Comparative Mitogenomic Analyses of Psectrocladius (Diptera: Chironomidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Collection and DNA Extraction
2.2. Sequencing, Assembly, and Annotation
2.3. Composition Analyses, RSCU, and Evolutionary Rate
2.4. Phylogenetic Analyses
3. Results
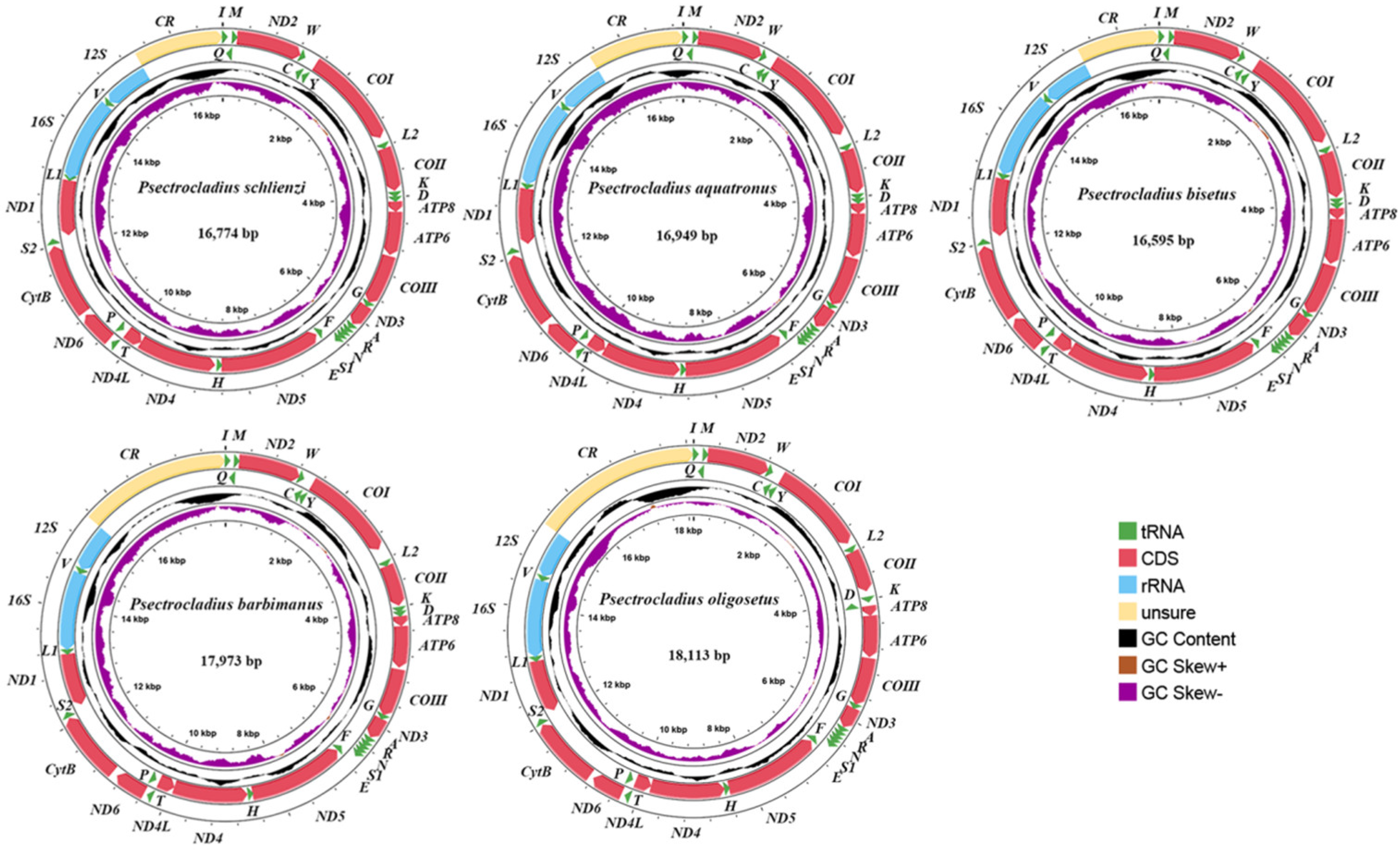
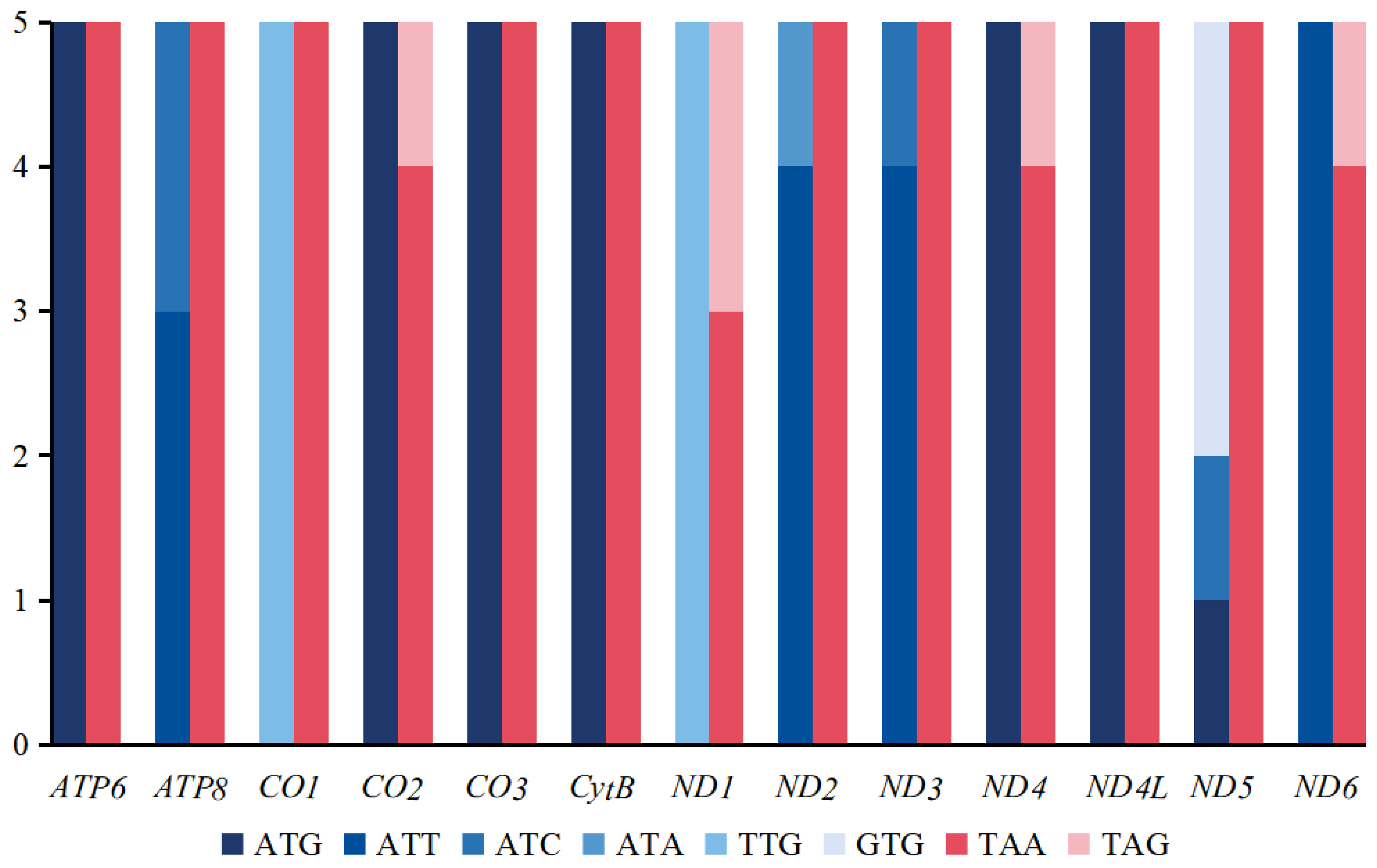
3.1. Mitogenomic Organization
3.2. Nucleotide Composition
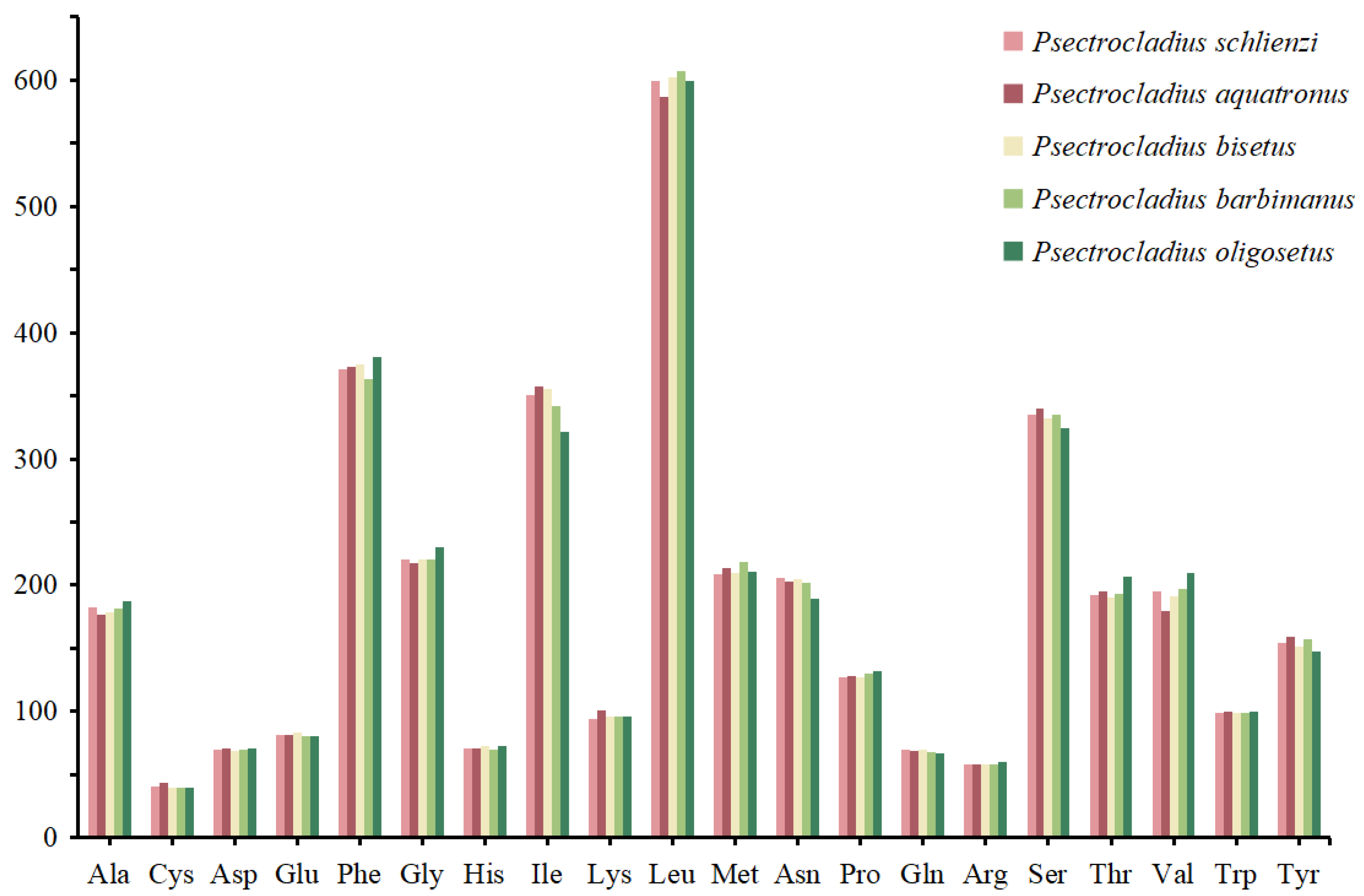
3.3. Codon Usage of PCGs
3.4. Substitution Rates and Nucleotide Diversity
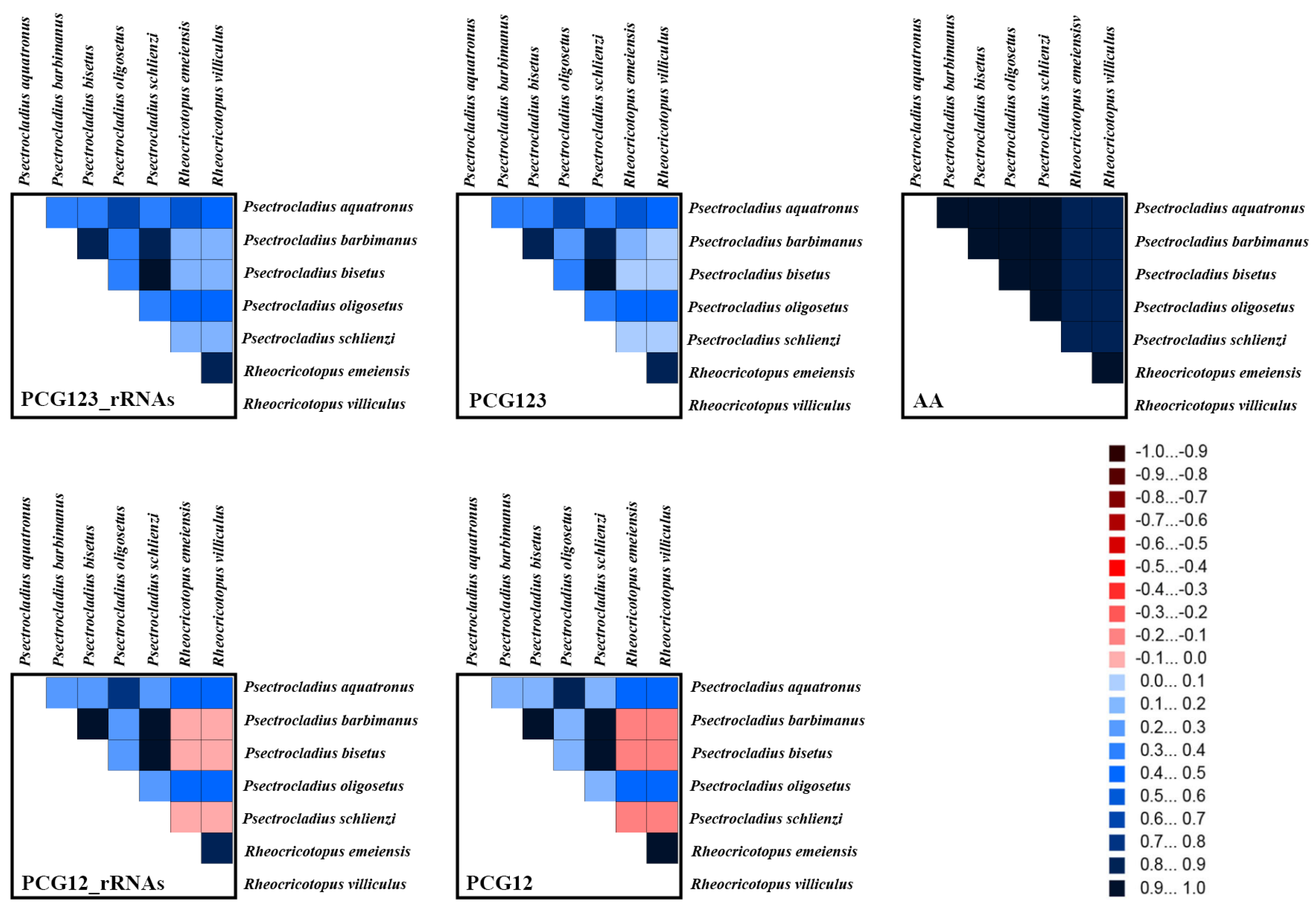
3.5. Heterogeneity Analyses of Mitogenomes
3.6. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Huang, J.; Wu, H. Mitogenomes provide insights into the phylogeny of Mycetophilidae (Diptera: Sciaroidea). Gene 2021, 783, 145564. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [PubMed]
- Song, N.; Li, X.; Yin, X.; Li, X.; Yin, J.; Pan, P. The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships. Sci. Rep. 2019, 9, 17765. [Google Scholar] [CrossRef]
- Krosch, M.; Cranston, P.; Bryant, L.; Strutt, F.; Mccluen, S. Towards a dated molecular phylogeny of the Tanypodinae (Chironomidae, Diptera). Invertebr. Syst. 2017, 31, 302–316. [Google Scholar] [CrossRef]
- Zheng, C.-G.; Liu, Z.; Zhao, Y.-M.; Wang, Y.; Bu, W.-J.; Wang, X.-H.; Lin, X.-L. First report on mitochondrial gene rearrangement in non-biting midges, revealing a synapomorphy in Stenochironomus Kieffer (Diptera: Chironomidae). Insects 2022, 13, 115. [Google Scholar] [CrossRef]
- Sari, A.; Duran, M.; Sen, A.; Bardakci, F. Investigation of Chironomidae (Diptera) relationships using mitochondrial COI gene. Biochem. Syst. Ecol. 2015, 59, 229–238. [Google Scholar] [CrossRef]
- Silva, F.L.D.; Ekrem, T. Phylogenetic relationships of nonbiting midges in the subfamily Tanypodinae (Diptera: Chironomidae) inferred from morphology. Syst. Entomol. 2016, 41, 73–92. [Google Scholar] [CrossRef]
- Ma, Z.; Li, R.; Zhu, B.; Zheng, X.; Zhou, C. Comparative mitogenome analyses of subgenera and species groups in Epeorus (Ephemeroptera: Heptageniidae). Insects 2022, 13, 599. [Google Scholar] [CrossRef]
- Ekrem, T.; Willassen, E. Exploring Tanytarsini relationships (Diptera: Chironomidae) using mitochondrial COII gene sequences. Insect Syst. Evol. 2004, 35, 263–276. [Google Scholar] [CrossRef]
- Sari, A.; Duran, M.; Bardakci, F. Discrimination of Orthocladiinae species (Diptera: Chironomidae) by using cytochrome c oxidase subunit I. Acta Zool. Bulg. 2012, 4, 73–80. [Google Scholar]
- Lin, X.L.; Zhao, Y.M.; Yan, L.P.; Liu, W.B.; Bu, W.J.; Wang, X.H.; Zheng, C.G. Mitogenomes provide new insights into the evolutionary history of Prodiamesinae (Diptera: Chironomidae). Zool. Scr. 2022, 51, 119–132. [Google Scholar] [CrossRef]
- Li, S.Y.; Chen, M.H.; Sun, L.; Wang, R.H.; Li, C.H.; Gresens, S.; Li, Z.; Lin, X.L. New mitogenomes from the genus Cricotopus (Diptera: Chironomidae, Orthocladiinae): Characterization and phylogenetic implications. Arch. Insect Biochem. Physiol. 2024, 115, e22067. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Shin, S.C. Complete mitochondrial genome of the Antarctic midge Parochlus steinenii (Diptera: Chironomidae). Mitochondrial DNA A 2016, 27, 3475–3476. [Google Scholar] [CrossRef]
- Andersen, T.; Sæther, O.; Cranston, P.; Epler, J. The larvae of Orthocladiinae (Diptera: Chironomidae) of the Holarctic region–Keys and diagnoses. Insect Syst. Evol. 2013, 66, 189–385. [Google Scholar] [CrossRef]
- Chimeno, C.; Rulik, B.; Manfrin, A.; Kalinkat, G.; Hölker, F.; Baranov, V. Facing the infinity: Tackling large samples of challenging Chironomidae (Diptera) with an integrative approach. PeerJ 2023, 11, e15336. [Google Scholar] [CrossRef]
- Ge, X.; Wang, C.; Pei, W.; Tang, Y.; Liu, W.; Yan, C. New descriptions of the larval and pupal stages of Orthocladius nitidoscutellatus and Psectrocladius nevalis from Xizang, China (Diptera, Chironomidae). Biodivers. Data J. 2024, 12, e121952. [Google Scholar] [CrossRef]
- Cranston, P.S.; Hardy, N.B.; Morse, G.E. A dated molecular phylogeny for the Chironomidae (Diptera). Syst. Entomol. 2012, 37, 172–188. [Google Scholar] [CrossRef]
- Baranov, V.; Lin, X.L.; Huebner, J.; Chimeno, C. Uncovering the hidden diversity of non-biting midges (Diptera, Chironomidae) from central Namibia, using morphology and DNA barcodes. Afr. Invertebr. 2024, 65, 15–36. [Google Scholar] [CrossRef]
- Lei, T.; Zheng, X.; Song, C.; Jin, H.; Chen, L.; Qi, X. Limited Variation in Codon Usage across Mitochondrial Genomes of Non-Biting Midges (Diptera: Chironomidae). Insects 2024, 15, 752. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.-G.; Zhu, X.-X.; Yan, L.-P.; Yao, Y.; Bu, W.-J.; Wang, X.-H.; Lin, X.-L. First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ 2021, 9, e11294. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.; Yiu, S.-M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Delbarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Kück, P.; Meid, S.A.; Groß, C.; Wägele, J.W.; Misof, B. AliGROOVE–visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, Version 1.4.4; Computer Program and Documentation Distributed by the Author. Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 13 April 2025).
- Xiao, X.-R.; Chen, M.-H.; Li, S.-Y.; Guo, B.-X.; Zhang, Y.; Zhang, Z.-C.; Qiao, Y.-J.; Lin, X.-L. Comparative Mitogenomic Analyses of Tanypodinae (Diptera: Chironomidae). Insects 2025, 16, 203. [Google Scholar] [CrossRef]
- Li, S.Y.; Zhao, Y.M.; Guo, B.X.; Li, C.H.; Sun, B.J.; Lin, X.L. Comparative Analysis of Mitogenomes of Chironomus (Diptera: Chironomidae). Insects 2022, 13, 1164. [Google Scholar] [CrossRef]



| Species | Sample ID | Life Stage | Sampling Metadata | GenBank Accession | Reference |
|---|---|---|---|---|---|
| Psectrocladius schlienzi | NAM65 | Adult male | Xigaze, Xizang, China, 29.317° N, 88.934° E, 22 July 2014, leg. X.-L. Lin | PV132385 | this study |
| Psectrocladius barbimanus | XL4313 | Adult male | Nagqu, Xizang, China, 31.733° N, 87.899° E, 6 September 2019, leg. E.-L. Zhang | PV132387 | this study |
| Psectrocladius aquatronus | NX001 | Adult male | Wuzhong, Ningxia, China, 37.737° N, 107.352° E, 27 April 2018, leg. L. Kang | PV132386 | this study |
| Psectrocladius bisetus | NX009 | Adult male | Wuzhong, Ningxia, China, 37.87° N, 107.57° E, 7 May 2018, leg. L. Kang | PV132389 | this study |
| Psectrocladius oligosetus | XL3277 | Larva | Jiuzhaigou, Sichuan, China, 33.228° N, 103.897° E, 12 July 2019, leg. X.-Y. Ge | PV132388 | this study |
| Rheocricotopus emeiensis | XL1426 | Adult male | Leshan, Sichuan, China, 29.574° N, 103.356° E, 23 July 2017, leg. C. Song | OP006246 | [13] |
| Rheocricotopus villiculus | ZJ497 | Adult male | Hangzhou, Zhejiang, China, 30.322° N, 119.442° E, 22 July 2019, leg. X.-L. Lin | MW373526 | [22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.-Y.; Xiao, X.-R.; Zhang, Y.; Zhang, Z.-C.; Zhang, D.-S.; Liu, Z.; Lin, X.-L. Comparative Mitogenomic Analyses of Psectrocladius (Diptera: Chironomidae). Insects 2025, 16, 420. https://doi.org/10.3390/insects16040420
Chen X-Y, Xiao X-R, Zhang Y, Zhang Z-C, Zhang D-S, Liu Z, Lin X-L. Comparative Mitogenomic Analyses of Psectrocladius (Diptera: Chironomidae). Insects. 2025; 16(4):420. https://doi.org/10.3390/insects16040420
Chicago/Turabian StyleChen, Xue-Yao, Xiu-Ru Xiao, Yan Zhang, Zhi-Chao Zhang, Dong-Sheng Zhang, Zheng Liu, and Xiao-Long Lin. 2025. "Comparative Mitogenomic Analyses of Psectrocladius (Diptera: Chironomidae)" Insects 16, no. 4: 420. https://doi.org/10.3390/insects16040420
APA StyleChen, X.-Y., Xiao, X.-R., Zhang, Y., Zhang, Z.-C., Zhang, D.-S., Liu, Z., & Lin, X.-L. (2025). Comparative Mitogenomic Analyses of Psectrocladius (Diptera: Chironomidae). Insects, 16(4), 420. https://doi.org/10.3390/insects16040420






