cVEP Training Data Validation—Towards Optimal Training Set Composition from Multi-Day Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Stimulus Presentation
2.3. Recordings
2.4. Hardware and Software for Data Analysis
2.5. CCA-Based Spatial Filter Design and Template Generation
2.6. Classification
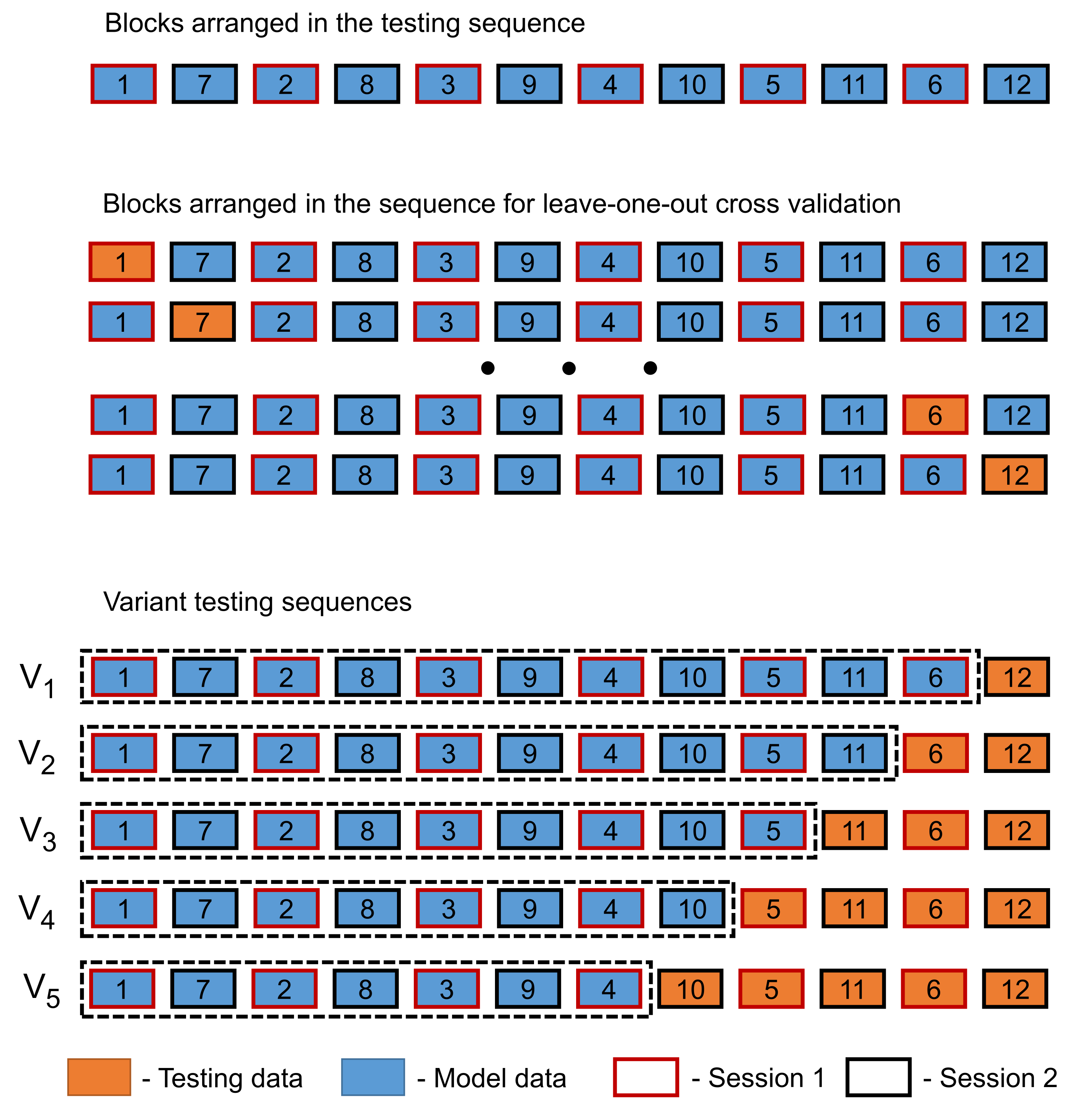
2.7. Procedure
- Variant 1 (), where block was used for testing and blocks – in order were used for model building.
- Variant 2 (), where blocks and were used for testing and blocks – in order without were used for model building.
- Variant 3 (), where blocks and were used for testing and blocks – in order without were used for model building.
- Variant 4 (), where blocks and were used for testing and blocks – in order without and were used for model building.
- Variant 5 (), where blocks and were used for testing and blocks – in order without and were used for model building.
- Step 1—model
- Step 2—model
- Step 3—model
- Step 4—model
- Step 5—model
- Step 6—model
- Step 7—model
- Step 8—model
- Step 9—model
- Step 10—model
- Step 11—model
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Brunner, C.; Birbaumer, N.; Blankertz, B.; Guger, C.; Kübler, A.; Mattia, D.; Millán, J.d.R.; Miralles, F.; Nijholt, A.; Opisso, E.; et al. BNCI Horizon 2020: Towards a roadmap for the BCI community. Brain-Comput. Interfaces 2015, 2, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Abiri, R.; Borhani, S.; Sellers, E.W.; Jiang, Y.; Zhao, X. A comprehensive review of EEG-based brain–computer interface paradigms. J. Neural Eng. 2019, 16, 011001. [Google Scholar] [CrossRef] [PubMed]
- Stegman, P.; Crawford, C.S.; Andujar, M.; Nijholt, A.; Gilbert, J.E. Brain–computer interface software: A review and discussion. IEEE Trans. Hum.-Mach. Syst. 2020, 50, 101–115. [Google Scholar] [CrossRef]
- Rashid, M.; Sulaiman, N.; PP Abdul Majeed, A.; Musa, R.M.; Bari, B.S.; Khatun, S. Current status, challenges, and possible solutions of EEG-based brain-computer interface: A comprehensive review. Front. Neurorobot. 2020, 14, 25. [Google Scholar] [CrossRef]
- Rezeika, A.; Benda, M.; Stawicki, P.; Gembler, F.; Saboor, A.; Volosyak, I. Brain-Computer Interface Spellers: A Review. Brain Sci. 2018, 8, 57. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; He, D.; Li, C.; Qi, S. Brain–Computer Interface Speller Based on Steady-State Visual Evoked Potential: A Review Focusing on the Stimulus Paradigm and Performance. Brain Sci. 2021, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cagigal, V.; Thielen, J.; Santamaría-Vázquez, E.; Pérez-Velasco, S.; Desain, P.; Hornero, R. Brain–computer interfaces based on code-modulated visual evoked potentials (c-VEP): A literature review. J. Neural Eng. 2021, 18, 061002. [Google Scholar] [CrossRef] [PubMed]
- Volosyak, I.; Rezeika, A.; Benda, M.; Gembler, F.; Stawicki, P. Towards solving of the Illiteracy phenomenon for VEP-based brain-computer interfaces. Biomed. Phys. Eng. Express 2020, 6, 035034. [Google Scholar] [CrossRef]
- Spüler, M.; Rosenstiel, W.; Bogdan, M. Online Adaptation of a C-VEP Brain-Computer Interface(BCI) Based on Error-Related Potentials and Unsupervised Learning. PLoS ONE 2012, 7, e51077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakanishi, M.; Wang, Y.; Jung, T.P. Session-to-Session Transfer in Detecting Steady-State Visual Evoked Potentials with Individual Training Data. In Foundations of Augmented Cognition: Neuroergonomics and Operational Neuroscience; Schmorrow, D.D., Fidopiastis, C.M., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 253–260. [Google Scholar]
- Nakanishi, M.; Wang, Y.; Chen, X.; Wang, Y.T.; Gao, X.; Jung, T.P. Enhancing Detection of SSVEPs for a High-Speed Brain Speller Using Task-Related Component Analysis. IEEE Trans. Biomed. Eng. 2017, 65, 104–112. [Google Scholar] [CrossRef]
- Yuan, P.; Chen, X.; Wang, Y.; Gao, X.; Gao, S. Enhancing performances of SSVEP-based brain–computer interfaces via exploiting inter-subject information. J. Neural Eng. 2015, 12, 046006. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Wang, Z.; Wang, B.; Lao, K.F.; Rosa, A.; Xu, P.; Jung, T.P.; Chen, C.P.; Wan, F. Inter-and Intra-Subject Transfer Reduces Calibration Effort for High-Speed SSVEP-Based BCIs. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 2123–2135. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, Y.; Li, Y.; Chen, S.; Zhou, W. Inter-and intra-subject template-based multivariate synchronization index using an adaptive threshold for SSVEP-based BCIs. Front. Neurosci. 2020, 14, 717. [Google Scholar] [CrossRef] [PubMed]
- Gembler, F.; Stawicki, P.; Rezeika, A.; Benda, M.; Volosyak, I. Exploring Session-to-Session Transfer for Brain-Computer Interfaces based on Code-Modulated Visual Evoked Potentials. In Proceedings of the 2020 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Toronto, ON, Canada, 11–14 October 2020; pp. 1505–1510. [Google Scholar]
- Saha, S.; Ahmed, K.I.U.; Mostafa, R.; Hadjileontiadis, L.; Khandoker, A. Evidence of variabilities in EEG dynamics during motor imagery-based multiclass brain–computer interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 26, 371–382. [Google Scholar] [CrossRef]
- Saha, S.; Baumert, M. Intra-and inter-subject variability in EEG-based sensorimotor brain computer interface: A review. Front. Comput. Neurosci. 2020, 13, 87. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Tao, Q.; Peng, W.; Zhang, T.; Si, Y.; Zhang, Y.; Yi, C.; Biswal, B.; Yao, D.; Xu, P. Inter-subject P300 variability relates to the efficiency of brain networks reconfigured from resting-to task-state: Evidence from a simultaneous event-related EEG-fMRI study. NeuroImage 2020, 205, 116285. [Google Scholar] [CrossRef]
- Wei, C.S.; Nakanishi, M.; Chiang, K.J.; Jung, T.P. Exploring human variability in steady-state visual evoked potentials. In Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Miyazaki, Japan, 7–10 October 2018; pp. 474–479. [Google Scholar]
- Tanaka, H. Group task-related component analysis (gTRCA): A multivariate method for inter-trial reproducibility and inter-subject similarity maximization for EEG data analysis. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef]
- Zerafa, R.; Camilleri, T.; Falzon, O.; Camilleri, K.P. To Train or Not to Train? A Survey on Training of Feature Extraction Methods for SSVEP-Based BCIs. J. Neural Eng. 2018, 15, 051001. [Google Scholar] [CrossRef]
- Stawicki, P.; Rezeika, A.; Volosyak, I. Effects of Training on BCI. In International Work-Conference on Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 2021; pp. 69–80. [Google Scholar]
- Wong, C.M.; Wang, Z.; Rosa, A.C.; Chen, C.P.; Jung, T.P.; Hu, Y.; Wan, F. Transferring Subject-Specific Knowledge Across Stimulus Frequencies in SSVEP-Based BCIs. IEEE Trans. Autom. Sci. Eng. 2021, 18, 552–563. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Y.; Wang, F.; Cao, L.; Zhou, W.; Wang, Z.; Chen, S. Cross-Subject Assistance: Inter-and Intra-Subject Maximal Correlation for Enhancing the Performance of SSVEP-Based BCIs. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 517–526. [Google Scholar] [CrossRef]
- Nagel, S.; Spüler, M. Modelling the brain response to arbitrary visual stimulation patterns for a flexible high-speed brain-computer interface. PLoS ONE 2018, 13, e0206107. [Google Scholar] [CrossRef] [Green Version]
- Thielen, J.; Marsman, P.; Farquhar, J.; Desain, P. From full calibration to zero training for a code-modulated visual evoked potentials for brain–computer interface. J. Neural Eng. 2021, 18, 056007. [Google Scholar] [CrossRef]
- Bin, G.; Gao, X.; Wang, Y.; Li, Y.; Hong, B.; Gao, S. A High-Speed BCI Based on Code Modulation VEP. J. Neural Eng. 2011, 8, 025015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, S.Q.; Wang, H.; Zhang, Z. Data Analytics in Steady-State Visual Evoked Potential-Based Brain–Computer Interface: A Review. IEEE Sens. J. 2020, 21, 1124–1138. [Google Scholar] [CrossRef]
- Zhuang, X.; Yang, Z.; Cordes, D. A Technical Review of Canonical Correlation Analysis for Neuroscience Applications. Hum. Brain Mapp. 2020, 41, 3807–3833. [Google Scholar] [CrossRef]
- Gembler, F.; Stawicki, P.; Saboor, A.; Volosyak, I. Dynamic Time Window Mechanism for Time Synchronous VEP-Based BCIs—Performance Evaluation with a Dictionary-Supported BCI Speller Employing SSVEP and c-VEP. PLoS ONE 2019, 14, e0218177. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, Y.; Gao, S.; Jung, T.P.; Gao, X. Filter Bank Canonical Correlation Analysis for Implementing a High-Speed SSVEP-Based Brain-Computer Interface. J. Neural Eng. 2015, 12, 046008. [Google Scholar] [CrossRef] [PubMed]
- Renton, A.I.; Mattingley, J.B.; Painter, D.R. Optimising non-invasive brain-computer interface systems for free communication between naïve human participants. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Gembler, F.W.; Rezeika, A.; Benda, M.; Volosyak, I. Five shades of grey: Exploring quintary m-sequences for more user-friendly c-vep-based bcis. Comput. Intell. Neurosci. 2020, 2020, 7985010. [Google Scholar] [CrossRef] [Green Version]
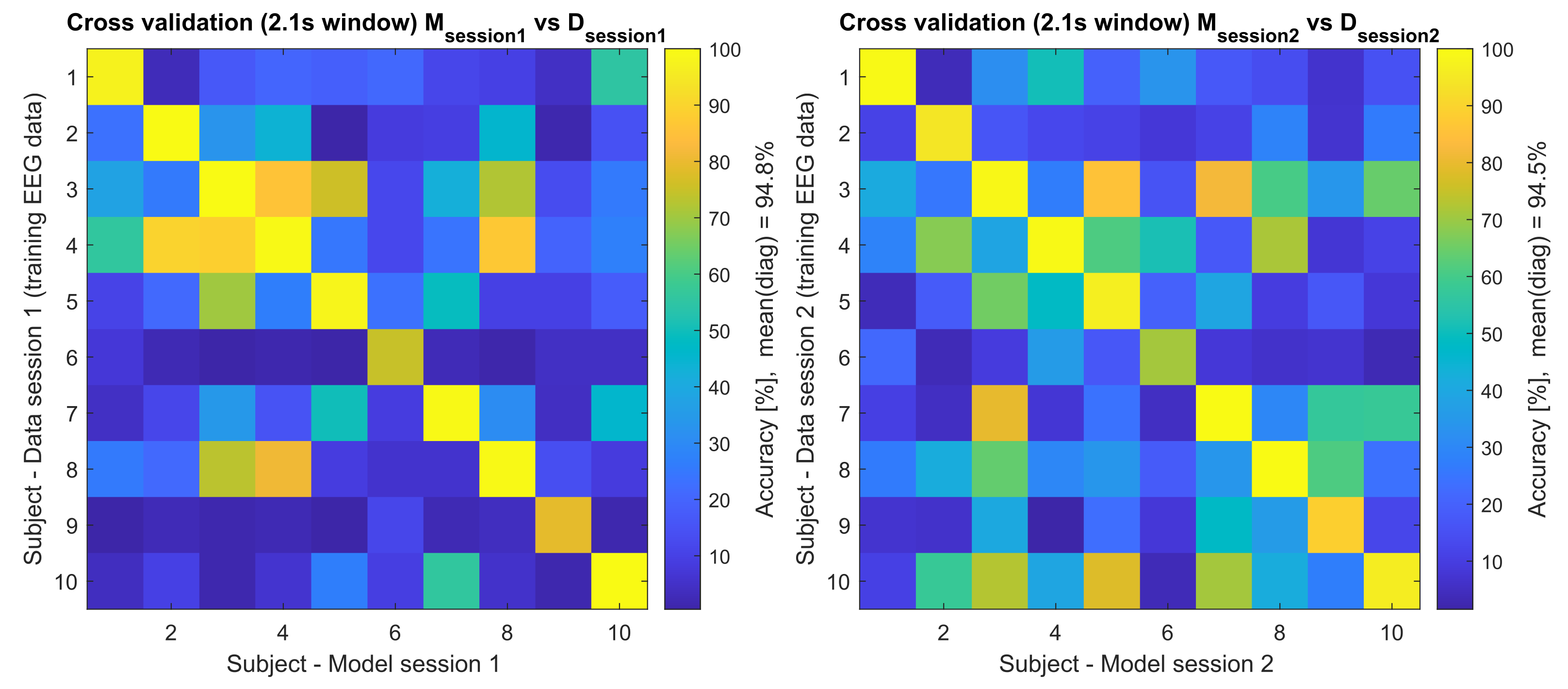
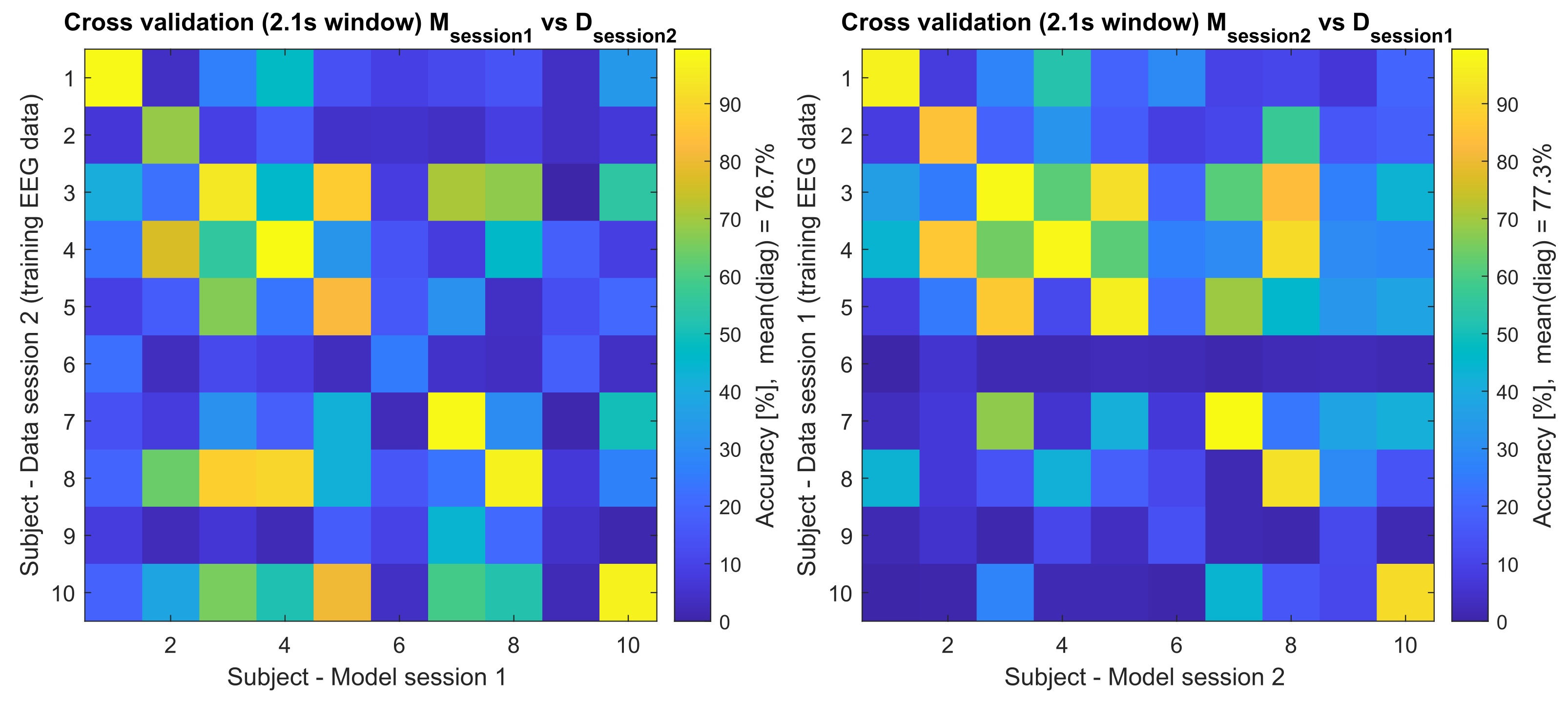
| Accuracy | Model Session 1 | Model Session 2 | ||
|---|---|---|---|---|
| Subject | Data Session 1 | Data Session 2 | Data Session 1 | Data Session 2 |
| 1 | 97.40 | 98.96 | 96.88 | 99.48 |
| 2 | 100 | 68.75 | 84.90 | 94.27 |
| 3 | 100 | 94.27 | 98.96 | 98.96 |
| 4 | 99.48 | 99.48 | 98.44 | 99.48 |
| 5 | 98.44 | 82.29 | 96.35 | 96.88 |
| 6 | 75.00 | 25.52 | 02.08 | 70.83 |
| 7 | 99.48 | 98.96 | 99.48 | 100 |
| 8 | 99.48 | 96.88 | 92.71 | 100 |
| 9 | 79.17 | 04.69 | 11.98 | 89.06 |
| 10 | 100 | 96.88 | 91.67 | 96.35 |
| Mean | 94.84 | 76.67 | 77.34 | 94.53 |
| SD | 09.45 | 34.21 | 37.39 | 09.00 |
| ITR | Model Session 1 | Model Session 2 | ||
| Subject | Data Session 1 | Data Session 2 | Data Session 1 | Data Session 2 |
| 1 | 90.91 | 94.16 | 89.9 | 95.37 |
| 2 | 96.77 | 49.47 | 70.44 | 85.15 |
| 3 | 96.77 | 85.15 | 94.16 | 94.16 |
| 4 | 95.37 | 95.37 | 93.03 | 95.37 |
| 5 | 93.03 | 66.75 | 88.9 | 89.90 |
| 6 | 57.10 | 09.50 | 00 | 51.95 |
| 7 | 95.37 | 94.16 | 95.37 | 96.77 |
| 8 | 95.37 | 89.90 | 82.49 | 96.77 |
| 9 | 62.51 | 00.10 | 02.14 | 76.64 |
| 10 | 96.77 | 89.90 | 80.78 | 88.90 |
| Mean | 88.00 | 67.45 | 69.72 | 87.10 |
| SD | 15.03 | 36.13 | 36.95 | 13.88 |
| Acc. | Single Blocks of Testing Sequence | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
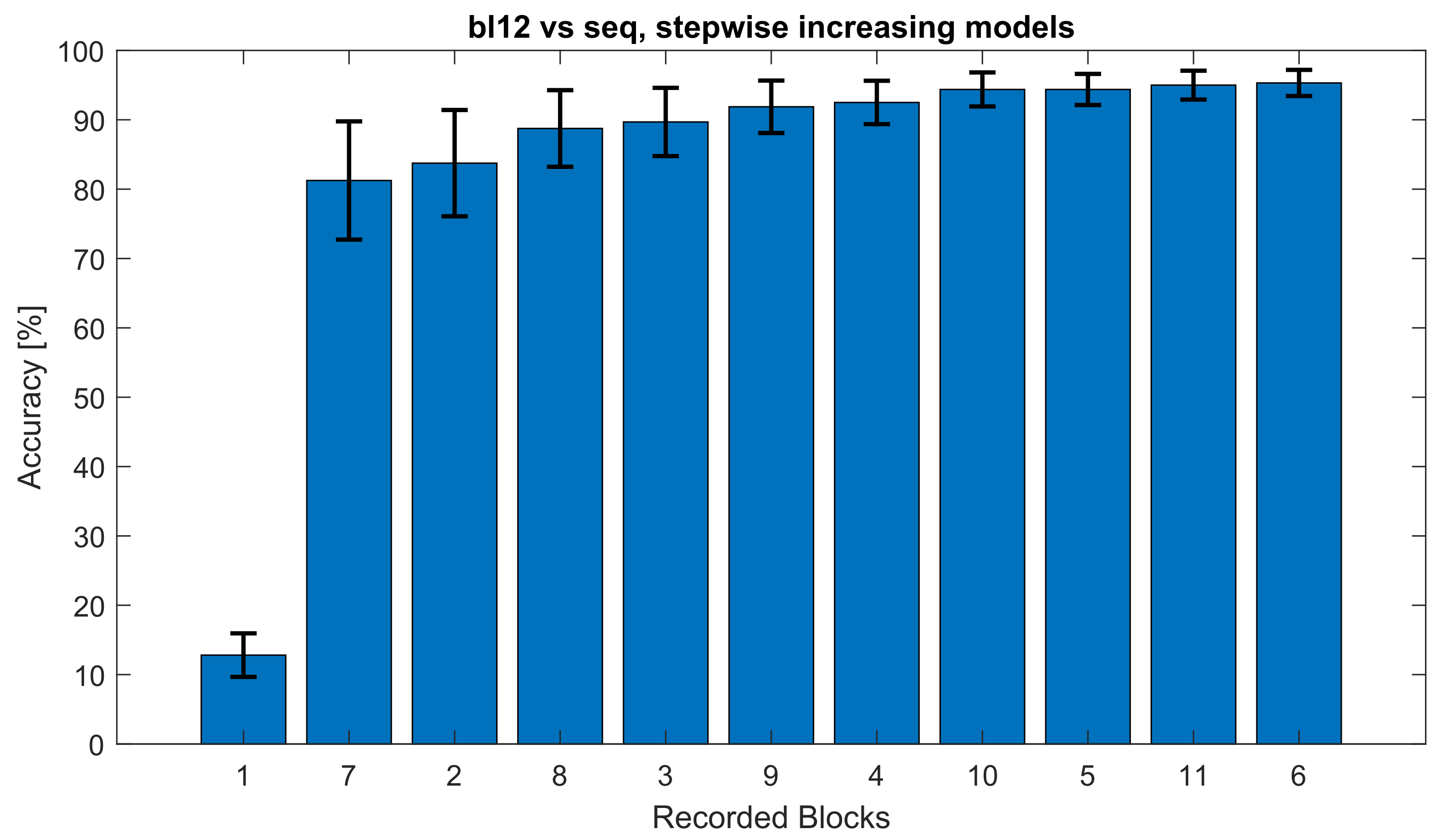
| Step | 1 | 7 | 2 | 8 | 3 | 9 | 4 | 10 | 5 | 11 | 6 | 12 | Mean | SD |
| 1 | 13.44 | 13.13 | 19.69 | 15.63 | 21.25 | 11.88 | 18.44 | 12.50 | 22.81 | 10.63 | 16.56 | 12.81 | 15.73 | 3.35 |
| 2 | 83.13 | 75.00 | 83.75 | 89.69 | 83.44 | 82.81 | 82.81 | 84.06 | 84.69 | 79.69 | 81.56 | 81.25 | 82.66 | 2.19 |
| 3 | 85.63 | 86.25 | 86.25 | 90.31 | 86.25 | 85.94 | 87.19 | 89.38 | 85.94 | 82.81 | 87.50 | 83.75 | 86.43 | 1.44 |
| 4 | 88.13 | 89.06 | 88.13 | 90.94 | 88.44 | 89.38 | 89.38 | 91.56 | 87.81 | 87.50 | 85.94 | 88.75 | 88.75 | 1.09 |
| 5 | 89.06 | 90.31 | 88.44 | 95.31 | 88.13 | 90.00 | 92.81 | 90.94 | 90.63 | 88.44 | 88.44 | 89.69 | 90.18 | 1.51 |
| 6 | 90.31 | 92.19 | 90.00 | 96.88 | 90.31 | 90.00 | 92.50 | 92.81 | 92.50 | 91.56 | 88.13 | 91.88 | 91.59 | 1.54 |
| 7 | 90.31 | 93.75 | 90.63 | 95.94 | 90.94 | 92.81 | 93.13 | 94.69 | 94.38 | 92.50 | 90.63 | 92.50 | 92.68 | 1.43 |
| 8 | 93.13 | 93.75 | 92.81 | 95.63 | 93.13 | 92.81 | 93.75 | 94.69 | 93.44 | 92.81 | 90.94 | 94.38 | 93.44 | 0.83 |
| 9 | 93.13 | 94.06 | 91.88 | 95.94 | 93.75 | 93.75 | 95.00 | 95.31 | 94.06 | 92.81 | 91.25 | 94.38 | 93.78 | 1.02 |
| 10 | 93.44 | 94.06 | 94.38 | 96.25 | 93.13 | 93.44 | 96.25 | 94.06 | 95.63 | 92.50 | 91.56 | 95.00 | 94.14 | 1.13 |
| 11 | 93.13 | 94.38 | 93.13 | 96.56 | 92.81 | 94.06 | 96.25 | 95.31 | 96.25 | 93.44 | 92.50 | 95.31 | 94.43 | 1.26 |
| SD | Single Blocks of Testing Sequence | |||||||||||||
| Step | 1 | 7 | 2 | 8 | 3 | 9 | 4 | 10 | 5 | 11 | 6 | 12 | ||
| 1 | 6.92 | 8.94 | 23.25 | 11.79 | 22.43 | 12.83 | 23.91 | 10.21 | 26.23 | 10.54 | 22.87 | 10.46 | ||
| 2 | 33.79 | 34.86 | 32.93 | 18.81 | 31.84 | 29.4 | 29.4 | 27.02 | 28.81 | 34.37 | 31.06 | 28.41 | ||
| 3 | 28.65 | 25.48 | 28.65 | 19.51 | 26.85 | 26.69 | 25.66 | 21.61 | 28.08 | 29.98 | 22.63 | 25.55 | ||
| 4 | 24.15 | 19.78 | 24.77 | 18.72 | 22.44 | 19.72 | 22.59 | 18.58 | 26.49 | 22.92 | 25.1 | 18.41 | ||
| 5 | 23.95 | 20.75 | 24.7 | 10.65 | 23.24 | 18.85 | 12.85 | 17.27 | 20.62 | 21.45 | 21.95 | 16.41 | ||
| 6 | 21.52 | 17.26 | 19.7 | 6.91 | 19.62 | 19.08 | 13.6 | 15.8 | 17.07 | 16.01 | 21.13 | 12.6 | ||
| 7 | 20.96 | 13.34 | 18.16 | 9.78 | 18.37 | 13.59 | 12.66 | 11.6 | 11.67 | 12.94 | 17.37 | 10.44 | ||
| 8 | 15.22 | 13.34 | 14.44 | 10.74 | 14.49 | 13.74 | 11.41 | 13.59 | 13.3 | 13.01 | 16.76 | 8.18 | ||
| 9 | 15.92 | 12.36 | 16.35 | 11.79 | 13.26 | 11.69 | 8.98 | 11.62 | 11.55 | 13.01 | 16.46 | 7.48 | ||
| 10 | 13.78 | 12.36 | 11.1 | 10.81 | 14.49 | 12.45 | 6.56 | 12.8 | 7.68 | 12.94 | 15.45 | 6.94 | ||
| 11 | 14.64 | 11.39 | 13.72 | 8.77 | 15.17 | 10.36 | 6.56 | 10.65 | 6.56 | 10.97 | 13.91 | 6.29 | ||
| Acc | |||||
|---|---|---|---|---|---|
| P1 | 100 | 98.44 | 97.92 | 98.44 | 98.75 |
| P2 | 93.75 | 98.44 | 96.88 | 97.66 | 98.13 |
| P3 | 96.88 | 100 | 100 | 100 | 100 |
| P4 | 100 | 98.44 | 98.96 | 98.44 | 98.75 |
| P5 | 93.75 | 96.88 | 96.88 | 98.44 | 96.25 |
| P6 | 71.88 | 70.31 | 68.75 | 68.75 | 66.25 |
| P7 | 100 | 100 | 100 | 100 | 100 |
| P8 | 100 | 98.44 | 98.96 | 99.22 | 99.38 |
| P9 | 75.0 | 67.19 | 70.83 | 75.0 | 80.0 |
| P10 | 93.75 | 96.88 | 96.88 | 97.66 | 98.13 |
| Mean | 92.50 | 92.500 | 92.61 | 93.36 | 93.56 |
| SD | 10.44 | 12.58 | 12.09 | 11.45 | 11.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stawicki, P.; Volosyak, I. cVEP Training Data Validation—Towards Optimal Training Set Composition from Multi-Day Data. Brain Sci. 2022, 12, 234. https://doi.org/10.3390/brainsci12020234
Stawicki P, Volosyak I. cVEP Training Data Validation—Towards Optimal Training Set Composition from Multi-Day Data. Brain Sciences. 2022; 12(2):234. https://doi.org/10.3390/brainsci12020234
Chicago/Turabian StyleStawicki, Piotr, and Ivan Volosyak. 2022. "cVEP Training Data Validation—Towards Optimal Training Set Composition from Multi-Day Data" Brain Sciences 12, no. 2: 234. https://doi.org/10.3390/brainsci12020234
APA StyleStawicki, P., & Volosyak, I. (2022). cVEP Training Data Validation—Towards Optimal Training Set Composition from Multi-Day Data. Brain Sciences, 12(2), 234. https://doi.org/10.3390/brainsci12020234







