Family-Based Whole-Exome Analysis of Specific Language Impairment (SLI) Identifies Rare Variants in BUD13, a Component of the Retention and Splicing (RES) Complex
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
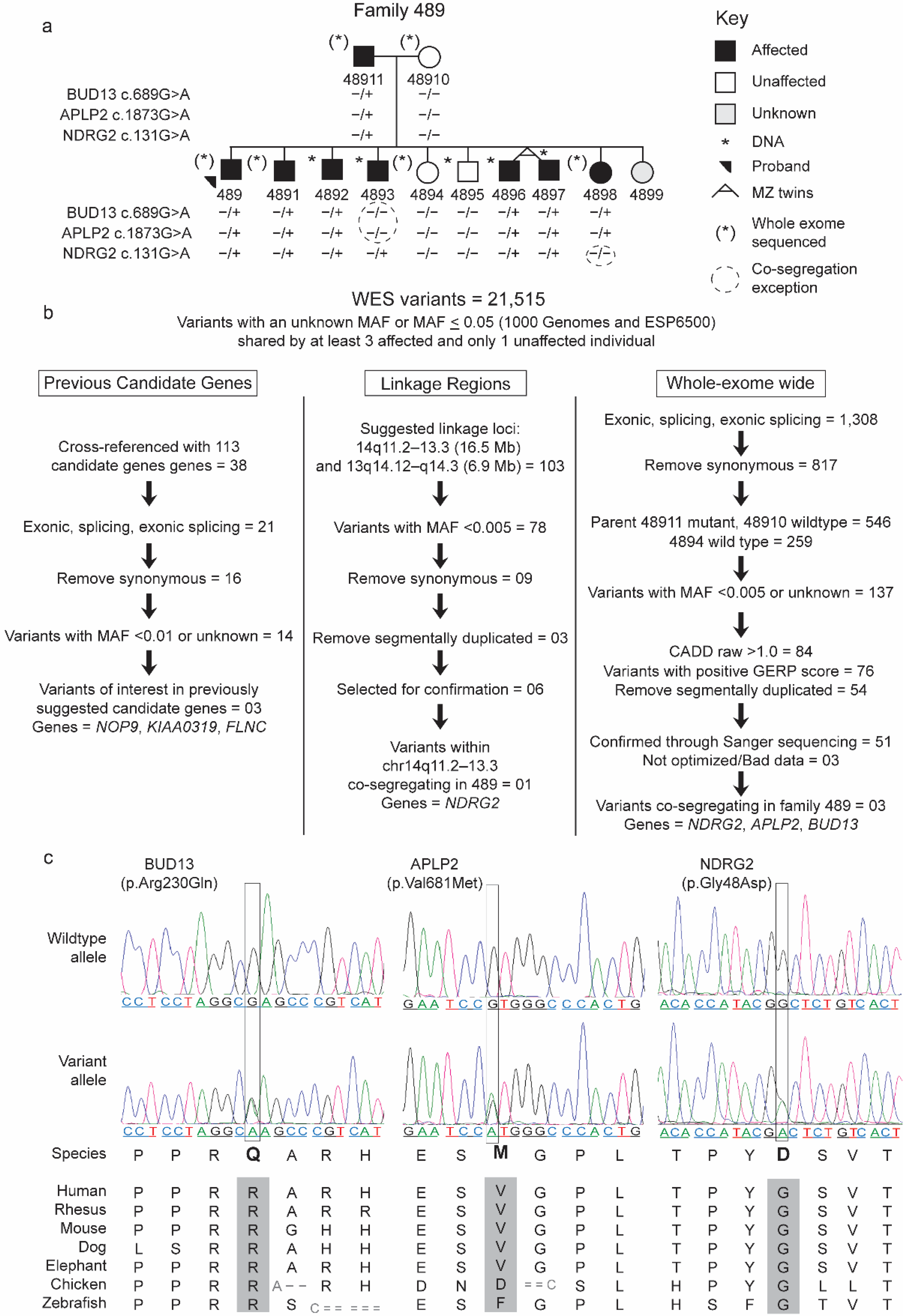
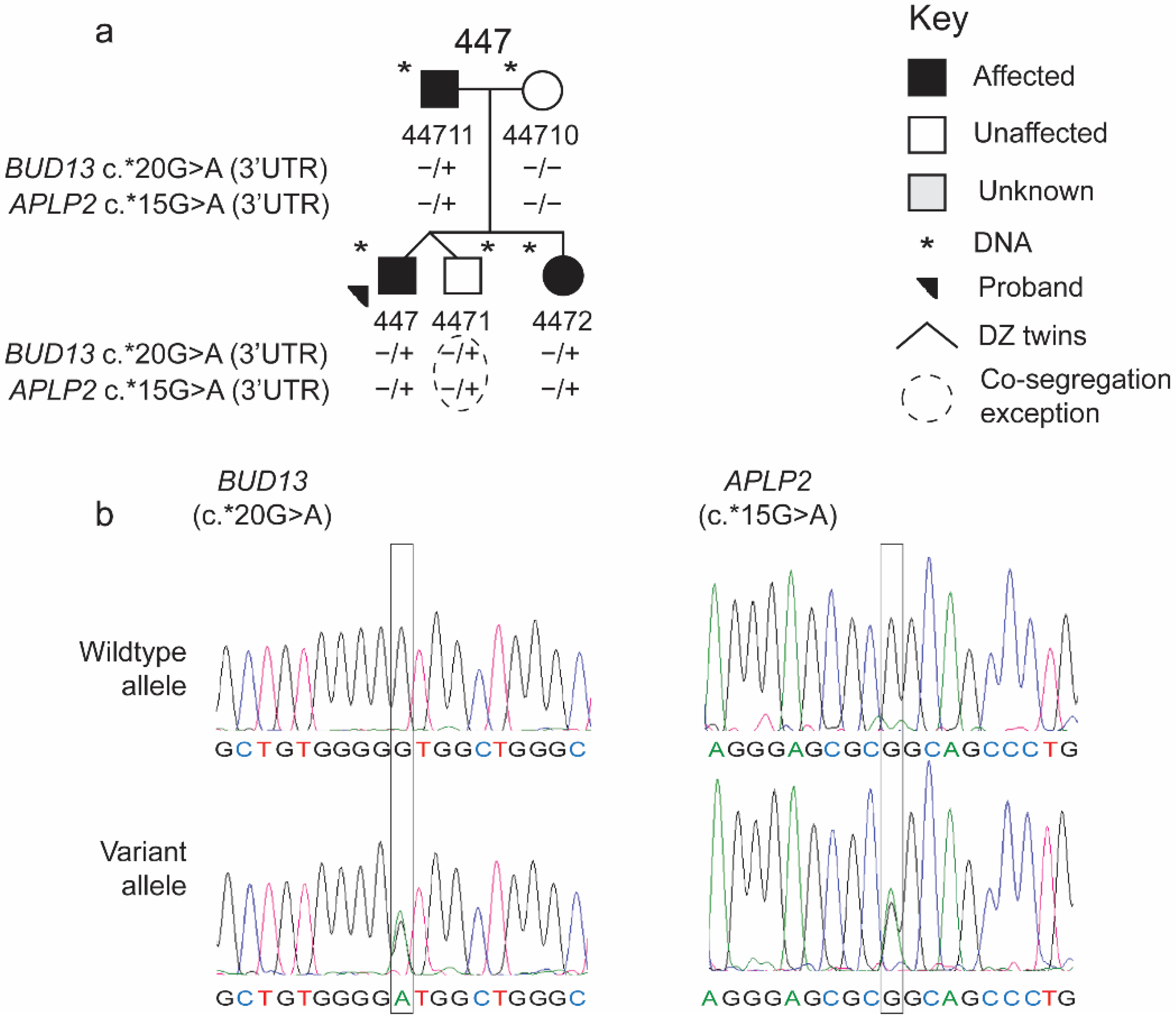
2.1.1. Family 489
2.1.2. Additional Participants
2.1.3. Phenotype
2.2. Genetic Analyses
2.2.1. DNA Collection and Preparation
2.2.2. Whole-Exome Sequencing and Data Analysis
2.2.3. Prioritization of Rare Variants in the WES
2.2.4. Identification of Candidate Genes, Confirmation, and Significance Testing
| Fam 489 Variants | Additional Variants | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Previous Candidates | Co-Segregating | NDRG2 | APLP2 | BUD13 | ||||||||||||||||
| Evidence Class | Evidence | KIAA0319 rs113411083 | FLNC rs202223616 | NOP9 rs183868211 | NDRG2 rs11552412 | APLP2 rs370970986 | BUD13 rs139478949 | rs779725845 | rs1063201 | chr11:129992279 | rs201861910 | rs35585096 | rs116087150 | rs1467808735 | rs144776650 | rs11216131 | rs1427011653 | rs145410701 | rs61730763 | rs145906707 |
| Genetic | MAF ≤ 0.05 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Co-segregation | − | − | − | + | + | + | − | − | − | + | − | − | − | − | − | − | − | − | + | |
| ≥1 proband | − | − | + | − | − | − | − | − | − | − | + | − | − | + | − | − | − | − | + | |
| Informatic | Positive GERP Score | + | + | + | + | + | + | + | − | + | − | + | + | + | − | + | − | − | + | + |
| Total # of damaging in silico scores | 2 | 1 | 0 | 4 | 2 | 4 | 2 | 0 | 2 | NA | 2 | 4 | 4 | 1 | 5 | 0 | 0 | 3 | NA | |
| HOPE output/AA change | ||||||||||||||||||||
| Size | ∧ | ∧ | ∨ | ∧ | ∧ | ∨ | ∧ | ∧ | ∧ | NA | ∧ | ∨ | ∨ | ∨ | ∨ | ∧ | ∨ | ∧ | NA | |
| >Hydrophobic | + | − | + | + | − | − | + | − | − | NA | + | + | + | − | + | − | + | − | NA | |
| Charge change | pos | neg | pos | neu | NC | pos | NC | NC | neg | NA | NC | pos | neu | pos | pos | NC | NC | NC | NA | |
| to | to | to | to | to | to | to | to | to | to | |||||||||||
| neu | pos | neu | neg | neu | pos | neu | neg | neu | neu | |||||||||||
| Causality | P | B | B | P | P | P | P | B | P | NA | P | P | P | B | P | B | B | P | NA | |
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tomblin, J.B.; Records, N.L.; Buckwalter, P.; Zhang, X.; Smith, E.; O’Brien, M. Prevalence of Specific Language Impairment in Kindergarten Children. J. Speech Lang. Hear. Res. 1997, 40, 1245–1260. [Google Scholar] [CrossRef]
- Norbury, C.F.; Gooch, D.; Wray, C.; Baird, G.; Charman, T.; Simonoff, E.; Vamvakas, G.; Pickles, A. The impact of nonverbal ability on prevalence and clinical presentation of language disorder: Evidence from a population study. J. Child Psychol. Psychiatry 2016, 57, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- National Institute on Deafness and Other Communication Disorders. Specific Language Impairment. Available online: https://www.nidcd.nih.gov/health/specific-language-impairment (accessed on 10 November 2017).
- Rice, M.L. Overlooked by Public Health: Specific Language Impairment. Open Access Government. 2017. Available online: https://www.openaccessgovernment.org/overlooked-public-healthspecific-language-impairment/34474/ (accessed on 26 October 2021).
- Brownlie, E.B.; Graham, E.; Bao, L.; Koyama, E.; Beitchman, J.H. Language disorder and retrospectively reported sexual abuse of girls: Severity and disclosure. J. Child Psychol. Psychiatry 2017, 58, 1114–1121. [Google Scholar] [CrossRef]
- Conti-Ramsden, G.; Botting, N. Social Difficulties and Victimization in Children With SLI at 11 Years of Age. J. Speech Lang. Hear. Res. 2004, 47, 145–161. [Google Scholar] [CrossRef]
- Bishop, D.V.M.; North, T.; Donlan, C. Genetic basis of specific language impairment: Evidence from a twin study. Dev. Med. Child Neurol. 1995, 37, 56–71. [Google Scholar] [CrossRef]
- Rice, M.L.; Haney, K.R.; Wexler, K. Family Histories of Children with SLI Who Show Extended Optional Infinitives. J. Speech Lang. Hear. Res. 1998, 41, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.L.; Wexler, K. Toward Tense as a Clinical Marker of Specific Language Impairment in English-Speaking Children. J. Speech Lang. Hear. Res. 1996, 39, 1239–1257. [Google Scholar] [CrossRef]
- Rice, M.L.; Taylor, C.L.; Zubrick, S.R.; Hoffman, L.; Earnest, K.K. Heritability of Specific Language Impairment and Nonspecific Language Impairment at Ages 4 and 6 Years Across Phenotypes of Speech, Language, and Nonverbal Cognition. J. Speech Lang. Hear. Res. 2020, 63, 793–813. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.L.; Zubrick, S.R.; Taylor, C.L.; Hoffman, L.; Gayán, J. Longitudinal Study of Language and Speech of Twins at 4 and 6 Years: Twinning Effects Decrease, Zygosity Effects Disappear, and Heritability Increases. J. Speech Lang. Hear. Res. 2018, 61, 79–93. [Google Scholar] [CrossRef]
- SLI Consortium. A Genomewide Scan Identifies Two Novel Loci Involved in Specific Language Impairment. Am. J. Hum. Genet. 2002, 70, 384–398. [Google Scholar] [CrossRef]
- SLI Consortium. Highly Significant Linkage to the SLI1 Locus in an Expanded Sample of Individuals Affected by Specific Language Impairment. Am. J. Hum. Genet. 2004, 74, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, C.W.; Flax, J.F.; Logue, M.W.; Vieland, V.J.; Bassett, A.S.; Tallal, P.; Brzustowicz, L.M. A Major Susceptibility Locus for Specific Language Impairment Is Located on 13q21. Am. J. Hum. Genet. 2002, 71, 45–55. [Google Scholar] [CrossRef]
- Newbury, D.F.; Winchester, L.; Addis, L.; Paracchini, S.; Buckingham, L.-L.; Clark, A.; Cohen, W.; Cowie, H.; Dworzynski, K.; Everitt, A.; et al. CMIP and ATP2C2 Modulate Phonological Short-Term Memory in Language Impairment. Am. J. Hum. Genet. 2009, 85, 264–272. [Google Scholar] [CrossRef]
- Bartlett, C.W.; Flax, J.F.; Logue, M.W.; Smith, B.J.; Vieland, V.J.; Tallal, P.; Brzustowicz, L.M. Examination of potential overlap in autism and language loci on chromosomes 2, 7, and 13 in two independent samples ascertained for specific language impairment. Hum. Hered. 2004, 57, 10–20. [Google Scholar] [CrossRef]
- Andres, E.M.; Earnest, K.K.; Smith, S.D.; Rice, M.L.; Raza, M.H. Pedigree-Based Gene Mapping Supports Previous Loci and Reveals Novel Suggestive Loci in Specific Language Impairment. J. Speech Lang. Hear. Res. 2020, 63, 4046–4061. [Google Scholar] [CrossRef]
- Nudel, R.; Simpson, N.H.; Baird, G.; O’Hare, A.; Conti-Ramsden, G.; Bolton, P.F.; Hennessy, E.R.; SLI Consortium; Ring, S.M.; Davey Smith, G.D.; et al. Genome-wide association analyses of child genotype effects and parent-of-origin effects in specific language impairment. Genes Brain Behav. 2014, 13, 418–429. [Google Scholar] [CrossRef]
- Andres, E.M.; Hafeez, H.; Yousaf, A.; Riazuddin, S.; Rice, M.L.; Basra, M.A.R.; Raza, M.H. A genome-wide analysis in consanguineous families reveals new chromosomal loci in specific language impairment (SLI). Eur. J. Hum. Genet. 2019, 27, 1274–1285. [Google Scholar] [CrossRef]
- Truong, D.T.; Shriberg, L.D.; Smith, S.D.; Chapman, K.L.; Scheer-Cohen, A.R.; DeMille, M.M.C.; Adams, A.K.; Nato, A.Q.; Wijsman, E.M.; Eicher, J.D.; et al. Multipoint genome-wide linkage scan for nonword repetition in a multigenerational family further supports chromosome 13q as a locus for verbal trait disorders. Hum. Genet. 2016, 135, 1329–1341. [Google Scholar] [CrossRef]
- Villanueva, P.; Newbury, D.F.; Jara, L.; De Barbieri, Z.; Mirza, G.; Palomino, H.M.; Fernández, M.A.; Cazier, J.-B.; Monaco, A.P.; Palomino, H. Genome-wide analysis of genetic susceptibility to language impairment in an isolated Chilean population. Eur. J. Hum. Genet. 2011, 19, 687–695. [Google Scholar] [CrossRef]
- Villanueva, P.; Nudel, R.; Hoischen, A.; Fernández, M.A.; Simpson, N.H.; Gilissen, C.; Reader, R.H.; Jara, L.; Echeverry, M.M.; Francks, C.; et al. Exome Sequencing in an Admixed Isolated Population Indicates NFXL1 Variants Confer a Risk for Specific Language Impairment. PLoS Genet. 2015, 11, e1004925. [Google Scholar] [CrossRef]
- Villanueva, P.; De Barbieri, Z.; Palomino, H.M.; Palomino, H. High prevalence of specific language impairment in Robinson Crusoe Island. A possible founder effect. Rev. Med. Chile 2008, 136, 186–192. [Google Scholar] [PubMed]
- Nudel, R. An investigation of NFXL1, a gene implicated in a study of specific language impairment. J. Neurodev. Disord. 2016, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Mountford, H.S.; Villanueva, P.; Fernández, M.A.; De Barbieri, Z.; Cazier, J.-B.; Newbury, D.F. Candidate gene variant effects on language disorders in Robinson Crusoe Island. Ann. Hum. Biol. 2019, 46, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.S.; Reader, R.H.; Hoischen, A.; Veltman, J.A.; Simpson, N.H.; Francks, C.; Newbury, D.F.; Fisher, S.E. Next-generation DNA sequencing identifies novel gene variants and pathways involved in specific language impairment. Sci. Rep. 2017, 7, 46105. [Google Scholar] [CrossRef] [PubMed]
- Centanni, T.M.; Green, J.R.; Iuzzini-Seigel, J.; Bartlett, C.W.; Hogan, T.P. Evidence for the multiple hits genetic theory for inherited language impairment: A case study. Front. Genet. 2015, 6, 272. [Google Scholar] [CrossRef][Green Version]
- Mountford, H.S.; Newbury, D.F. The genomic landscape of language: Insights into evolution. J. Lang. Evol. 2017, 3, 49–58. [Google Scholar] [CrossRef]
- Rice, M.L.; Tager-Flusberg, H. Language Phenotypes. In Neurophenotypes; Jagaroo, V., Santangelo, S.L., Eds.; Innovations in Cognitive Neuroscience; Springer: Boston, MA, USA, 2016; pp. 227–243. [Google Scholar]
- Rice, M.L.; Hoffman, L. Predicting Vocabulary Growth in Children With and Without Specific Language Impairment: A Longitudinal Study From 2;6 to 21 Years of Age. J. Speech Lang. Hear. Res. 2015, 58, 345–359. [Google Scholar] [CrossRef]
- Rice, M.L.; Wexler, K.; Cleave, P.L. Specific Language Impairment as a Period of Extended Optional Infinitive. J. Speech Lang. Hear. Res. 1995, 38, 850–863. [Google Scholar] [CrossRef]
- Rice, M.L.; Smith, S.D.; Gayán, J. Convergent genetic linkage and associations to language, speech and reading measures in families of probands with Specific Language Impairment. J. Neurodev. Disord. 2009, 1, 264–282. [Google Scholar] [CrossRef][Green Version]
- MacArthur, D.G.; Manolio, T.A.; Dimmock, D.P.; Rehm, H.L.; Shendure, J.; Abecasis, G.R.; Adams, D.R.; Altman, R.B.; Antonarakis, S.E.; Ashley, E.A.; et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Wechsler, D. Wechsler Intelligence Scale for Children, 3rd ed.; The Psychological Corporation: San Antonio, TX, USA, 1991. [Google Scholar]
- Wechsler, D. Adult Intelligence Scale-Third Edition; The Psychological Corporation: San Antonio, TX, USA, 1997. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Soranzo, N. The end of the start for population sequencing. Nature 2015, 526, 52–53. [Google Scholar] [CrossRef]
- NHLBI Exome Sequencing Project (ESP) Home Page. Available online: https://evs.gs.washington.edu/EVS/ (accessed on 18 December 2020).
- Guerra, J.; Cacabelos, R. Genomics of speech and language disorders. J. Transl. Genet. Genom. 2019. [Google Scholar] [CrossRef]
- Ceroni, F.; Simpson, N.H.; Francks, C.; Baird, G.; Conti-Ramsden, G.; Clark, A.; Bolton, P.F.; Hennessy, E.R.; Donnelly, P.; Bentley, D.R.; et al. Homozygous microdeletion of exon 5 in ZNF277 in a girl with specific language impairment. Eur. J. Hum. Genet. 2014, 22, 1165–1171. [Google Scholar] [CrossRef]
- Catts, H.W.; Adlof, S.M.; Weismer, S.E. Language Deficits in Poor Comprehenders: A Case for the Simple View of Reading. J. Speech Lang. Hear. Res. 2006, 49, 278–293. [Google Scholar] [CrossRef]
- Catts, H.W.; Adlof, S.M.; Hogan, T.P.; Weismer, S.E. Are Specific Language Impairment and Dyslexia Distinct Disorders? J. Speech Lang. Hear. Res. 2005, 48, 1378–1396. [Google Scholar] [CrossRef]
- Adlof, S.M.; Hogan, T.P. If We Don’t Look, We Won’t See: Measuring Language Development to Inform Literacy Instruction. Policy Insights Behav. Brain Sci. 2019, 6, 210–217. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M.; Stone, E.A.; Asimenos, G.; NISC Comparative Sequencing Program; Green, E.D.; Batzoglou, S.; Sidow, A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005, 15, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.R.; Nelson, S.F. Calculating the statistical significance of rare variants causal for Mendelian and complex disorders. BMC Med. Genom. 2018, 11, 53. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Ramensky, V.; Bork, P.; Sunyaev, S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002, 30, 3894–3900. [Google Scholar] [CrossRef]
- Sunyaev, S.R.; Eisenhaber, F.; Rodchenkov, I.V.; Eisenhaber, B.; Tumanyan, V.G.; Kuznetsov, E.N. PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. Des. Sel. 1999, 12, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Venselaar, H.; Beek, T.A.H.T.; Kuipers, R.K.P.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Neale, M.C.; Cardon, L.R. Methodology for Genetic Studies of Twins and Families; Springer Science & Business Media: Dordect, South Holland, The Netherlands, 2013. [Google Scholar]
- Riazuddin, S.; Hussain, M.; Razzaq, A.; Iqbal, Z.; Shahzad, M.; Polla, D.L.; Song, Y.; Van Beusekom, E.; Khan, A.A.; Tomas-Roca, L.; et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry 2016, 22, 1604–1614. [Google Scholar] [CrossRef]
- Peter, B.; Wijsman, E.M.; Nato, A.Q., Jr.; University of Washington Center for Mendelian, G.; Matsushita, M.M.; Chapman, K.L.; Stanaway, I.B.; Wolff, J.; Oda, K.; Gabo, V.B.; et al. Genetic Candidate Variants in Two Multigenerational Families with Childhood Apraxia of Speech. PLoS ONE 2016, 11, e0153864. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, K.A.; Frinton, E.; Nudel, R.; Chan, M.T.M.; Thompson, P.; Hayiou-Thomas, M.E.; Talcott, J.B.; Stein, J.; Monaco, A.P.; Hulme, C.; et al. Further evidence for a parent-of-origin effect at the NOP9 locus on language-related phenotypes. J. Neurodev. Disord. 2016, 8, 24. [Google Scholar] [CrossRef]
- Gifford, C.A.; Ranade, S.S.; Samarakoon, R.; Salunga, H.T.; de Soysa, T.Y.; Huang, Y.; Zhou, P.; Elfenbein, A.; Wyman, S.K.; Bui, Y.K.; et al. Oligogenic inheritance of a human heart disease involving a genetic modifier. Science 2019, 364, 865–870. [Google Scholar] [CrossRef]
- Rylaarsdam, L.E.; Guemez-Gamboa, A. Genetic Causes and Modifiers of Autism Spectrum Disorder. Front. Cell. Neurosci. 2019, 13, 385. [Google Scholar] [CrossRef] [PubMed]
- Katsanis, N.; Ansley, S.J.; Badano, J.L.; Eichers, E.R.; Lewis, R.A.; Hoskins, B.E.; Scambler, P.J.; Davidson, W.S.; Beales, P.L.; Lupski, J.R. Triallelic Inheritance in Bardet-Biedl Syndrome, a Mendelian Recessive Disorder. Science 2001, 293, 2256–2259. [Google Scholar] [CrossRef]
- Eichers, E.R.; Lewis, R.A.; Katsanis, N.; Lupski, J.R. Triallelic inheritance: A bridge between Mendelian and multifactorial traits. Ann. Med. 2004, 36, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Kousi, M.; Katsanis, N. Genetic Modifiers and Oligogenic Inheritance. Cold Spring Harb. Perspect. Med. 2015, 5, a017145. [Google Scholar] [CrossRef]
- Devanna, P.; Chen, X.S.; Ho, J.; Gajewski, D.; Smith, S.D.; Gialluisi, A.; Francks, C.; Fisher, S.E.; Newbury, D.F.; Vernes, S.C. Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol. Psychiatry 2017, 23, 1375–1384. [Google Scholar] [CrossRef]
- Devanna, P.; Van De Vorst, M.; Pfundt, R.; Gilissen, C.; Vernes, S.C. Genome-wide investigation of an ID cohort reveals de novo 3′UTR variants affecting gene expression. Hum. Genet. 2018, 137, 717–721. [Google Scholar] [CrossRef]
- Enard, W.; Przeworski, M.; Fisher, S.E.; Lai, C.S.; Wiebe, V.; Kitano, T.; Monaco, A.P.; Paabo, S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 2002, 418, 869–872. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.S.L.; Fisher, S.E.; Hurst, J.A.; Vargha-Khadem, F.; Monaco, A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 2001, 413, 519–523. [Google Scholar] [CrossRef]
- Raza, M.H.; Mattera, R.; Morell, R.; Sainz, E.; Rahn, R.; Gutierrez, J.; Paris, E.; Root, J.; Solomon, B.; Brewer, C.; et al. Association between Rare Variants in AP4E1, a Component of Intracellular Trafficking, and Persistent Stuttering. Am. J. Hum. Genet. 2015, 97, 715–725. [Google Scholar] [CrossRef]
- Kazemi, N.; Estiar, M.A.; Fazilaty, H.; Sakhinia, E. Variants in GNPTAB, GNPTG and NAGPA genes are associated with stutterers. Gene 2018, 647, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.H.; Domingues, C.E.F.; Webster, R.; Sainz, E.; Paris, E.; Rahn, R.; Gutierrez, J.; Chow, H.M.; Mundorff, J.; Kang, C.-S.; et al. Mucolipidosis types II and III and non-syndromic stuttering are associated with different variants in the same genes. Eur. J. Hum. Genet. 2016, 24, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Drayna, D. A role for inherited metabolic deficits in persistent developmental stuttering. Mol. Genet. Metab. 2012, 107, 276–280. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reader, R.H.; Covill, L.E.; Nudel, R.; Newbury, D.F. Genome-Wide Studies of Specific Language Impairment. Curr. Behav. Neurosci. Rep. 2014, 1, 242–250. [Google Scholar] [CrossRef][Green Version]
- Thomson, E.; Rappsilber, J.; Tollervey, D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 2007, 13, 2165–2174. [Google Scholar] [CrossRef]
- Gialluisi, A.; Newbury, D.F.; Wilcutt, E.G.; Olson, R.K.; DeFries, J.C.; Brandler, W.M.; Pennington, B.F.; Smith, S.D.; Scerri, T.S.; Simpson, N.H.; et al. Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014, 13, 686–701. [Google Scholar] [CrossRef]
- Jangi, M.; Boutz, P.L.; Paul, P.; Sharp, P.A. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014, 28, 637–651. [Google Scholar] [CrossRef]
- Frankiw, L.; Majumdar, D.; Burns, C.; Vlach, L.; Moradian, A.; Sweredoski, M.J.; Baltimore, D. BUD13 Promotes a Type I Interferon Response by Countering Intron Retention in Irf7. Mol. Cell 2019, 73, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.P.; Moreno-Mateos, M.A.; Gohr, A.; Miao, L.; Chan, S.H.; Irimia, M.; Giraldez, A.J. RES complex is associated with intron definition and required for zebrafish early embryogenesis. PLoS Genet. 2018, 14, e1007473. [Google Scholar] [CrossRef]
- Servetti, M.; Pisciotta, L.; Tassano, E.; Cerminara, M.; Nobili, L.; Boeri, S.; Rosti, G.; Lerone, M.; Divizia, M.T.; Ronchetto, P.; et al. Neurodevelopmental Disorders in Patients With Complex Phenotypes and Potential Complex Genetic Basis Involving Non-Coding Genes, and Double CNVs. Front. Genet. 2021, 12, 732002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; You, Y.; Wu, Y.; Zhang, Y.; Wang, M.; Song, Y.; Liu, X.; Kou, C. Association of BUD13 polymorphisms with metabolic syndrome in Chinese population: A case-control study. Lipids Health Dis. 2017, 16, 127. [Google Scholar] [CrossRef][Green Version]
- Lin, E.; Kuo, P.-H.; Liu, Y.-L.; Yang, A.C.; Kao, C.-F.; Tsai, S.-J. Association and interaction of APOA5, BUD13, CETP, LIPA and health-related behavior with metabolic syndrome in a Taiwanese population. Sci. Rep. 2016, 6, 36830. [Google Scholar] [CrossRef]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E.J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Li, H.D.; Funk, C.C.; McFarland, K.; Dammer, E.B.; Allen, M.; Carrasquillo, M.M.; Levites, Y.; Chakrabarty, P.; Burgess, J.D.; Wang, X.; et al. Integrative functional genomic analysis of intron retention in human and mouse brain with Alzheimer’s disease. Alzheimer’s Dement. 2021, 17, 984–1004. [Google Scholar] [CrossRef]
- Fu, L.; Shi, Z.; Luo, G.; Tu, W.; Wang, X.; Fang, Z.; Li, X. Multiple microRNAs regulate human FOXP2 gene expression by targeting sequences in its 3′ untranslated region. Mol. Brain 2014, 7, 71. [Google Scholar] [CrossRef]
- Weil, D.; Piton, A.; Lessel, D.; Standart, N. Mutations in genes encoding regulators of mRNA decapping and translation initiation: Links to intellectual disability. Biochem. Soc. Trans. 2020, 48, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.L. Toward epigenetic and gene regulation models of specific language impairment: Looking for links among growth, genes, and impairments. J. Neurodev. Disord. 2012, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Belkadi, A.; Bolze, A.; Itan, Y.; Cobat, A.; Vincent, Q.B.; Antipenko, A.; Shang, L.; Boisson, B.; Casanova, J.-L.; Abel, L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA 2015, 112, 5473–5478. [Google Scholar] [CrossRef]
- Yap, K.; Lim, Z.Q.; Khandelia, P.; Friedman, B.; Makeyev, E.V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012, 26, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Blencowe, B.J. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 2015, 87, 14–27. [Google Scholar] [CrossRef]
- Thompson, M.; Bixby, R.; Dalton, R.; Vandenburg, A.; Calarco, J.A.; Norris, A.D. Splicing in a single neuron is coordinately controlled by RNA binding proteins and transcription factors. eLife 2019, 8, e46726. [Google Scholar] [CrossRef]
| Gene | Genomic Position (hg19) | c.DNA Variant | AA Change | rsID | IDs of SLI Probands with Variant n = 175 | MAF in gnomAD | In Silico Prediction Scores | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glob | Euro | SIFT | Poly Phen-2 | Mutation assessor | PROVEAN | Mutation Taster | ||||||
| KIAA0319 | Chr6: | c.2164G > A | p.Arg722Trp | rs113411083 | NA | 0.00275 | 0.0047 | 0.068 | 0.998 | 1.495 | −3.28 | 101 |
| 24566953 | (T) | prob D | (low) | D | P | |||||||
| FLNC | Chr7: | c.6808G > A | p.Glu2270Lys | rs202223616 | NA | 0.00073 | 0.00168 | 1 | 0.371 | 1.23 | −2.44 | 56 |
| 128494547 | (T) | B | (low) | N | DC | |||||||
| NOP9 | Chr14: | c.62G > C | p.Arg21Pro | rs183868211 | 346, 353, 355, 411, 472 | 0.00936 | 0.02304 | 0.147 | 0.01 | 2.39 | −0.94 | 103 |
| 24769222 | (T) | B | (med) | N | P | |||||||
| Gene | Genomic Position (hg19) | c.DNA Variant | AA Change | rsID | IDs of SLI Probands with Variant n = 175 | MAF in gnomAD | In Silico Prediction Scores | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glob | Euro | SIFT | Poly Phen-2 | Mutation Assessor | PROVEAN | Mutation Taster | ||||||
| BUD13 | Chr11: | c.689G > A | p.Arg230Glu | rs139478949 | NA | 0.00002 | 0.00005 | 0.013 | 0.934 | 3.405 | −2.51 | 43 |
| 116633616 | (D) | poss D | (med) | D | DC | |||||||
| APLP2 | Chr11: | c.2041G > A | p.Val681Met | rs370970986 | NA | 0.00002 | 0.00002 | 0.39 | 0.94 | 1.1 | −0.01 | 21 |
| 130011820 | (D) | poss D | (low) | N | P | |||||||
| NDRG2 | Chr14: | c.143G > A | p.Gly48Asp | rs11552412 | NA | NA | NA | 0 | 1.00 | 3.445 | −6.06 | 94 |
| 21490631 | (D) | prob D | (med) | D | DC | |||||||
| Genomic Position (hg19) Chr11 | c.DNA | AA Change | rsID | IDs of SLI Probands with Variant n = 175 | MAF in gnomAD | In Silico Prediction Scores | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glob | Euro | SIFT | Poly Phen-2 | Mutation Assessor | PROVEAN | Mutation Taster | |||||
| 129991652 | c.660T > G | p.Asp220Glu | rs1063201 | 434 | 0.00006 | 0.00002 | 0.736 | 0 | 0.255 | −0.59 | 45 |
| (T) | B | (neutral) | N | P | |||||||
| 129992279 | c.793G > A | p.Glu265Lys | NA | 463 | NA | NA | 0.022 | 0 | 0.55 | −1.54 | 56 |
| (D) | B | (neutral) | N | DC | |||||||
| 130013358 | c.*15G > A | 3′UTR | rs201861910 | 447 | 0.001221 | 0.001972 | NA | NA | NA | NA | NA |
| Genomic Position (hg19) Chr11 | c.DNA | AA Change | rsID | IDs of SLI Probands with Variant n = 175 | MAF in gnomAD | In Silico Prediction Scores | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glob | Euro | SIFT | Poly Phen-2 | Mutation Assessor | PROVEAN | Mutation Taster | |||||
| 116643617 | c.64C > A | p.Ala22Ser | rs35585096 | 337, 455, 483, 405 | 0.023 | 0.000 | 0.112 | 0.578 | 2.44 | −0.81 | 99 |
| (D) | poss D | (med) | N | P | |||||||
| 116633875 | c.430G > A | p.Arg144Cys | rs116087150 | 49324 | 0.000 | 0.000 | 0.045 | 1 | 2.81 | −4.15 | 180 |
| (D) | prob D | (med) | D | DC | |||||||
| 116633787 | c.518T > C | p.Asp173Gly | rs1467808735 | 360 | 0.000 | 0.000 | 0.013 | 1 | 3.27 | −4.47 | 94 |
| (D) | prob D | (med) | D | DC | |||||||
| 116633724 | c.581C > T | p.Arg194His | rs144776650 | 384, 422, 484 | 0.003 | 0.005 | 0.06 | 0.091 | 1.725 | −3.73 | 29 |
| (T) | B | (low) | D | P | |||||||
| 116633580 | c.725C > A | p.Arg242Ile | rs11216131 | 500 | 0.001 | 0.001 | 0.002 | 0.999 | 3.58 | −4.43 | 97 |
| (D) | prob D | (high) | D | DC | |||||||
| 116633425 | c.880C > G | p.Ala294Pro | rs1427011653 | 201 | NA | NA | 0.231 | 0.002 | 2.395 | −0.75 | 27 |
| (T) | B | (med) | N | P | |||||||
| 116633353 | c.952A > T | p.Tyr318Asn | rs145410701 | 438 | 0.001 | 0.000 | 0.33 | 0.138 | 2.045 | −1.26 | 142 |
| (T) | B | (med) | N | P | |||||||
| 116631482 | c.1223G > A | p.Pro408Leu | rs61730763 | 427 | 0.003 | 0.000 | 0.023 | 0.275 | 2.63 | −7.04 | 98 |
| (D) | B | (med) | D | DC | |||||||
| 116619178 | c.*20G > A | 3′UTR | rs145906707 | 431, 447 | 0.003 | 0.003 | NA | NA | NA | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andres, E.M.; Earnest, K.K.; Zhong, C.; Rice, M.L.; Raza, M.H. Family-Based Whole-Exome Analysis of Specific Language Impairment (SLI) Identifies Rare Variants in BUD13, a Component of the Retention and Splicing (RES) Complex. Brain Sci. 2022, 12, 47. https://doi.org/10.3390/brainsci12010047
Andres EM, Earnest KK, Zhong C, Rice ML, Raza MH. Family-Based Whole-Exome Analysis of Specific Language Impairment (SLI) Identifies Rare Variants in BUD13, a Component of the Retention and Splicing (RES) Complex. Brain Sciences. 2022; 12(1):47. https://doi.org/10.3390/brainsci12010047
Chicago/Turabian StyleAndres, Erin M., Kathleen Kelsey Earnest, Cuncong Zhong, Mabel L. Rice, and Muhammad Hashim Raza. 2022. "Family-Based Whole-Exome Analysis of Specific Language Impairment (SLI) Identifies Rare Variants in BUD13, a Component of the Retention and Splicing (RES) Complex" Brain Sciences 12, no. 1: 47. https://doi.org/10.3390/brainsci12010047
APA StyleAndres, E. M., Earnest, K. K., Zhong, C., Rice, M. L., & Raza, M. H. (2022). Family-Based Whole-Exome Analysis of Specific Language Impairment (SLI) Identifies Rare Variants in BUD13, a Component of the Retention and Splicing (RES) Complex. Brain Sciences, 12(1), 47. https://doi.org/10.3390/brainsci12010047







