Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression
Abstract
1. Introduction
2. Results
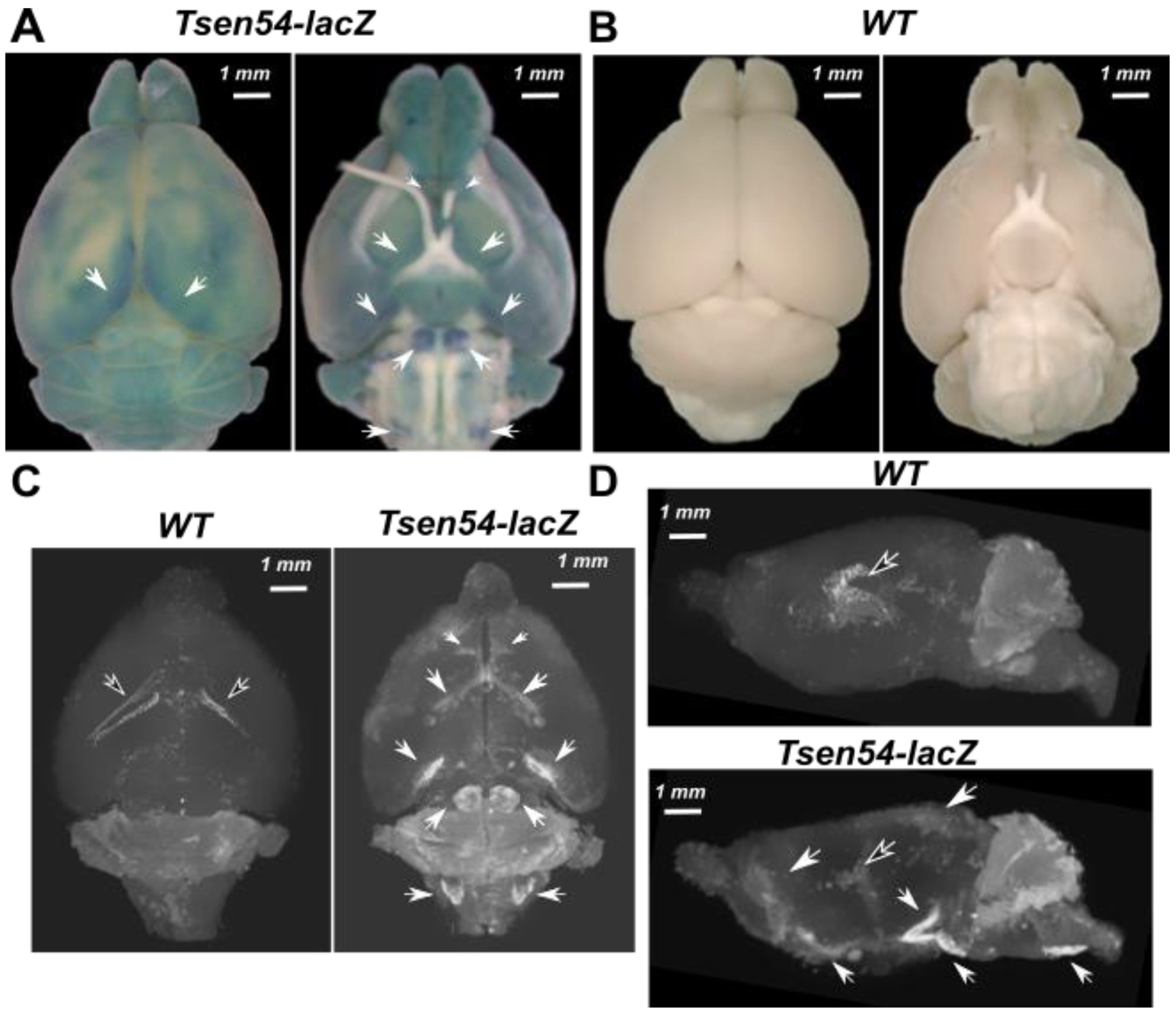
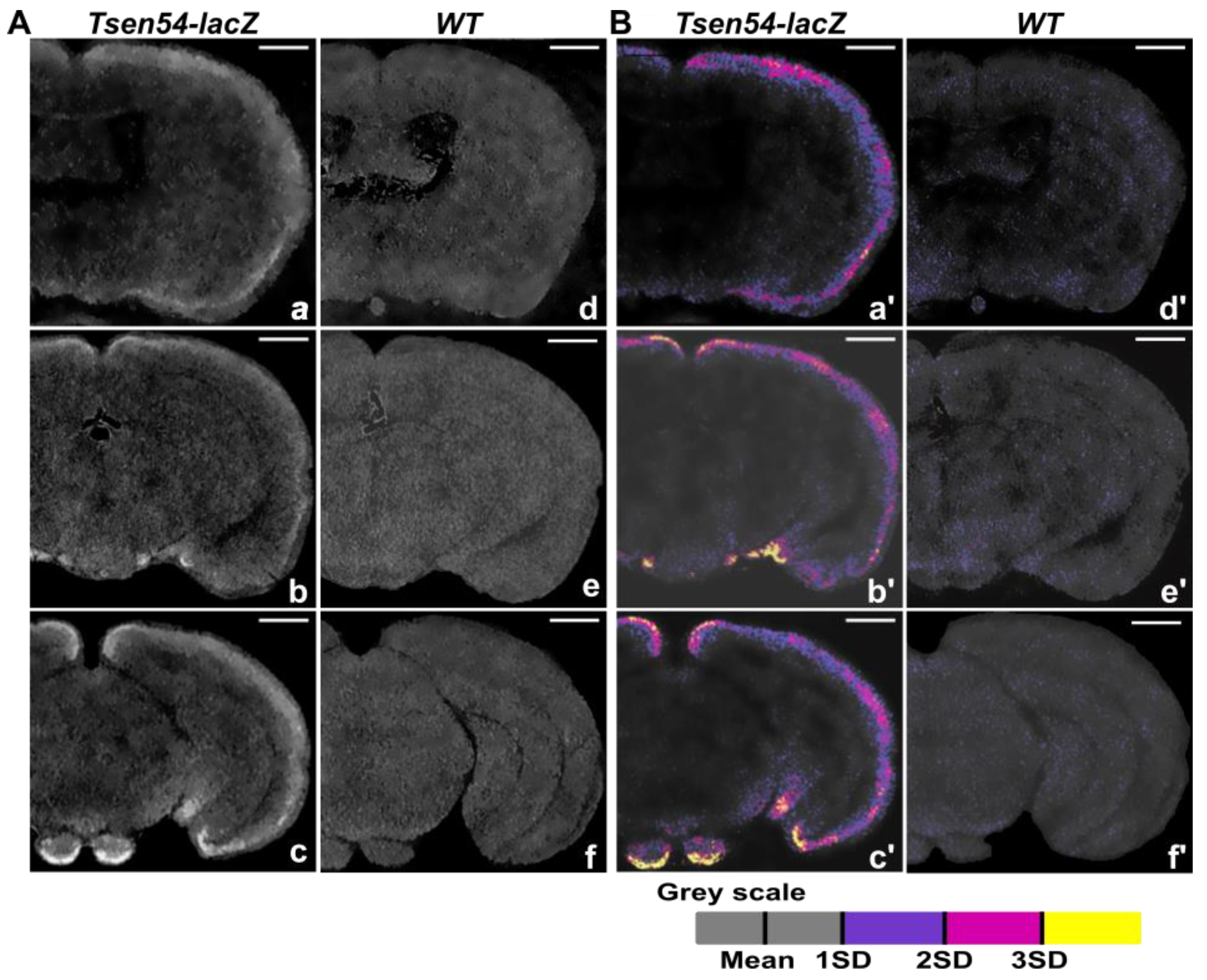
2.1. Three-Dimensional Imaging of Tsen54-lacZ Reporter Gene Expression in Whole-Mount Mouse Brain by X-ray
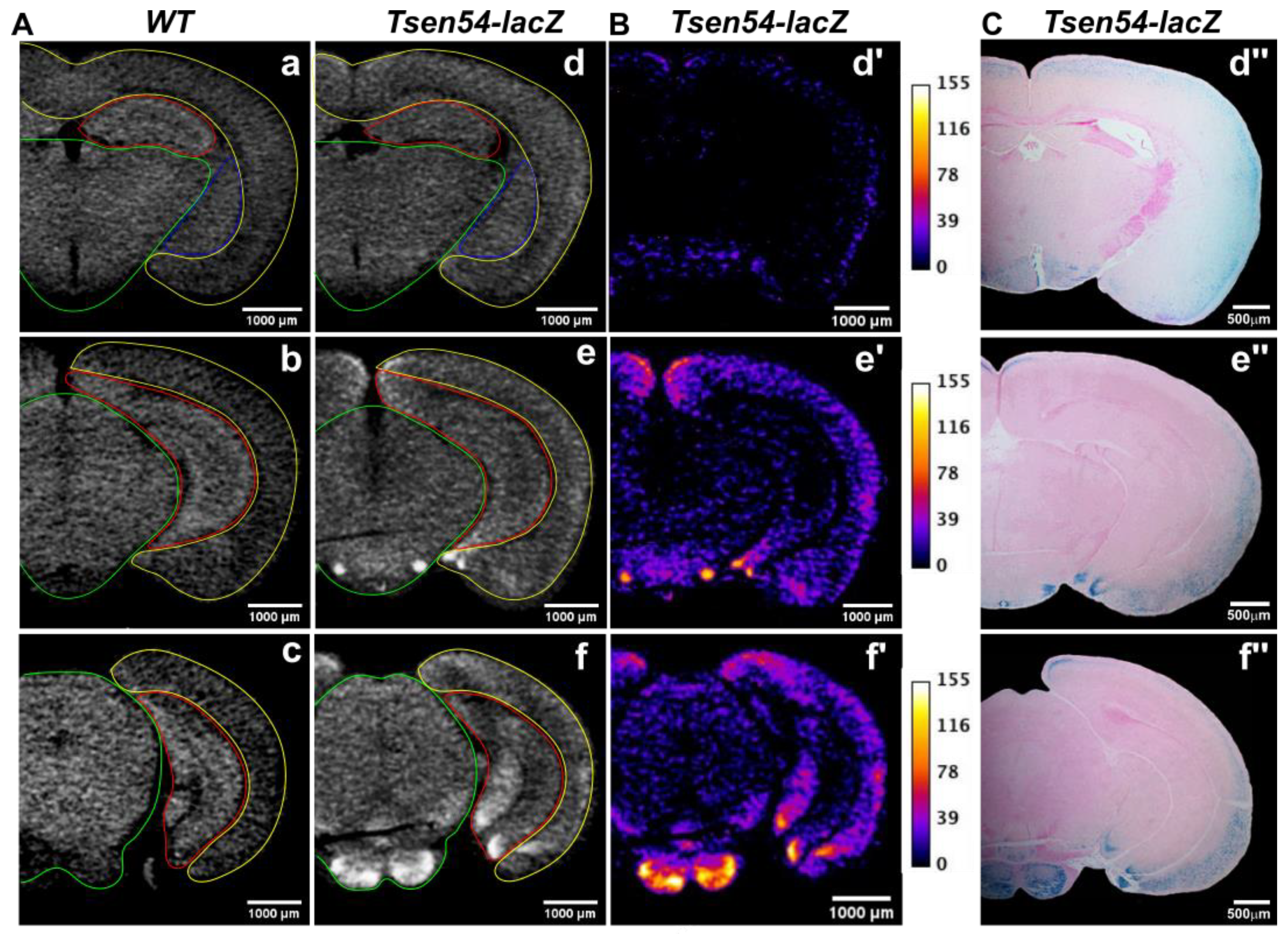
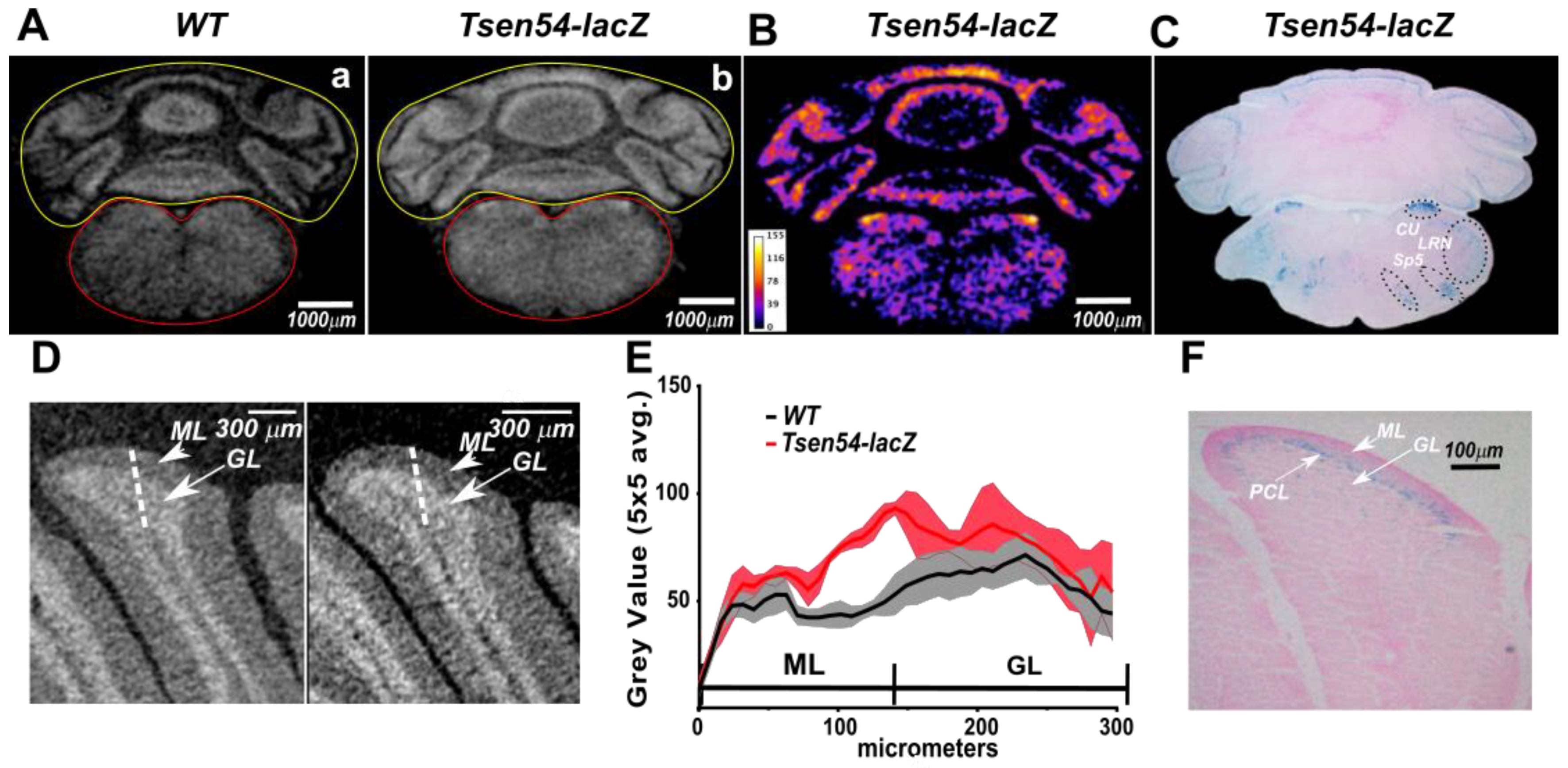
2.2. Comparative Analysis of Virtual 2D Micro-CT and Histological Sections in the X-gal/FeCN Tained Brains
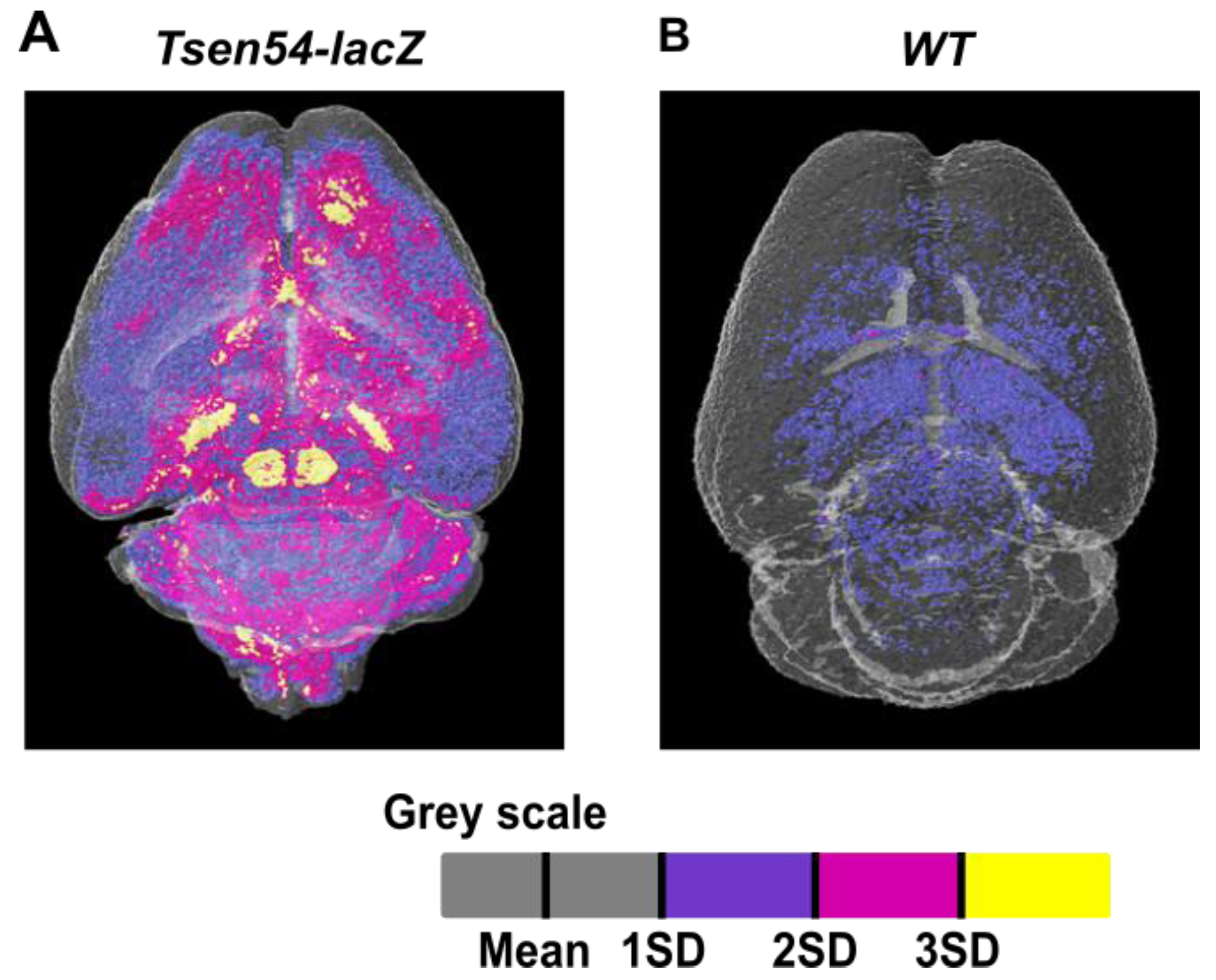
2.3. Relative Quantitative Analysis of the Tsen54-lacZ Gene Activity in Murine Brain
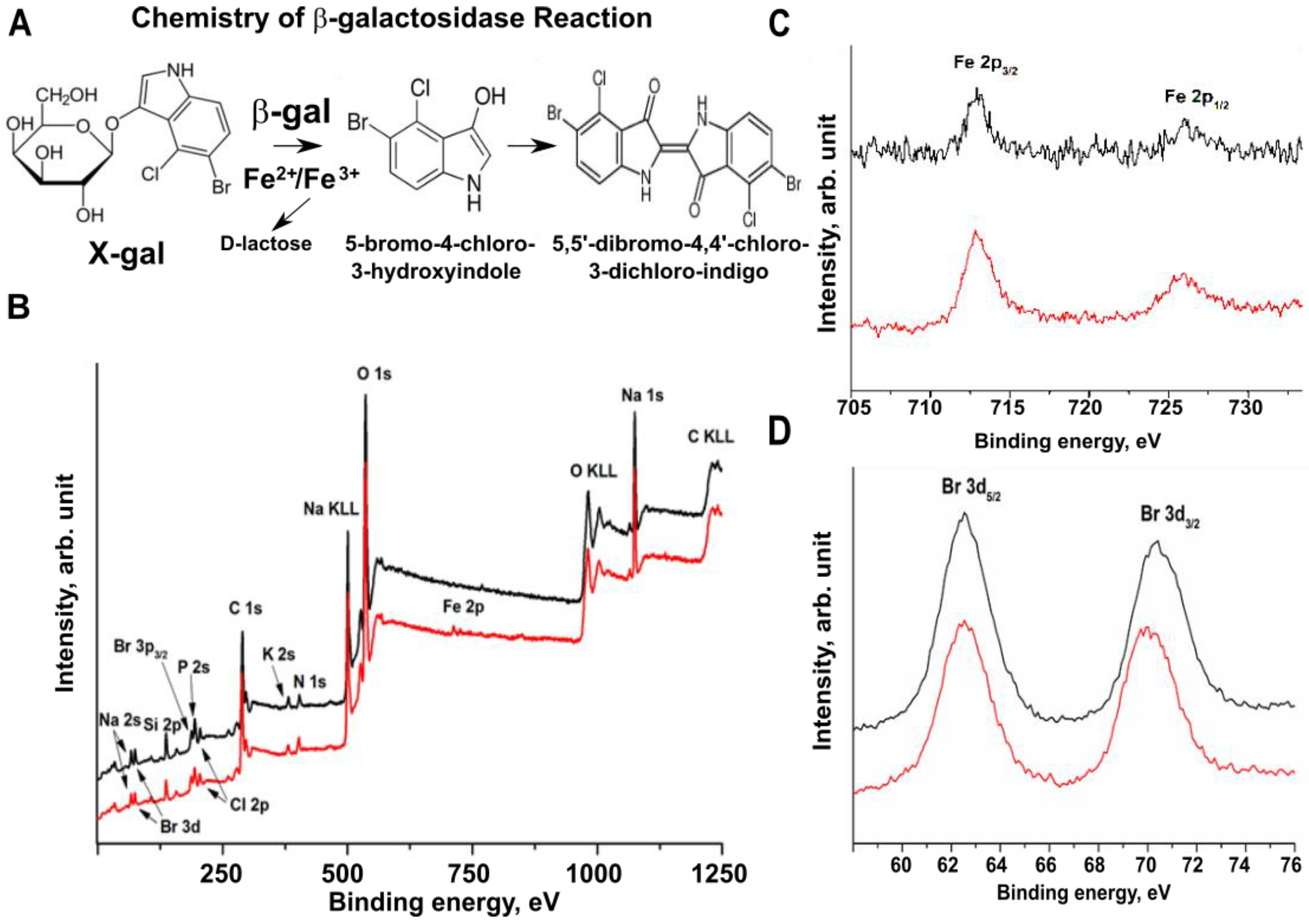
2.4. Bromine Atoms in the β-gal/X-gal/FeCN Reaction Product Are the Radiodense Substrates for X-ray Imaging
2.5. Three-Dimensional Map of the Brain Regions Characterized by Specific Tsen54-lacZ Reporter Gene Expression
3. Discussion
4. Material and Methods
4.1. Ethics Statement
4.2. Animals
4.3. Processing for Micro-CT
4.4. Micro-CT Imaging
4.5. Light Microscopy
4.6. XPS Analysis of β-Galactosidase Reaction Products In Vitro
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Streicher, J.; Donat, M.A.; Strauss, B.; Spörle, R.; Schughart, K.; Müller, G.B. Computer-based three-dimensional visualization of developmental gene expression. Nat. Genet. 2000, 25, 147–152. [Google Scholar] [CrossRef]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef]
- Knowles, D.W.; Biggin, M.D. Building quantitative, three-dimensional atlases of gene expression and morphology at cellular resolution. Wiley Interdiscip Rev. Dev. Biol. 2013, 2, 767–779. [Google Scholar] [CrossRef]
- Ju, T.; Warren, J.; Carson, J.; Bello, M.; Kakadiaris, I.; Chiu, W.; Thaller, C.; Eichele, G. 3D volume reconstruction of a mouse brain from histological sections using warp filtering. J. Neurosci. Methods 2006, 156, 84–100. [Google Scholar] [CrossRef][Green Version]
- Brandle, K. A new method for aligning histological serial sections for three-dimensional reconstruction. Comput. Biomed. Res. 1989, 22, 52–62. [Google Scholar] [CrossRef]
- Weninger, W.J.; Streicher, J.; Muller, G.B. 3-dimensional reconstruction of histological serial sections using a computer. Wien. Klin. Wochenschr. 1996, 108, 515–520. [Google Scholar]
- Weninger, W.J.; Meng, S.; Streicher, J.; Müller, G.B. A new episcopic method for rapid 3-D reconstruction: Applications in anatomy and embryology. Anat. Embryol. 1998, 197, 341–348. [Google Scholar] [CrossRef]
- Denk, W.; Strickler, J.H.; Webb, W.W. Two-photon laser scanning fluorescence microscopy. Science 1990, 248, 73–76. [Google Scholar] [CrossRef]
- Helmchen, F.; Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2005, 2, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Micheva, K.D.; Smith, S.J. Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 2007, 55, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Gong, H.; Zhang, B.; Wang, Q.; Yan, C.; Wu, J.; Liu, Q.; Zeng, S.; Luo, Q. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 2010, 330, 1404–1408. [Google Scholar] [CrossRef]
- Quintana, L.; Sharpe, J. Optical projection tomography of vertebrate embryo development. Cold Spring Harb Protoc. 2011, 6, 586–594. [Google Scholar] [CrossRef][Green Version]
- Botcherby, E.J.; Smith, C.W.; Kohl, M.M.; Debàrre, D.; Booth, M.J.; Juškaitis, R.; Paulsen, O.; Wilson, T. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl. Acad. Sci. USA 2012, 109, 2919–2924. [Google Scholar] [CrossRef]
- Kuwajima, T.; Sitko, A.A.; Bhansali, P.; Jurgens, C.; Guido, W.; Mason, C. ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 2013, 140, 1364–1368. [Google Scholar] [CrossRef]
- Chung, K.; Wallace, J.; Kim, S.Y.; Kalyanasundaram, S.; Andalman, A.S.; Davidson, T.J.; Mizabekov, J.; Zalocusky, K.A.; Mattis, J.; Denisin, A.K.; et al. Structural and molecular interrogation of intact biological systems. Nature 2013, 497, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.T.; Fujimoto, S.; Imai, T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013, 16, 1154–1161. [Google Scholar] [CrossRef]
- Susaki, E.A.; Tainaka, K.; Perrin, D.; Kishino, F.; Tawara, T.; Watanabe, T.M.; Yokoyama, C.; Onoe, H.; Eguchi, M.; Yamaguchi, S.; et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 2014, 157, 726–739. [Google Scholar] [CrossRef]
- Tomer, R.; Lovett-Barron, M.; Kauvar, I.; Andalman, A.; Burns, V.M.; Sankaran, S.; Grosenick, L.; Broxton, M.; Yang, S.; Deisseroth, K. SPED light sheet microscopy: Fast mapping of biological system structure and function. Cell 2015, 163, 1796–17806. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Yang, Z.; Li, X. Tissue clearing technique: Recent progress and biomedical applications. Review. J. Anat. 2020, 238, 489–507. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gaviro, M.V.; Sanderson, D.; Ripoll, J.; Desco, M. Biomedical applications of tissue clearing and three-dimensional imaging in health and disease. iScience 2020, 23, 101432. [Google Scholar] [CrossRef] [PubMed]
- Concilio, S.C.; Russell, S.J.; Peng, K.W. A brief review of reporter gene imaging in oncolytic virotherapy and gene therapy. Mol. Ther. Oncolytics 2021, 10, 98–109. [Google Scholar] [CrossRef]
- Mezzanotte, L.; van’t Root, M.; Karatas, H.; Goun, E.A.; Löwik, C. In vivo molecular bioluminescence imaging: New tools and applications. Trends Biotechnol. 2017, 35, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M.; Kohrt, D.; Florentine, C.M.; Grossel, M.J. A firefly luciferase dual color bioluminescence reporter assay using two substrates to simultaneously monitor two gene expression events. Sci. Rep. 2018, 8, 5990. [Google Scholar] [CrossRef] [PubMed]
- Nemes, B.; Bölcskei, K.; Kecskés, A.; Kormos, V.; Gaszner, B.; Aczél, T.; Hegedüs, D.; Pintér, E.; Helyes, Z.; Sándor, Z. Human Somatostatin SST 4 Receptor Transgenic Mice: Construction and Brain Expression Pattern Characterization. Int. J. Mol. Sci. 2021, 22, 3758. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Woodroofe, C.C.; Wood, M.G.; Que, I.; Van’t Root, M.; Shi, C.; Kirkland, T.A.; Encell, L.P.; Wood, K.V.; Löwik, C.; et al. Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun. 2018, 9, 132. [Google Scholar]
- Neues, F.; Goerlich, R.; Renn, J.; Beckmann, F.; Epple, M. Skeletal deformations in medaka (Oryzias latipes) visualized by synchrotron radiation micro-computer tomography (SRmicroCT). J. Struct. Biol. 2007, 160, 236–240. [Google Scholar] [CrossRef]
- Neues, F.; Epple, M. X-ray microcomputer tomography for the study of biomineralized endo- and exoskeletons of animals. Chem. Rev. 2008, 108, 4734–4741. [Google Scholar] [CrossRef]
- Saito, S.; Murase, K. Ex vivo imaging of mouse brain using micro-CT with non-ionic iodinated contrast agent: A comparison with myelin staining. Br. J. Radiol. 2012, 85, e973–e978. [Google Scholar] [CrossRef]
- Wong, M.D.; Spring, S.; Henkelman, R.M. Structural stabilization of tissue for embryo phenotyping using micro-CT with iodine staining. PLoS ONE 2014, 8, e84321. [Google Scholar] [CrossRef]
- Ermakova, O.; Orsini, T.; Gambadoro, A.; Chiani, F.; Tocchini-Valentini, G.P. Three-dimensional microCT imaging of murine embryonicdevelopment from immediate post-implantation to organogenesis: Application for phenotyping analysis of early embryonic lethality in mutant animals. Mamm. Genome 2018, 29, 245–259. [Google Scholar] [CrossRef]
- Hsu, C.W.; Kalaga, S.; Akoma, U.; Rasmussen, T.L.; Christiansen, A.E.; Dickinson, M.E. High resolution imaging of mouse embryos and neonates with X-ray micro-computed tomography. Curr. Protoc. Mouse Biol. 2019, 9, e63. [Google Scholar] [CrossRef] [PubMed]
- Busse, M.; Müller, M.; Kimm, M.A.; Ferstl, S.; Allner, S.; Achterhold, K.; Herzen, J.; Pfeiffer, F. Three-dimensional virtual histology enabled through cytoplasm-specific X-ray stain for microscopic and nanoscopic computed tomography. Proc. Natl. Acad. Sci. USA. 2018, 115, 2293–2298. [Google Scholar] [CrossRef] [PubMed]
- Kastner, D.B.; Kharazia, V.; Nevers, R.; Smyth, C.; Astudillo-Maya, D.A.; Williams, G.M.; Yang, Z.; Holobetz, C.M.; Santina, L.D.; Parkinson, D.Y.; et al. Scalable method for micro-CT analysis enables large scale quantitative characterization of brain lesions and implants. Sci. Rep. 2020, 10, 20851. [Google Scholar] [CrossRef]
- Khimchenko, A.; Deyhle, H.; Schulz, G.; Schweighauser, G.; Hench, J.; Chicherova, N.; Bikis, C.; Hieber, S.E.; Müller, B. Extending two-dimensional histology into the third dimension through conventional micro computed tomography. Neuroimage 2016, 139, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Saccomano, M.; Albers, J.; Tromba, G.; Dobrivojević-Radmilović, M.; Gajović, S.; Alves, F.; Dullin, C. Synchrotron inline phase contrast µCT enables detailed virtual histology of embedded soft-tissue samples with and without staining. J. Synchrotron. Rad. 2018, 25, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Chiani, F.; Orsini, T.; Gambadoro, A.; Pasquini, M.; Putti, S.; Cirilli, M.; Ermakova, O.; Tocchini-Valentini, G.P. Functional loss of Ccdc1 51 leads to hydrocephalus in a mouse model of primary ciliary dyskinesia. Dis. Model. Mech. 2019, 12, 8. [Google Scholar] [CrossRef]
- Massimi, L.; Pieroni, N.; Maugeri, L.; Fratini, M.; Brun, F.; Burkeeva, I.; Santamaria, G.; Medici, V.; Poloni, T.E.; Balducci, C.; et al. Assessment of plaque morphology in Alzheimer’s mouse cerebellum using three-dimensional X-ray phase-contrast virtual histology. Sci Rep. 2020, 10, 1123. [Google Scholar] [CrossRef]
- Cedola, A.; Bravin, A.; Burkeeva, I.; Fratini, M.; Pacurenau, A.; Mittone, A.; Massimi, L.; Cloetens, P.; Coan, P.; Campi, G.; et al. X-ray phase contrast tomography reveals early vascular alterations and neuronal loss in a multiple sclerosis model. Sci. Rep. 2017, 7, 5890. [Google Scholar] [CrossRef]
- Kuan, A.T.; Phelps, J.S.; Thomas, L.A.; Nguyen, T.M.; Han, J.; Chen, C.L.; Azevedo, A.W.; Tuthill, J.C.; Funke, J.; Cloetens, P.; et al. Dense neuronal reconstruction through X-ray holographic nano-tomography. Nat. Neurosci. 2020, 23, 1637–1643. [Google Scholar] [CrossRef]
- Schulz, G.; Weitkamp, T.; Zanette, I.; Pfeiffer, F.; Beckmann, F.; David, C.; Rutishauser, S.; Reznikova, E.; Müller, B. High-resolution tomographic imaging of a human cerebellum: Comparison of absorption and grating-based phase contrast. J. R. Soc. Interface 2010, 7, 1665–1676. [Google Scholar] [CrossRef]
- Dyer, E.L.; Gray Roncal, W.; Prasad, J.A.; Fernandes, H.L.; Gürsoy, D.; De Andrade, V.; Fezzaa, K.; Xiao, X.; Vogelstein, J.T.; Jacobsen, C.; et al. Quantifying mesoscale neuroanatomy using X-ray microtomography. eNeuro 2017, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.C.; Araujo, B.H.S.; Dias, C.S.B. High-resolution synchrotron-based X-ray microtomography as a tool to unveil the three-dimensional neuronal architecture of the brain. Sci. Rep. 2018, 8, 12074. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Kimm, M.A.; Ferstl, S.; Allner, S.; Achterhold, K.; Herzen, J.; Pfeiffer, F.; Busse, M. Nucleus-specific X-ray stain for 3D virtual histology. Sci. Rep. 2018, 8, 17855. [Google Scholar] [CrossRef]
- Dullin, C.; Ufartes, R.; Larsson, E.; Martin, S.; Lazzarini, M.; Tromba, G.; Missbach-Guentner, J.; Pinkert-Leetsch, D.; Katschinski, D.M.; Alves, F. μCT of ex-vivo stained mouse hearts and embryos enables a precise match between 3D virtual histology, classical histology and immunochemistry. PLoS ONE 2017, 12, e0170597. [Google Scholar] [CrossRef]
- Töpperwien, M.; Markus, A.; Alves, F.; Salditt, T. Contrast enhancement for visualizing neuronal cytoarchitecture by propagation-based x-ray phase-contrast tomography. Neuroimage 2019, 199, 70–80. [Google Scholar] [CrossRef]
- Shahmoradian, S.H.; Tsai, E.H.R.; Diaz, A.; Guizar-Sicarios, M.; Raabe, J.; Spycher, L.; Britschgi, M.; Ruf, A.; Stahlberg, H.; Holler, M. Three-dimensional imaging of biological tissue by cryo X-ray ptychography. Sci. Rep. 2017, 7, 6291. [Google Scholar] [CrossRef]
- Bayguinov, P.O.; Fisher, M.R.; Fitzpatrick, J.A. Assaying three-dimensional cellular architecture using X-ray tomographic and correlated imaging approaches. J. Biol. Chem. 2020, 295, 15782–15793. [Google Scholar] [CrossRef] [PubMed]
- Metscher, B.D.; Müller, G.B. MicroCT for molecular imaging: Quantitative visualization of complete three-dimensional distributions of gene products in embryonic limbs. Dev. Dyn. 2011, 240, 2301–2308. [Google Scholar] [CrossRef]
- Hoshi, M.; Reginensi, A.; Joens, M.S.; Fitzpatrick, J.A.J.; McNeill, H.; Jain, S. Reciprocal Spatiotemporally Controlled Apoptosis Regulates Wolffian Duct Cloaca Fusion. J. Am. Soc. Nephrol. 2018, 29, 775–783. [Google Scholar] [CrossRef]
- Casadaban, M.J.; Cohen, S.N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980, 138, 179–207. [Google Scholar] [CrossRef]
- Chilvers, K.F.; Perry, J.D.; James, A.L.; Reed, R.H. Synthesis and evaluation of novel fluorogenic substrates for the detection of bacterial beta-galactosidase. J. Appl. Microbiol. 2001, 91, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.H.; Zeng, Q.; Shah, K.; Dong-Eog, K.; Schellingerhout, D.; Weissleder, R. In vivo imaging of beta-galactosidase activity using far red fluorescent switch. Cancer Res. 2004, 64, 1579–1583. [Google Scholar] [CrossRef] [PubMed]
- Louie, A.Y.; Huber, M.M.; Ahrens, E.T.; Rothbacher, U.; Moats, R.; Jacobs, R.E.; Fraser, S.E.; Meade, T.J. In vivo visualization of gene expression using magnetic resonance imaging. Nat. Biotechnol. 2000, 18, 321–325. [Google Scholar] [CrossRef] [PubMed]
- West, D.B.; Pasumarthi, R.K.; Baridon, B.; Djan, E.; Trainor, A.; Griffey, S.M.; Engelhard, E.K.; Rapp, J.; Li, B.; de Jong, P.J.; et al. A lacZ reporter gene expression atlas for 313 adult KOMP mutant mouse lines. Genome Res. 2015, 25, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.M., Jr.; Spencer, A.H. Molecular and neuroimaging findings in pontocerebellar hypoplasia type 2 (PCH2): Is prenatal diagnosis possible? Am. J. Med. Genet. A 2010, 152, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Rudnik-Schöneborn, S.; Barth, P.G.; Zerres, K. Pontocerebellar hypoplasia. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Namavar, Y.; Barth, P.G.; Kasher, P.R.; van Ruissen, F.; Brockmann, K.; Bernert, G.; Writzl, K.; Ventura, K.; Cheng, E.Y.; Ferriero, D.M.; et al. Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain 2011, 134, 143–156. [Google Scholar] [CrossRef]
- Namavar, Y.; Chitayat, D.; Barth, P.G.; van Ruissen, F.; de Wissel, M.B.; Poll-The, B.T.; Silver, R.; Baas, F. TSEN54 mutations cause pontocerebellar hypoplasia type 5. Eur J. Hum. Genet. 2011, 19, 724–726. [Google Scholar] [CrossRef][Green Version]
- Cassandrini, D.; Biancheri, R. Pontocerebellar hypoplasia: Clinical, pathologic, and genetic studies. Neurology 2010, 75, 1459–1464. [Google Scholar] [CrossRef]
- Van Dijk, T.; Baas, F.; Barth, P.G.; Poll-The, B.T. What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet. J. Rare Dis. 2018, 13, 92. [Google Scholar] [CrossRef]
- Paushkin, S.V.; Patel, M.; Furia, B.S.; Peltz, S.W.; Trotta, C.R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 2004, 117, 311–321. [Google Scholar] [CrossRef]
- Budde, B.S.; Namavar, Y.; Barth, P.G.; Poll-The, B.T.; Nurnberg, G.; Becker, C.; van Ruissen, F.; Weterman, M.A.; Fluiter, K.; te Beek, E.T.; et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008, 40, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Breuss, M.W.; Sultan, T.; James, K.N.; Rosti, R.O.; Scott, E.; Musaev, D.; Furia, B.; Reis, A.; Sticht, H.; Al-Owain, M.; et al. Autosomal-recessive mutations in the tRNA splicing endonuclease subunit TSEN15 cause pontocerebellar hypoplasia and progressive microcephaly. Am. J. Hum. Genet. 2016, 99, 228–235. [Google Scholar] [CrossRef]
- Kasher, P.R.; Namavar, Y.; van Tijn, P.; Fluiter, K.; Sizarov, A.; Kamermans, M.; Grierson, A.J.; Zivkovic, D.; Baas, F. Impairment of the tRNA-splicing endonuclease subunit 54 (tsen54) gene causes neurological abnormalities and larval death in zebrafish models of pontocerebellar hypoplasia. Hum. Mol. Genet. 2011, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Pearson, B.; Wolf, P.L.; Vazquez, J. A comparative study of a series of new indolyl compounds to localize beta-galactosidase in tissues. Lab. Invest. 1963, 12, 1249–1259. [Google Scholar]
- Cotson, S.; Holt, S.J. Studies in enzyme cytochemistry. IV. Kinetics of aerial oxidation of indoxyl and some of its halogen derivatives. Proc. R. Soc. Lond. B Biol. Sci. 1958, 148, 506–519. [Google Scholar]
- Lojda, Z.; Kraml, J. Indigogenic methods for glycosidases. I. An improved method for beta-D-glucosidase and its application to localization studies on intestinal and renal enzymes. Histochemie 1970, 22, 347–361. [Google Scholar]
- Wiberg, E.; Wiberg, N. Inorganic Chemistry; Nils, W.E., Ed.; Academic Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Grosvenor, A.P.; Kobe, B.A.; McIntyre, N.S. Examination of the oxidation of iron by oxygen using X-ray photoelectron spectroscopy and QUASES. Surface Science 2004, 565, 151–162. [Google Scholar] [CrossRef]
- Paxinos, G.; Franklin, K. The Mouse Brain in Stereotaxic Coordinates, 2nd ed.; Paxinos, G., Franklin, K., Eds.; Academic Press: San Diego, CA, USA, 2004. [Google Scholar]
- Wang, X.; Zeng, W.; Yang, X.; Fang, C.; Han, Y.; Fei, P. Bi-channel image registration and deep-learning segmentation (BIRDS) for efficient, versatile 3D mapping of mouse brain. Elife 2021, 10, e63455. [Google Scholar] [CrossRef] [PubMed]
- Skarnes, W.C.; Rosen, B.; West, A.P.; Koutsourakis, M.; Bushell, W.; Iyer, V.; Mujica, A.O.; Thomas, M.; Harrow, J.; Cox, T.; et al. A conditional knockout resource for the genome-wide stud of mouse gene function. Nature 2011, 474, 337–342. [Google Scholar] [CrossRef]
- Bradley, A.; Anastassiadis, K.; Ayadi, A.; Battey, J.F.; Bell, C.; Birling, M.C.; Bottomley, J.; Brown, S.D.; Bürger, A.; Bult, C.J.; et al. The mammalian gene function resource: The international knockout mouse consortium. Mamm. Genome 2012, 23, 580–586. [Google Scholar] [CrossRef]
- Marcó, S.; Haurigot, V.; Bosch, F. In vivo gene therapy for mucopolysaccharidosis Type III (Sanfilippo Syndrome): A new treatment horizon. Hum. Gene Ther. 2019, 30, 1211–1221. [Google Scholar] [CrossRef]
- Meyer-Dilhet, G.; Courchet, J. In utero cortical electroporation of plasmids in the mouse embryo. STAR Protoc. 2020, 19, 100027. [Google Scholar] [CrossRef]
- Epstein, R.A. Parahippocampal and retrosplenial contributions to human spatial navigation. Trends Cogn Sci. 2008, 12, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Sugar, J.; Witter, M.P.; van Strien, N.M.; Cappaert, N.L. The retrosplenial cortex: Intrinsic connectivity and connections with the (para)hippocampal region in the rat. Front. Neuroinform. 2011, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Squire, L.R.; Stark, C.E.; Clark, R.E. The medial temporal lobe. Annu. Rev. Neurosci. 2004, 27, 279–306. [Google Scholar] [CrossRef] [PubMed]
- Dillingham, C.M.; Frizzati, A.; Nelson, A.J.; Vann, S.D. How do mammillary body inputs contribute to anterior thalamic function? Neurosci. Biobehav Rev. 2015, 54, 108–119. [Google Scholar] [CrossRef]
- Kratochwil, C.F.; Maheshwari, U.; Rijli, F.M. The long journey of pontine nuclei neurons: From rhombic lip to cortico-ponto-cerebellar circuitry. Front. Neur. Circuits 2017, 11, 33. [Google Scholar] [CrossRef]
- Di Pietro, C.; La Sala, G.; Matteoni, R.; Marazziti, D.; Tocchini-Valentini, G.P. Genetic ablation of Gpr37l1 delays tumor occurrence in Ptch1+/− mouse models of medulloblastoma. Exp. Neurol. 2019, 312, 33–42. [Google Scholar] [CrossRef]
| Anatomical Region | Anatomical Structure | Anatomical Sub-Structure | Color Code | REL | |
|---|---|---|---|---|---|
| Cortical plate | Isocortex | Secondary motor area Layer 1 |  |  |  |
| Cortical plate | Isocortex | Retrosplenial area, dorsal part |  |  |  |
| Cortical plate | Isocortex | Perirhinal area, Layer 1 |  |  |  |
| Cortical plate | Hippocampal formation | Dentate gyrus, Molecular cell layer (temporal lobe) |  | ||
| Cortical plate | Olfactory area | Nucleus of the lateral olfactory tract (NLOT) |  |  |  |
| Cortical plate | Olfactory area | Anterior olfactory nucleus (AON) |  |  |  |
| Cerebral nuclei | Striatum | Bed nucleus of the accessory olfactory tract (BA) |  |  |  |
| Interbrain | Hypothalamus | Lateral mammilary nucleus (LM) |  |  | |
| Interbrain | Hypothalamus | Lateral preoptic area (LPO) |  |  |  |
| Hindbrain | Medulla (Motor related) | Lateral reticular nucleus (LRN) |  |  | |
| Hindbrain | Medulla (Sensory related) | Cuneate nucleus (CU) |  |  | |
| Hindbrain | Medulla (Sensory related) | Spinal nucleus of trigeminal caudal part |  |  |  |
| Hindbrain | Pons (Motor related) | Pontine gray |  |  | |
| Cerebellum | Cerebral cortex | Vermal regions |  |  |  |
| Cerebellum | Cerebral cortex | Hemispheric regions |  |  |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ermakova, O.; Orsini, T.; Fruscoloni, P.; Chiani, F.; Gambadoro, A.; Putti, S.; Cirilli, M.; Mezzi, A.; Kaciulis, S.; Pasquini, M.; et al. Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression. Brain Sci. 2021, 11, 746. https://doi.org/10.3390/brainsci11060746
Ermakova O, Orsini T, Fruscoloni P, Chiani F, Gambadoro A, Putti S, Cirilli M, Mezzi A, Kaciulis S, Pasquini M, et al. Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression. Brain Sciences. 2021; 11(6):746. https://doi.org/10.3390/brainsci11060746
Chicago/Turabian StyleErmakova, Olga, Tiziana Orsini, Paolo Fruscoloni, Francesco Chiani, Alessia Gambadoro, Sabrina Putti, Maurizio Cirilli, Alessio Mezzi, Saulius Kaciulis, Miriam Pasquini, and et al. 2021. "Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression" Brain Sciences 11, no. 6: 746. https://doi.org/10.3390/brainsci11060746
APA StyleErmakova, O., Orsini, T., Fruscoloni, P., Chiani, F., Gambadoro, A., Putti, S., Cirilli, M., Mezzi, A., Kaciulis, S., Pasquini, M., Raspa, M., Scavizzi, F., & Tocchini-Valentini, G. P. (2021). Three-Dimensional X-ray Imaging of β-Galactosidase Reporter Activity by Micro-CT: Implication for Quantitative Analysis of Gene Expression. Brain Sciences, 11(6), 746. https://doi.org/10.3390/brainsci11060746









