dataset II for 40 epochs, and the best-performing model from the validation set is selected as the final model for testing. Because there were apparent differences between the MRI datasets collected for macaques and humans, the weights of all layers were updated on the pre-trained model.
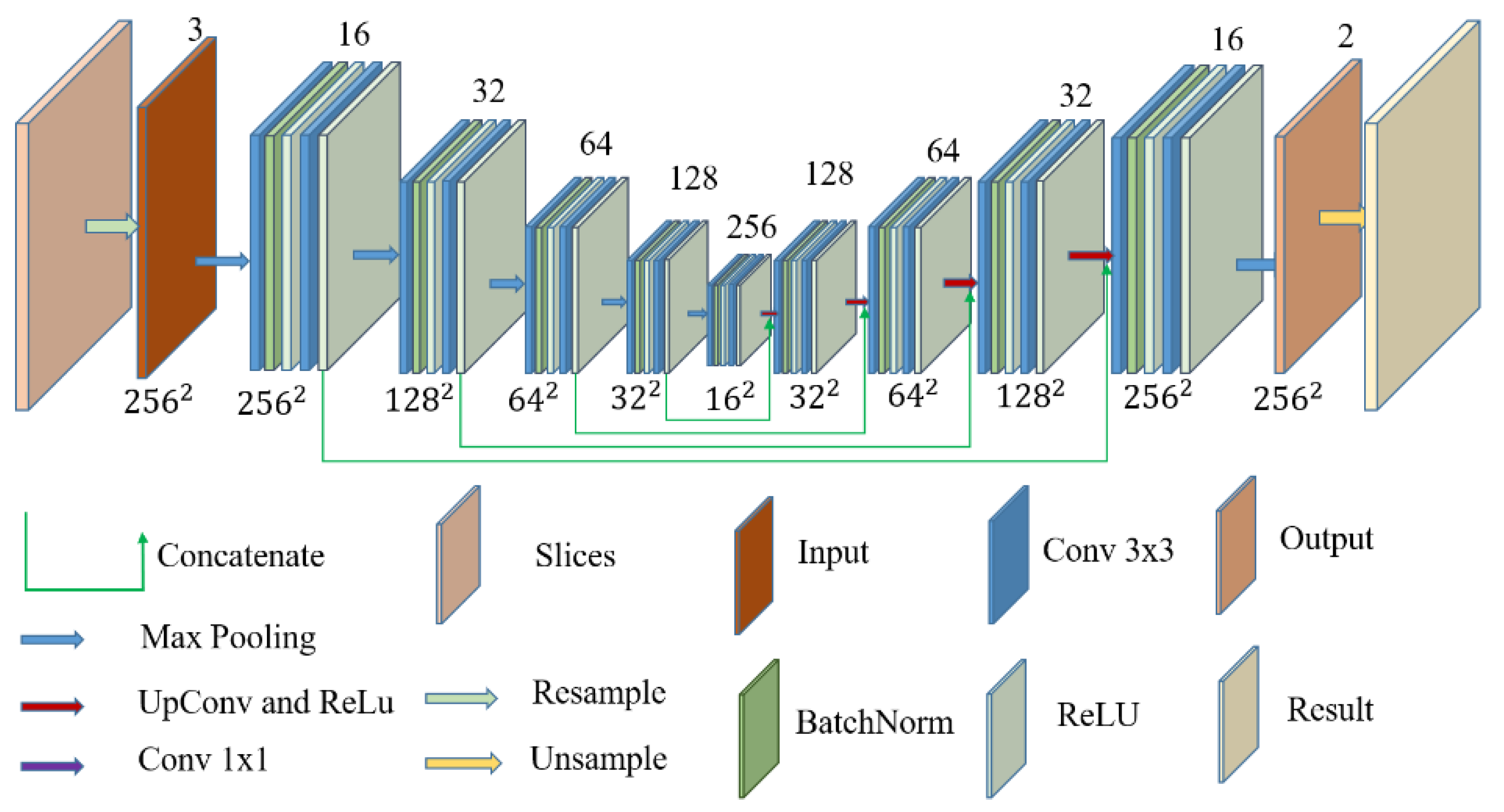
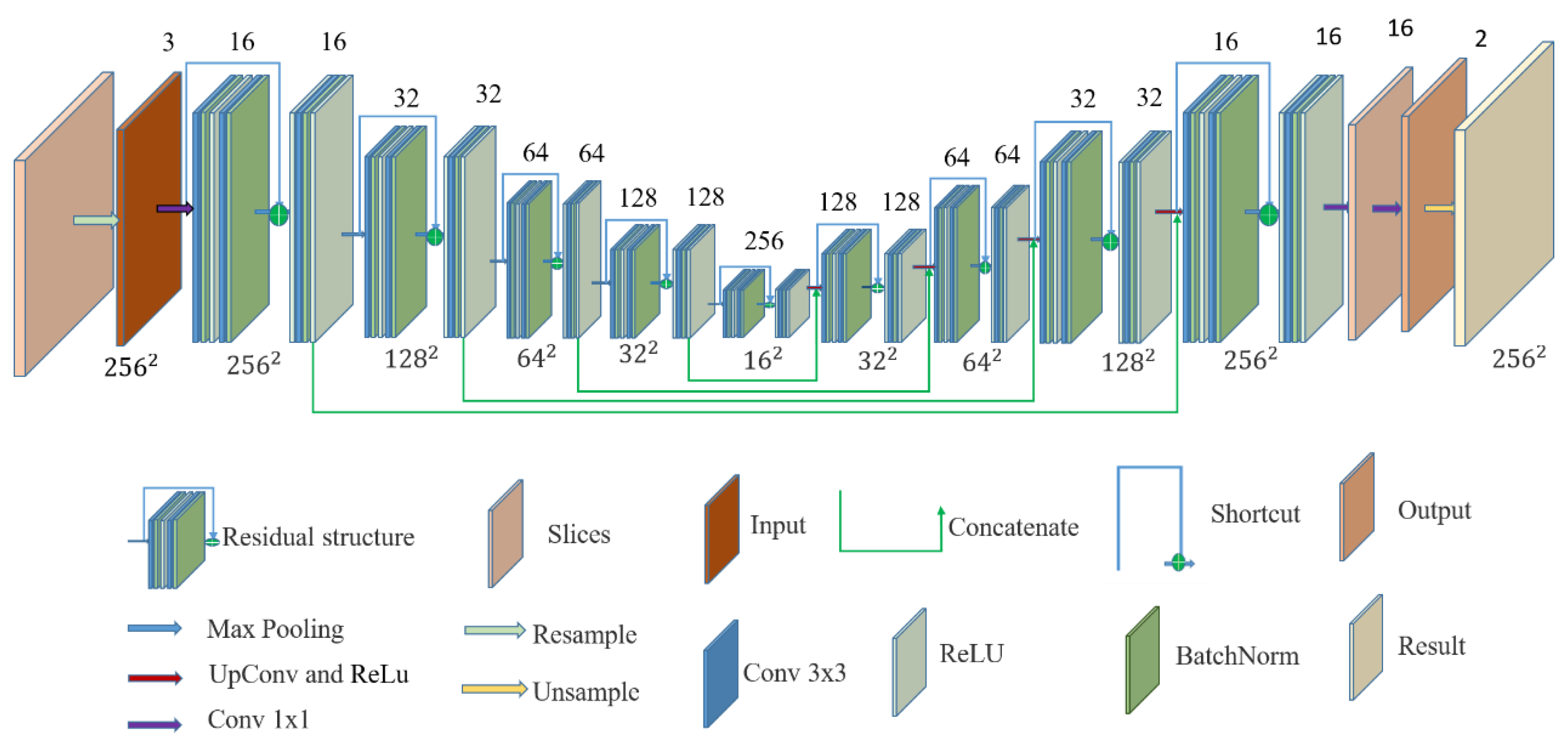
2.3. U-Net Structure
2.4. ResTLU-Net
2.5. Experiment and Evaluation
3. Results
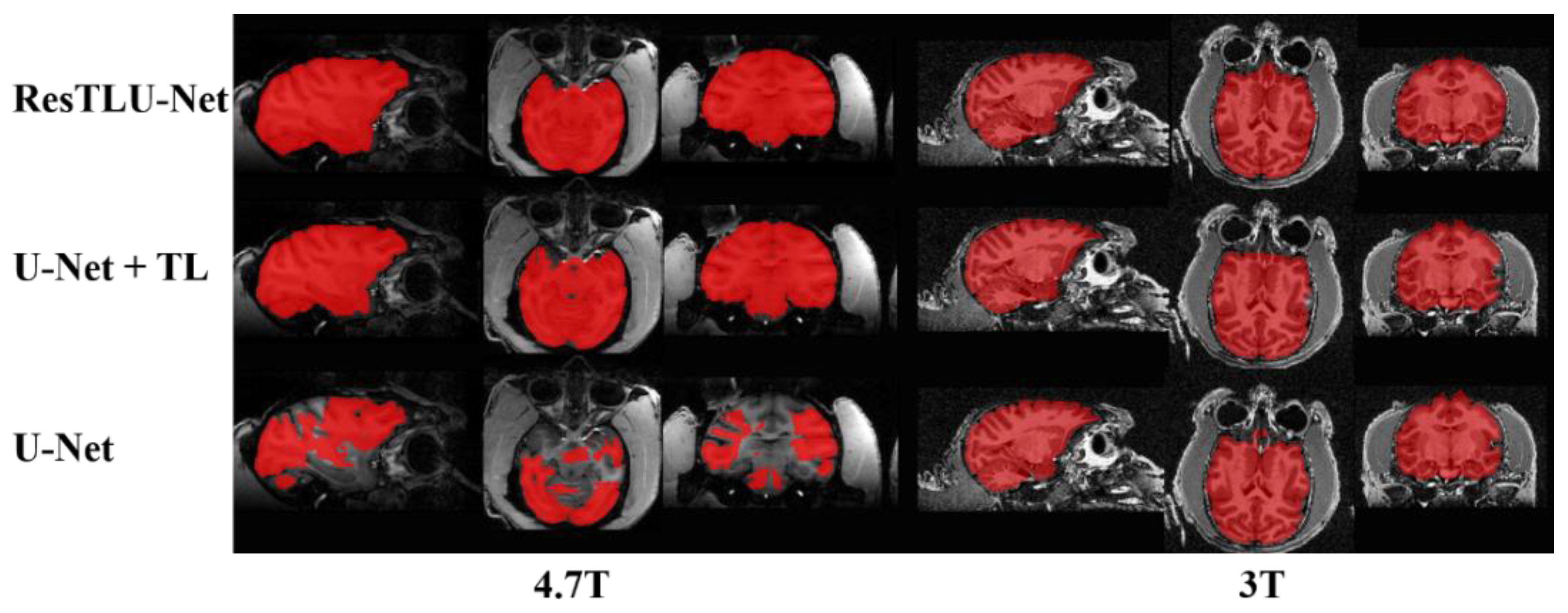
3.1. Comparsion of U-Net and ResTLU-Net
3.2. Verification of the Validity
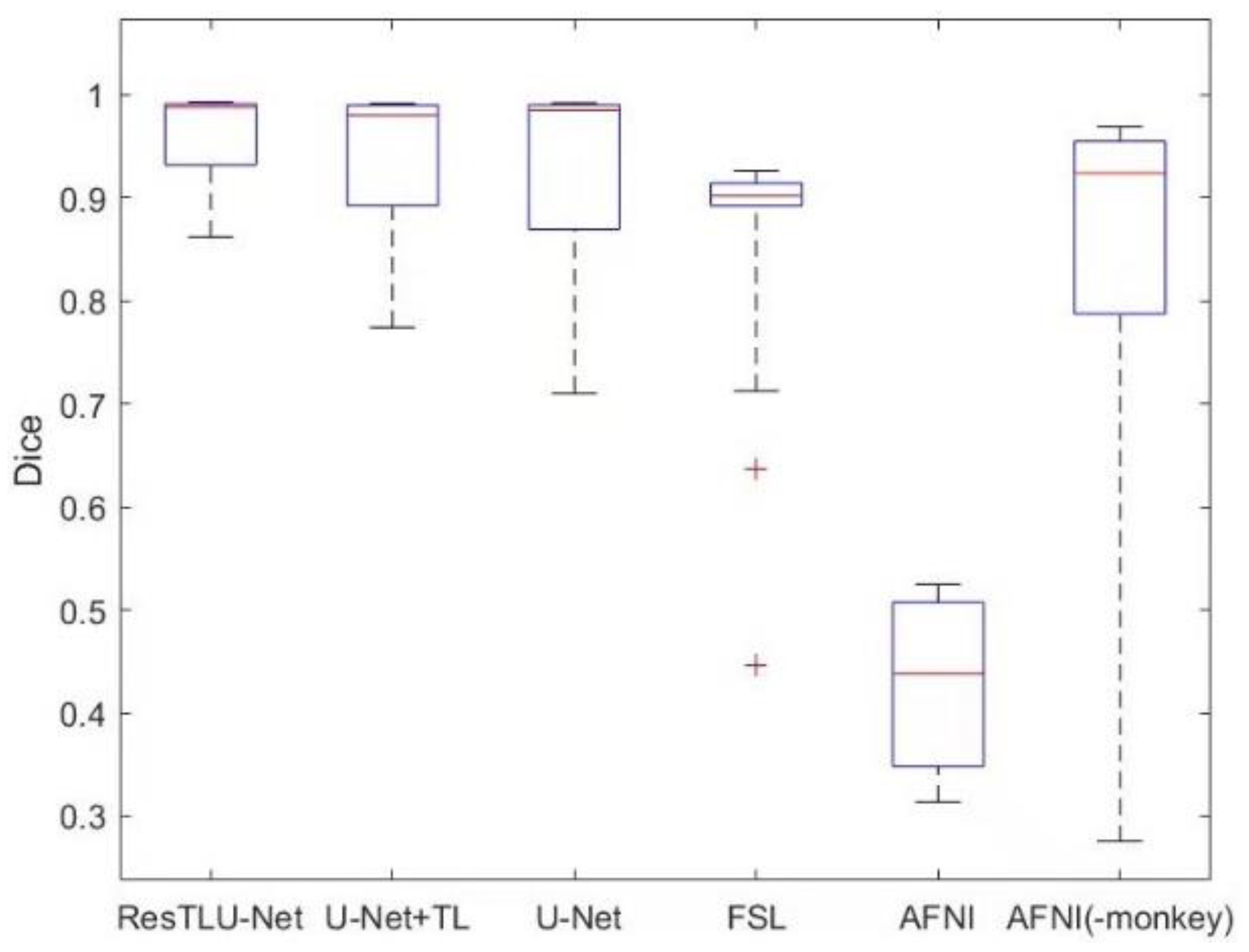
3.3. Comparison of ResTLU-Net and Other Brain Tissue Extraction Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lepage, C.; Wagstyl, K.; Jung, B.; Seidlitz, J.; Sponheim, C.; Ungerleider, L.; Wang, X.; Evans, A.C.; Messinger, A. CIVET-Macaque: An automated pipeline for MRI-based cortical surface generation and cortical thickness in macaques. NeuroImage 2021, 227, 117622. [Google Scholar] [CrossRef] [PubMed]
- Oler, J.A.; Fox, A.S.; Shelton, S.E.; Rogers, J.; Dyer, T.D.; Davidson, R.J.; Shelledy, W.; Oakes, T.R.; Blangero, J.; Kalin, N.H. Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature 2010, 466, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.S.; Oler, J.A.; Shackman, A.J.; Shelton, S.E.; Raveendran, M.; McKay, D.R.; Converse, A.K.; Alexander, A.; Davidson, R.J.; Blangero, J.; et al. Intergenerational neural mediators of early-life anxious temperament. Proc. Natl. Acad. Sci. USA 2015, 112, 9118–9122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donahue, C.J.; Glasser, M.F.; Preuss, T.M.; Rilling, J.K.; Van Essen, D.C. Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc. Natl. Acad. Sci. USA 2018, 115, E5183–E5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Nenning, K.-H.; Schwartz, E.; Hong, S.-J.; Vogelstein, J.T.; Fair, D.A.; Schroeder, C.E.; Margulies, D.S.; Smallwood, J.; Milham, M.P.; et al. Cross-species Functional Alignment Reveals Evolutionary Hierarchy Within the Connectome. NeuroImage 2019, 223, 117346. [Google Scholar] [CrossRef] [PubMed]
- Poo, M.-M. Whereto the mega brain projects? Natl. Sci. Rev. 2014, 1, 12–14. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wang, F.; Wu, Z.; Wang, L.; Zhang, C.; Shen, D.; Li, G. Mapping hemispheric asymmetries of the macaque cerebral cortex during early brain development. Hum. Brain Mapp. 2020, 41, 95–106. [Google Scholar] [CrossRef]
- Acosta-Cabronero, J.; Williams, G.B.; Pereira, J.M.; Pengas, G.; Nestor, P.J. The impact of skull-stripping and radio-frequency bias correction on grey-matter segmentation for voxel-based morphometry. NeuroImage 2008, 39, 1654–1665. [Google Scholar] [CrossRef]
- Glasser, M.F.; Sotiropoulos, S.N.; Wilson, J.A.; Coalson, T.S.; Fischl, B.; Andersson, J.L.; Xu, J.; Jbabdi, S.; Webster, M.; Polimeni, J.R.; et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 2013, 80, 105–124. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Sturgeon, D.; Ramirez, J.S.B.; Froudist-Walsh, S.; Margulies, D.S.; Schroeder, C.E.; Fair, D.A.; Milham, M.P. Interindividual Variability of Functional Connectivity in Awake and Anesthetized Rhesus Macaque Monkeys. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2019, 4, 543–553. [Google Scholar] [CrossRef]
- Tasserie, J.; Grigis, A.; Uhrig, L.; Dupont, M.; Amadon, A.; Jarraya, B. Pypreclin: An automatic pipeline for macaque functional MRI preprocessing. NeuroImage 2020, 207, 116353. [Google Scholar] [CrossRef] [PubMed]
- Segonne, F.; Dale, A.M.; Busa, E.; Glessner, M.; Salat, D.; Hahn, H.K.; Fischl, B. A hybrid approach to the skull stripping problem in MRI. NeuroImage 2004, 22, 1060–1075. [Google Scholar] [CrossRef] [PubMed]
- Puccio, B.; Pooley, J.P.; Pellman, J.S.; Taverna, E.C.; Craddock, R.C. The preprocessed connectomes project repository of manually corrected skull-stripped T1-weighted anatomical MRI data. Gigascience 2016, 5, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidlitz, J.; Sponheim, C.; Glen, D.; Ye, F.Q.; Saleem, K.S.; Leopold, D.A.; Ungerleider, L.; Messinger, A. A population MRI brain template and analysis tools for the macaque. NeuroImage 2018, 170, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002, 17, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. Fsl. NeuroImage 2012, 62, 782–790. [Google Scholar] [CrossRef] [Green Version]
- Cox, R.W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef]
- Vovk, A.; Cox, R.W.; Stare, J.; Suput, D.; Saad, Z.S. Segmentation priors from local image properties: Without using bias field correction, location-based templates, or registration. NeuroImage 2011, 55, 142–152. [Google Scholar] [CrossRef] [Green Version]
- Selvaganesan, K.; Whitehead, E.; DeAlwis, P.M.; Schindler, M.K.; Inati, S.; Saad, Z.S.; Ohayon, J.E.; Cortese, I.C.M.; Smith, B.; Steven, J.; et al. Robust, atlas-free, automatic segmentation of brain MRI in health and disease. Heliyon 2019, 5, e01226. [Google Scholar] [CrossRef] [Green Version]
- Desikan, R.S.; Segonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 2006, 31, 968–980. [Google Scholar] [CrossRef]
- Zhong, T.; Zhao, F.; Pei, Y.; Ning, Z.; Liao, L.; Wu, Z.; Niu, Y.; Wang, L.; Shen, D.; Zhang, Y.; et al. DIKA-Nets: Domain-invariant knowledge-guided attention networks for brain skull stripping of early developing macaques. NeuroImage 2021, 227, 117649. [Google Scholar] [CrossRef] [PubMed]
- Croxson, P.L.; Forkel, S.J.; Cerliani, L.; Thiebaut de Schotten, M. Structural Variability Across the Primate Brain: A Cross-Species Comparison. Cereb. Cortex 2018, 28, 3829–3841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopkins, W.D.; Meguerditchian, A.; Coulon, O.; Bogart, S.; Mangin, J.F.; Sherwood, C.C.; Grabowski, M.W.; Bennett, A.J.; Pierre, P.J.; Fears, S.; et al. Evolution of the central sulcus morphology in primates. Brain Behav. Evol. 2014, 84, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Henschel, L.; Conjeti, S.; Estrada, S.; Diers, K.; Fischl, B.; Reuter, M. FastSurfer—A fast and accurate deep learning based neuroimaging pipeline. NeuroImage 2020, 219, 117012. [Google Scholar] [CrossRef]
- Isensee, F.; Jger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. Automated design of deep learning methods for biomedical image segmentation. arXiv 2019, arXiv:1904.08128. [Google Scholar]
- Roy, S.; Butman, J.A.; Reich, D.S.; Calabresi, P.A.; Pham, D.L. Multiple Sclerosis Lesion Segmentation from Brain MRI via Fully Convolutional Neural Networks. arXiv 2018, arXiv:1803.09172. [Google Scholar]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X. An iterative convolutional neural network algorithm improves electron microscopy image segmentation. arXiv 2015, arXiv:1506.05849. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Wachinger, C.; Reuter, M.; Klein, T. DeepNAT: Deep convolutional neural network for segmenting neuroanatomy. NeuroImage 2018, 170, 434–445. [Google Scholar] [CrossRef]
- Chen, R.T.Q.; Rubanova, Y.; Bettencourt, J.; Duvenaud, D. Neural ordinary differential equations. arXiv 2018, arXiv:1806.07366. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICAI 2015; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Chapter 28; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Springer: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Zhao, G.; Liu, F.; Oler, J.A.; Meyerand, M.E.; Kalin, N.H.; Birn, R.M. Bayesian convolutional neural network based MRI brain extraction on nonhuman primates. NeuroImage 2018, 175, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Autio, J.A.; Glasser, M.F.; Ose, T.; Donahue, C.J.; Bastiani, M.; Ohno, M.; Kawabata, Y.; Urushibata, Y.; Murata, K.; Nishigori, K.; et al. Towards HCP-Style macaque connectomes: 24-Channel 3T multi-array coil, MRI sequences and preprocessing. NeuroImage 2020, 215, 116800. [Google Scholar] [CrossRef] [PubMed]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Advances in Neural Information Processing Systems 27, Proceedings of the Annual Conference on Neural Information Processing Systems 2014, Montreal, QC, Canada, 8–13 December 2014; Curran Associates: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- Ghafoorian, M.; Mehrtash, A.; Kapur, T.; Karssemeijer, N.; Marchiori, E.; Pesteie, M.; Guttmann, C.R.G.; de Leeuw, F.-E.; Tempany, C.M.; van Ginneken, B.; et al. Transfer Learning for Domain Adaptation in MRI: Application in Brain Lesion Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2017; Springer: Cham, Switzerland, 2017; pp. 516–524. [Google Scholar]
- Wang, X.; Li, X.H.; Cho, J.W.; Russ, B.E.; Rajamani, N.; Omelchenko, A.; Ai, L.; Korchmaros, A.; Sawiak, S.; Benn, R.A.; et al. U-net model for brain extraction: Trained on humans for transfer to non-human primates. NeuroImage 2021, 235, 118001. [Google Scholar] [CrossRef] [PubMed]
- Van Essen, D.C.; Ugurbil, K.; Auerbach, E.; Barch, D.; Behrens, T.E.; Bucholz, R.; Chang, A.; Chen, L.; Corbetta, M.; Curtiss, S.W.; et al. The Human Connectome Project: A data acquisition perspective. NeuroImage 2012, 62, 2222–2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milham, M.P.; Ai, L.; Koo, B.; Xu, T.; Amiez, C.; Balezeau, F.; Baxter, M.G.; Blezer, E.L.A.; Brochier, T.; Chen, A.; et al. An Open Resource for Non-human Primate Imaging. Neuron 2018, 100, 61–74.E2. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Xu, Z.; Taylor, G.; Studer, C.; Goldstein, T. Visualizing the Loss Landscape of Neural Nets. In Advances in Neural Information Processing Systems 31, Proceedings of the 32nd Annual Conference on Neural Information Processing Systems (NeurIPS 2018), Montreal, QC, Canada, 3–8 December 2018; Curran Associates: Red Hook, NY, USA, 2018; Volume 31. [Google Scholar]
- Emin Orhan, A.; Pitkow, X. Skip Connections Eliminate Singularities. arXiv 2017, arXiv:1701.09175. [Google Scholar]
- Veit, A.; Wilber, M.; Belongie, S. Residual Networks Behave Like Ensembles of Relatively Shallow Networks. In Advances in Neural Information Processing Systems 29, Proceedings of the 30th Annual Conference on Neural Information Processing Systems 2016, Barcelona, Spain, 5–10 December 2016; Curran Associates: Red Hook, NY, USA, 2016; Volume 29. [Google Scholar]
- Kleesiek, J.; Urban, G.; Hubert, A.; Schwarz, D.; Maier-Hein, K.; Bendszus, M.; Biller, A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. NeuroImage 2016, 129, 460–469. [Google Scholar] [CrossRef]



| Model | Dice | TPR | TNR | Jaccard | VOE | FNR | FPR | PPV |
|---|---|---|---|---|---|---|---|---|
| DenseNet | 0.8372 | 0.7812 | 0.9996 | 0.7737 | 0.2263 | 0.2184 | 0.0079 | 0.9914 |
| LinkNet | 0.8025 | 0.7415 | 0.9998 | 0.7383 | 0.2617 | 0.2583 | 0.0035 | 0.9962 |
| U-Net | 0.9283 | 0.8826 | 0.9998 | 0.8785 | 0.1215 | 0.1172 | 0.0043 | 0.9955 |
| U-Net + TL | 0.9413 | 0.8991 | 0.9998 | 0.8957 | 0.1043 | 0.1006 | 0.0037 | 0.9960 |
| ResTLU-Net | 0.9581 | 0.9281 | 0.9996 | 0.9232 | 0.0768 | 0.0716 | 0.0052 | 0.9945 |
| Model | CV1 | CV2 | CV3 | CV4 | CV5 |
|---|---|---|---|---|---|
| U-Net | 0.9559 ± 0.0158 | 0.6915 ± 0.0949 | 0.9427 ± 0.0097 | 0.9439 ± 0.0032 | 0.9580 ± 0.0167 |
| U-Net + TL | 0.9485 ± 0.0270 | 0.8409 ± 0.0741 | 0.9576 ± 0.0112 | 0.9419 ± 0.0175 | 0.9558 ± 0.0052 |
| ResTLU-Net | 0.9548 ± 0.0020 | 0.8742 ± 0.0670 | 0.9633 ± 0.0146 | 0.9410 ± 0.0031 | 0.9667 ± 0.0024 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Fei, H.; Abdu Nasher, S.N.; Xia, X.; Li, H. A Macaque Brain Extraction Model Based on U-Net Combined with Residual Structure. Brain Sci. 2022, 12, 260. https://doi.org/10.3390/brainsci12020260
Wang Q, Fei H, Abdu Nasher SN, Xia X, Li H. A Macaque Brain Extraction Model Based on U-Net Combined with Residual Structure. Brain Sciences. 2022; 12(2):260. https://doi.org/10.3390/brainsci12020260
Chicago/Turabian StyleWang, Qianshan, Hong Fei, Saddam Naji Abdu Nasher, Xiaoluan Xia, and Haifang Li. 2022. "A Macaque Brain Extraction Model Based on U-Net Combined with Residual Structure" Brain Sciences 12, no. 2: 260. https://doi.org/10.3390/brainsci12020260
APA StyleWang, Q., Fei, H., Abdu Nasher, S. N., Xia, X., & Li, H. (2022). A Macaque Brain Extraction Model Based on U-Net Combined with Residual Structure. Brain Sciences, 12(2), 260. https://doi.org/10.3390/brainsci12020260







