Simple Summary
As the largest subfamily within Pyralidae, Phycitinae comprises over 3300 known species worldwide. The considerable diversity and high incidence of homoplasy within Phycitinae present significant challenges for species identification when relying exclusively on morphological characters. Therefore, the integration of molecular data analysis is particularly crucial. In this study, we describe a new pyralid moth species, integrating both morphological and molecular evidence. We provide detailed illustrations of adults, wing venation, male and female genitalia, DNA barcodes, and the complete mitochondrial genome of the new species, and conduct a phylogenetic analysis based on mitochondrial genes to get a better understanding of the taxonomic status of the new species.
Abstract
The Nyctegretis seminigra sp. nov. from China is described. The new species is similar to N. triangulella, but can be distinguished by its bicolored forewing pattern and a pointed protrusion on the costa of the valva in males, as well as the signum location in the females. Detailed illustrations of adults, wing venation, and the male and female genitalia of the new species are provided. The DNA barcode of the new species has been successfully sequenced. Genetic divergence analyses among species within Nyctegretis are conducted using pairwise analysis, revealing that all included species exhibit highly species-specific DNA barcodes. The complete mitochondrial genomes of the new species and its related species, N. triangulella, are obtained through next-generation sequencing, followed by a comparative analysis. Furthermore, phylogenetic analyses based on mitogenomes through Maximum Likelihood and Bayesian Inference approaches are also performed to elucidate the taxonomic status of the new species.
1. Introduction
The genus Nyctegretis was established by Zeller [1], with Tinea achatinella Hübner designated as its type species. Roesler [2] systematically revised the Palaearctic species of Nyctegretis and synonymized Synallorema Gozmány and Trichorachia Hampson under the genus. Subsequently, Leraut [3,4] further synonymized Pseudopiesmopoda Roesler and Mesciniadia Hampson with Nyctegretis. It is characterized by forewing with stalked veins R3+R4+5, hindwing with fused M2 and M3, male genitalia featuring a narrow band-like transtilla, and female genitalia with a well-developed antrum and a signum formed by chitinous tubercles. Currently, the genus comprises 11 species [5], with a remarkably broad yet disjunct global distribution spanning the Afrotropical, Australasian, and Palaearctic regions. In China, two species have been recorded [6].
Mitogenomes have been used for phylogenetic analysis in several Lepidoptera groups [7,8]. In Phycitinae, mitogenomes of 25 species from 16 genera have been sequenced (GenBank, https://www.ncbi.nlm.nih.gov, accessed on 9 January 2025). However, most of these sequences lack detailed descriptions. As the largest subfamily within Pyralidae, Phycitinae comprises over 3300 known species worldwide [4,9]. It is obvious that the current mitochondrial genome data are insufficient to adequately represent such a diverse group.
In this study, we describe a new species, N. seminigra sp. nov., based on specimens collected from Guangxi, Hainan, and Yunnan, China. Diagnosis and illustrations of the adults and genitalia are provided for the new species. The complete mitogenomes of N. seminigra sp. nov. and N. triangulella are sequenced and annotated here. This study represents the first report on the mitochondrial genome data for this genus. These findings will contribute to a better understanding of the genus Nyctegretis, and provide valuable data for further phylogeny-based systematic studies.
2. Materials and Methods
2.1. Sample Collection
The specimens examined in this study were collected using 250 W high-pressure mercury lamps on a white sheet. Samples for DNA extraction were preserved in absolute ethanol and stored at −20 °C. The type specimens of N. seminigra sp. nov. are deposited in HAASM and NKU. Detailed depository information for each specimen is indicated in the systematic section.
2.2. Morphological Analyses
Genitalia were dissected according to Li [10]. Images of adults and genitalia were taken using Leica M205A and Leica DM750 microscopes, respectively, coupled with Leica Application Suite 4.6 software (Leica, Wetzlar, Germany). Morphological terminology follows Roesler [2] and Slamka [11].
2.3. DNA Extraction and Sequencing
DNA extraction and mitochondrial cox1 gene amplification were performed following our previous study [12]. The final 658 bp barcode region of cox1 was registered in GenBank (accession numbers: PV156261–PV156270). Two alcohol-preserved specimens were sent to BGI Tech Solutions Co., Ltd. (Shenzhen, China) for DNA extraction and library preparation. Genome sequencing was conducted on the DNBSEQ platform using 150 bp paired-end reads to generate high-quality sequencing data. The complete mitogenomes of N. seminigra sp. nov. and N. triangulella have been submitted to GenBank (accession numbers: PV021108 and PV021107, respectively).
2.4. DNA Barcode Analysis
Seventy-one cox1 sequences from six species of Nyctegretis were used to calculate interspecific and intraspecific divergences. Ten of these sequences were generated using Sanger sequencing, while two sequences were derived from mitochondrial genome data obtained through next-generation sequencing in this study, and the remaining sequences were retrieved from the BOLD System. The BIN numbers are as follows: N. cullinanensis (AEX2771, AEW9294); N. infractalis (AAX3960); N. lineana (AAE5583); N. triangulella (ACN1315); N. ruminella (ACJ7106). Genetic distances were calculated under the Kimura 2-parameter model [13] using MEGA X.
2.5. Mitogenome Analysis
Complete mitogenomes were assembled using MitoZ v3.6 [14]. Annotations were performed using Geneious Prime R11 [15] and MITOS (Galaxy 24.2.rc1) [16]. Nucleotide composition, amino acid usage frequency, and relative synonymous codon usage (RSCU) of the protein-coding genes (PCGs) were calculated using PhyloSuite v1.2.3 [17,18]. Mitogenome maps were depicted using Blast Ring Image Generator (BRIG) [19]. Fifty-nine mitogenomes of Pyralidae (as ingroups) and two mitogenomes of Crambidae (as outgroup sequences) were used for phylogenetic analysis using PhyloSuite v1.2.3.
3. Results
3.1. Systematics
3.1.1. Genus Nyctegretis Zeller, 1848
Type species: Tinea achatinella Hübner, [1824], by monotypy.
Mesciniadia Hampson in Ragonot and Hampson, 1901. Type species: Nephopteryx [sic] infractalis Walker, 1864, by monotypy.
Trichorachia Hampson, 1930. Type species: Trichorachia leonine Hampson, 1930, by monotypy.
Synallorema Gozmány, 1958. Type species: Nyctegretis triangulella Ragonot, 1901, by monotypy.
Pseudopiesmopoda Roesler, 1982. Type species: Pseudopiesmopoda malgassicola Roesler, 1982, by monotypy.
Diagnosis. Species of the genus Nyctegretis share several characteristic head features with those of other genera in the tribe Phycitini: the head is covered with rough scales; the male antenna bears appressed scales dorsally and is pubescent ventrally, lacking sinus in base of the flagellum, while the female antenna is filiform; the labial palpus is slender throughout, extending upward beyond the vertex; and the maxillary palpus is short and slightly flattened. However, Nyctegretis can be distinguished from related genera by its wing venation and structures of the male and female genitalia: the forewing has long-stalked R3 and R4+5 and stalked or fused M2 and M3; the hindwing has fused M2 and M3 shortly stalked with CuA1; the transtilla is narrow and band-like and the relatively wide valva has a rod-like costa that is usually protruding at the end in the male genitalia; the antrum is usually well developed, and the signum (absent in N. aenigmella) consists of a group of triangular or plate-like chitinous tubercles in the female genitalia.
Biology. The larvae of Nyctegretis lineana feed on a variety of plants, including Ononis, Artemisia, Antennaria, Gnaphalium, Sedum, Cytisus, and Trifolium [20]. The larvae of N. ruminella feed on various detritus rather than on green leaves [21]. The host plants of the remaining species within this genus are currently unknown.
Remarks. Gozmány [22] established the genus Synallorema based on the monotypic species Nyctegretis triangulella Ragonot (originally described by Ragonot in Ragonot and Hampson, 1901: 29 [23]), which was known to be found in Japan, Hungary, and eastern Austria. Roesler [2,24] treated Synallorema as a synonym of Nyctegretis, a view followed by Du et al. [6]. However, Fletcher and Nye [25] mistakenly treated Homoeosoma triangulella Hampson (described by Hampson in Ragonot and Hampson, 1901: 256) from “Sou-tcheou” (Suzhou, Jiangsu Province, China) as its type species and thereby regarded Synallorema to be a valid genus name. In agreement with Roesler’s perspective, we hereby treat Synallorema as a synonym of Nyctegretis.
Checklist and Distributions of the Species in Nyctegretis
- 1.
- Nyctegretis aenicta (Turner, 1913) [26]
- Ecbletodes aenicta Turner, 1913: 120. Type locality: Australia (Northern Queensland).
- Distribution: Australia.
- 2.
- Nyctegretis aenigmella Leraut, 2002 [27]
- Nyctegretis aenigmella Leraut, 2002: 148. Type locality: France (Corse).
- Distribution: France.
- 3.
- Nyctegretis cullinanensis Balinsky, 1991 [28]
- Nyctegretis cullinanensis Balinsky, 1991: 111. Type locality: South Africa (Transvaal).
- Distribution: Kenya, South Africa.
- 4.
- Nyctegretis inclinella Ragonot, 1888 [29,30,31]
- Nyctegretis inclinella Ragonot, 1888: 32. Type locality: South Africa (Eastern Cape).
- Distribution: Madagascar, Mozambique, South Africa.
- 5.
- Nyctegretis infractalis (Walker, 1864) [32,33]
- Nephopteryx infractalis Walker, 1864: 958. Type locality: Malaysia (Borneo).
- Distribution: Australia, Indonesia, Malaysia.
- 6.
- Nyctegretis leonina (Hampson, 1930) [34]
- Trichorachia leonina Hampson, 1930: 65. Type locality: Sierra Leone, Zimbabwe (South Rhodesia).
- Distribution: Sierra Leone, South Africa, Zimbabwe.
- 7.
- Nyctegretis lineana (Scopoli, 1786) [2,6,35,36,37,38]
- Phalaena lineana Scopol, 1786: 57. Type locality: Italy (Insubria).
- Tinea achatinella Hübner, 1824: pl. 68. Type locality: Europe.
- Nyctegretis achatinella var. griseella Caradja, 1910: 130. Type locality: France (Dax, Vernet), Kazakhstan (Oral).
- Nyctegretis calamitatella Roesler, 1973: 290. Type locality: China (Shanxi).
- Nyctegretis lineana katastrophella Roesler, 1970: 47. Type locality: Mongolia (Cojbalsan aimak)
- Distribution: Austria, China, England, Finland, Germany, Italy, Japan, Korea, Mongolia, Netherlands, Norway, Russia, and Spain.
- 8.
- Nyctegretis malgassicola (Roesler, 1982) [3,31,39]
- Pseudopiesmopoda malgassicola Roesler, 1982: 858. Type locality: Madagascar.
- Nyctegretis malgassicola insularis Leraut, 2019: 38. Type locality: Comoros (Grande Comore).
- Distribution: Comoros, Madagascar, Mascareignes.
- 9.
- Nyctegretis otoptila (Turner, 1913) [26]
- Ecbletodes otoptila Turner, 1913: 120. Type locality: Australia (Northern Territory).
- Distribution: Australia.
- 10.
- Nyctegretis ruminella La Harpe, 1860 [2,21,35,40]
- Nyctegretis ruminella La Harpe, 1860: 397. Type locality: Italy (Sicily).
- Distribution: Bulgaria, France, Gibraltar, Greece, Italy, Malta, North Africa, Romania, Russia, Spain, and Turkey.
- 11.
- Nyctegretis triangulella Ragonot, 1901 [2,6,35,36,37,38]
- Nyctegretis triangulella Ragonot, in Ragonot and Hampson, 1901: 29. Type locality: Japan.
- Nyctegretis impossibilella Roesler, 1969: 205. Type locality: Greece (Platamon).
- Distribution: Austria, China, Czech Republic, England, Germany, Greece, Hungary, Iraq, Iran, Israel, Italy, Japan, Romania, Russia, Slovakia, and Turkey.
- 12.
- Nyctegretis seminigra Yang, Zhou and Ren, sp. nov.
- Type locality: China (Guangxi, Hainan, Yunnan).
- Distribution: China (Guangxi, Hainan, Yunnan).
Key to the species of Nyctegretis
| 1. Forewing with distinct antemedial line and postmedial line …………………………….2 |
| - Forewing with antemedial line and postmedial line indistinct or absent ……………....7 |
| 2. Underside of male forewing with oblique ridge of long hair from origin of R4 to M1 before termen [34] …………………………………………………………………..N. leonina |
| - Underside of male forewing without oblique ridge of long hair ……………….………..3 |
| 3. Signum band-like ……………………………………………………………………………...4 |
| - Signum raindrop-shaped or oval …………………………………………………………….6 |
| 4. Costa of valva without protrusion [30,31] ……………………………………...N. inclinella |
| - Costa of valva with a protrusion ………………….…………………………………………5 |
| 5. Subapical protrusion on costa of valva small and pointed; signum located in the anterior half of the corpus bursa ……………………………………..N. seminigra sp. nov. |
| - Subapical protrusion on costa of valve large, split into a stocky extension and a curved spine; signum located in the posterior half of the corpus bursa [2,35] …...N. triangulella |
| 6. Uncus rounded at apex; signum raindrop-shaped [2,21] ……….…………...N. ruminella |
| - Uncus narrowed and pointed at apex; signum oval [2,41] ……………………...N. lineana |
| 7. Forewing with a white costal band …………………………………………………………8 |
| - Forewing without white costal band …………………………………….………………...10 |
| 8. Forewing with basal and external area yellow, medial area dark brown; signum absent [27] ………………………………………………………………………..N. aenigmella |
| - Forewing gray or yellowish brown; signum present ……………………………………...9 |
| 9. Costa of valva clubbed; signum triangular [28] …………………………..N. cullinanensis |
| - Costa of valva bifurcated; signum oval [39];………………………………..N. malgassicola |
| 10. Hindwing normal, not arched at base of costa [26] …….……………………..N. aenicta |
| - Hindwing arched at base of costa, with a tuft of long hair ……………………………..11 |
| 11. Forewing with CuA1 stalked with CuA2 [26] ………………………………....N. otoptila |
| - Forewing with CuA1 separated from CuA2 [33] ……………………………..N. infractalis |
3.1.2. Nyctegretis seminigra Yang, Zhou, and Ren, sp. nov.
Zoobank: urn:lsid:zoobank.org:act:CA7E2E67-D432-47B0-88B3-679C987760BD
Type material. Holotype ♂, CHINA, Hainan Province, Wuzhishan, alt. 580 m, 11.IV.2013, leg. Ying-Dang Ren, Xiao-Guang Liu (HAASM). Paratypes: Hainan Province: 8♂1♀, same data as holotype, except dated 11–12. IV.2013 (HAASM); 1♀, Wuzhishan, alt. 700 m, 19.V.2007, leg. Zhi-Wei Zhang, Wei-Chun Li, genitalia slide No. LJY11392; 3♂, Jianfengling, alt. 960 m, 13–15.IV.2013, leg. Ying-Dang Ren, Xiao-Guang Liu (HAASM); 1♂, Diaoluoshan, alt. 900 m, 9.IV.2013, leg. Ying-Dang Ren, Xiao-Guang Liu (HAASM); 1♂, Wuzhishan City, Shuiman Town (18.90 N, 109.67 E), alt. 735 m, 25.VIII.2020, leg. Linlin Yang, genitalia slide No. DNAYLL18393 (HAASM); 1♂, Yinggeling, alt. 620 m, 28.III.2010, leg. Bingbing Hu, genitalia slide No. LJY10453. Guangxi Zhuang Autonomous Region: 1♀, Chongzuo City, Longzhou County, Pona, Nonggang Nature Reserve (22.49 N, 106.95 E), alt. 160 m, 19.VIII.2020, leg. Linlin Yang, genitalia slide No. DNAYLL18388 (HAASM). Yunnan Province: 3♂3♀, Honghe Prefecture, Pingbian County, Daweishan, alt. 1800 m, 6.XI.2010, leg. Bingbing Hu et al., genitalia slide Nos. DNAYLL18046m, WYQ14041f (NKU); 1♂, Puer City, Yunpanshan Town, Taiyanghe Nature Reserve, alt. 1450 m, 2.ix.2014, leg. Zhenguo Zhang, genitalia slide No. DNAYLL18041 (NKU); 1♀, Puer City, Yunpanshan Town, Taiyanghe Nature Reserve, alt. 1450 m, 3.VI.2014, leg. Zhenguo Zhang (NKU); 1♂, Puer City, Yunpanshan Town, Taiyanghe Nature Reserve, alt. 1580 m, 6.viii.2020, leg. Linlin Yang, genitalia slide No. DNAYLL18399 (HAASM).
Diagnosis. Externally, the new species can be easily distinguished from its relatives by the bicolored forewing pattern. The genitalia structures of the new species closely resemble those of N. triangulella, but can be differentiated from the latter by the costa of the valva with a small, pointed protrusion subapically in the male, and the signum located in the anterior half of the bursa in the female. In N. triangulella, the costa of the valva is unequally split at the terminal end into a stocky extension and a narrowly curved spine in the male genitalia, and the signum is located in the posterior half of the bursa in the female genitalia.
Description. Adult (Figure 1). Wingspan 14.0 mm in holotype, 13.5–15.0 mm in male paratypes, 14.5–17.5 mm in female paratypes.
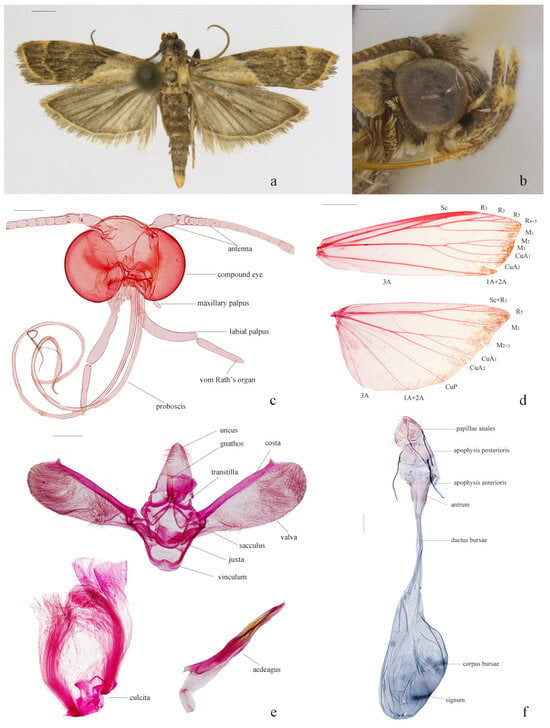
Figure 1.
Morphological features of Nyctegretis seminigra sp. nov. (a), adult, holotype, male; (b), head in lateral view, paratype, male; (c), dissected head, paratype, female; (d), wing venation, paratype, female; (e), male genitalia, paratype; (f), female genitalia, paratype. ((c,d,f), slide No. DNAYLL18393; (e), slide No. LJY10453; (a), scale bar = 1 mm; (b–e), scale bars = 0.25 mm).
Head (Figure 1b,c). Frons with appressed pale gray scales, shining silvery; vertex grayish brown, with dark-tipped, rough, forward-directed scales; ochreous-yellowish occiput. Antenna: scape 1.5-times as long as wide, with a blackish brown dorsal surface, pale yellow ventral surface; yellowish-brown flagellum, scales tipped with black. Labial palpus curved upwards, reaching above the vertex, segmental ratio about 1:1.8:1 in males, 1:1.5:1.2 in females; first segment pale yellow, tinted with a few dark scales, terminal two segments dark brown but grayish yellow at distal ends. Maxillary palpus yellowish brown, about 1/4 length of labial palpus, three-segmented, of nearly equal length. Thorax. Patagium, tegula, and thorax bronze, individual scales dark-tipped. Foreleg dark brown, except yellowish brown at apices of tarsomeres; mid- and hindleg predominantly grayish yellow, interspersed with blackish-brown markings, tarsomeres dark brown, margined with grayish white at apices.
Wings (Figure 1a,d). Forewing with a bicolored pattern: basal area grayish yellow, with a few scattered blackish brown scales at base, along costa, and dorsum; medial and external areas blackish brown, faintly tinged with grayish yellow; antemedial line grayish yellow, hardly distinguishable from basal area, extending from near basal 2/5 of costa outwardly oblique to distal 2/5 of dorsum; postmedial line grayish yellow, slightly dentate and sinuate, extending from distal 1/4 of costa to distal 1/5 of dorsum; discal spots confluent into a grayish yellow line; terminal line yellowish brown; cilia dark gray. Hindwing scales grayish brown to dark gray; cilia grayish brown. Venation: forewing with R3 and R4+5 stalked for about 2/3 of their total length, R2 stalked R3+R4+5 about half length of R2, M2, and M3 stalked at basal 1/4 and terminating at the same point; hindwing with Sc+R1 and Rs stalked for about half length of Sc+R1, M2 and M3 fused and shortly stalked with CuA1.
Male genitalia (Figure 1e). Uncus triangular, length approximately 1.3 times its width, tapered to a slightly pointed apex, densely hirsute on distal third. Gnathos 3/4 length of uncus, of nearly equal width throughout, rod-like, truncate at apex. Transtilla slender, arched, band-like. Valva about three times length of width, narrow at base, gradually broadened to rounded apical margin; costa not reaching to the end of the valva, clubbed, with a pointed protrusion subapically; sacculus about 2/5 length of valva; a clasper-like, elevated chitin structure beyond apex of sacculus; Vinculum rounded trapezoidal, length about 2/3 of greatest width. Juxta U-shaped, middle part rounded quadrilateral, lateral lobes slender, fingerlike, about 2/3 length of gnathos. Aedeagus clavate, about 4/5 length of valva, base slightly broader than apex; with a thorn-like cornutus. Culcita one pair, simple, about 4/5 length of valva.
Female genitalia (Figure 1f). Papillae anales subtriangular, approximately twice as long as its width, apex rounded, sparsely covered with long setae. Eighth tergite about 3/4 as long as its width, anterior margin slightly concave in middle, posterior margin straight. Apophysis anterioris 1.3-times length of apophysis posterioris. Ostium bursae rather wide. Antrum strongly sclerotized, cup-shaped, approximately 3/4 as long as its width. Ductus bursae membraneous, gradually expanding into corpus bursae. Corpus bursae about 1.6-times as long as its width, elliptically rounded, densely covered with granules at the junction with ductus bursae. Signum represented by a band-like structure composed of numerous plate-like chitinous tubercles arranged obliquely, located in the anterior 1/5 to 1/2 region of corpus bursae. Ductus seminalis arising from near middle of corpus bursae, approximate to signum.
Biology. Larval host plant is unknown.
Distribution. China (Guangxi, Hainan, Yunnan).
Etymology. The species name is derived from the Latin prefix semi- (meaning half), and the Latin word nigrum (meaning black), referring to the forewing of the new species that has a blackish brown distal half (including the medial and external areas).
3.2. DNA Barcode Analysis
The genetic distance among six species of Nyctegretis was determined through pairwise analysis of 71 sequences. The results (Table 1) indicate that all included species possess highly species-specific DNA barcodes. The minimum interspecific divergences to the nearest species ranged from 7.16% to 9.82%, while the maximal intraspecific distances ranged from 0.46% to 2.58%. For N. seminigra sp. nov., the intraspecific genetic divergence among the six barcoded specimens ranged from 0 to 1.26%. Its genetically closest relatives are N. lineana and N. triangulella, with interspecific divergences of 9.66% to 10.36% and 9.73% to 11.71%, respectively.

Table 1.
Pairwise distance matrix of DNA barcodes calculated among seven species of Nyctegretis.
3.3. Mitogenome Analysis
The mitochondrial genome is 15,198 bp-long in N. seminigra sp. nov. and 15,205 bp-long in N. triangulella. The structure and composition of the mitochondrial genomes in both species are consistent with those of other pyraloid moths, containing a total of 37 genes: 13 PCGs, 22 tRNA genes, two rRNA genes; and one A+T rich region (Figure 2; Table S1). Among these genes, 23 are located on the majority strand (J-strand), whereas the remaining 14 genes are encoded on the minority strand (N-strand). Both mitogenomes are compact, with no gene rearrangements or gene losses. In N. seminigra sp. nov., there are eight gene overlaps ranging from 1 bp to 8 bp in length, with the largest overlap located between trnW and trnC. In contrast, N. triangulella exhibits ten gene overlaps ranging from 1 bp to 12 bp in length, and the largest overlap is found between rrnL and trnV. N. seminigra sp. nov. contains 14 gene spacers ranging from 2 bp to 44 bp in length, while N. triangulella has 12 intergenic spacers ranging from 2 bp to 42 bp in length. The largest intergenic spacer in both species is located between trnQ and nad2, consistent with observations from other studies on pyraloid moths [42,43].
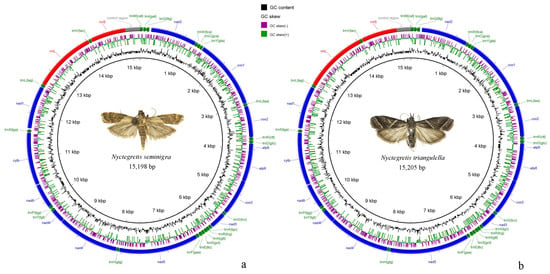
Figure 2.
Complete mitogenome maps. (a), Nyctegretis seminigra sp. nov.; (b), N. triangulella. Arrows indicate the orientation of gene transcription. GC skew is shown on the outer surface of the ring whereas GC content is shown on the inner surface. The anticodon of each tRNA gene is shown in parentheses.
3.3.1. Nucleotide Composition
The base composition of the genes in the mitogenomes of N. seminigra sp. nov. and N. triangulella is summarized in Table S1. The A+T content in the entire nucleotide sequence is 79.2% for N. seminigra sp. nov. and 80.4% for N. triangulella, indicating a clear AT bias in both species. This bias is consistent across all genetic elements. Both species exhibit negative AT skewness and negative GC skewness values throughout their entire sequences, suggesting a higher abundance of T over A and C over G. This pattern is consistent with the base composition observed in other groups within the superfamily Pyraloidea. In both species, the PCGs and A+T rich regions display negative AT skewness values, indicating a preference for T over A. In contrast, the tRNA and rRNA gene regions show positive AT skewness values, revealing a preference for A over T. The GC skewness values for the PCGs, tRNA genes, and rRNA genes are positive in both species, indicating a preference for C over G in these regions.
3.3.2. PCG Regions and Codon Usage
The combined lengths of PCGs in the two mitogenomes are 11,184 bp (N. seminigra sp. nov.) and 11,190 bp (N. triangulella), respectively (Table S2). Comparative analysis revealed that the majority of individual genes exhibited near-identical lengths between the two species, with one exception observed in nad2, displaying lengths of 1008 bp in N. seminigra sp. nov. mitogenome compared to 1014 bp in N. triangulella mitogenome. In both mitogenomes, atp8 is the shortest gene while nad5 is the longest (Table S1).
Comparative analysis of start and stop codon usage revealed consistent and divergent patterns between the two mitochondrial genomes. Most of the start codons are consistent between the two mitogenomes, with ATG used in five PCGs (atp6, cox3, nad4, nad4L, cytb), and ATT in another five PCGs (nad2, cox2, atp8, nad3, nad5); CGA is used as the start codon for cox1 and ATA is used for nad1. The start codon for nad6 differs between the N. seminigra sp. nov. mitogenome (ATA) and N. triangulella mitogenome (ATT). The most common identical stop codon in both the mitogenomes is TAA (in six PCGs: nad2, atp8, atp6, cox3, nad4L, cytb), followed by two incomplete T stop codon (cox2, nad5), one incomplete TA stop codon for nad4, and one TAG for nad3. For the remaining three PCGs, the stop codons differ between the two mitogenomes: nad6 and nad1 use TAG as their stop codon, and cox1 uses TAA in the N. seminigra sp. nov. mitogenome; meanwhile, nad6 and nad1 use TAA and cox1 has an incomplete T stop codon in the N. triangulella mitogenome.
The amino acids’ frequency and relative synonymous codon usage (RSCU) values for PCGs in the two mitogenomes are summarized in Table S3 and Figure S1. The most common amino acid in both mitogenomes is Ile, with a frequency of 12.21% in the N. seminigra sp. nov. mitogenome, and 12.31% in the N. triangulella mitogenome. The least frequently used amino acid is Cys, with respective frequencies of 0.78% and 0.86%. Codon-level analysis demonstrated a significant AT bias, evidenced by a preferential usage of NNA and NNU codons across both mitogenomes. In both the mitogenomes, 26 codons exhibit positive codon usage bias (RSCU > 1), indicating that these codons are used more frequently. In contrast, 36 codons in the N. seminigra sp. nov. mitogenome and 35 codons in the N. triangulella mitogenome show a negative codon usage bias (RSCU < 1). Among these, two codons, CUG (Leu1) and CGC (Arg), are used only in the N. seminigra sp. nov. mitogenome, while CCG (Pro) is used only in the N. triangulella mitogenome; one codon, AGG (Ser1) is never used in either mitogenome.
3.3.3. tRNA Genes, rRNA Genes, and the A+T Rich Region
The mitochondrial tRNA genes are AT-rich, with N. seminigra sp. nov. mitogenome exhibiting a 80.4% AT-content versus 81.3% in the N. triangulella mitogenome, accompanied by positive AT-skew (0.011 and 0.015) and GC-skew (0.184 and 0.166) values, respectively (Table S2). The relative positions and anticodon sequences of tRNAs in both mitogenomes are identical, spanning 63–71 bp in length. The cloverleaf secondary structures (Figures S2 and S3) of the tRNA genes are generally similar between the two mitogenomes. Both mitogenomes lack the DHU arm in trnS1 and TΨC loop in trnI. In addition to the typical Watson–Crick base pairs (A-U and G-C), a small number of U-U mismatches are observed in the TΨC arm of trnE and anticodon arm of trnS2 in both mitogenomes. The N. seminigra sp. nov. mitogenome exhibits additional U-U mismatches localized in the anticodon arm of trnQ and trnR.
The mitochondrial rRNA genes are also AT-rich: 84.5% in the N. seminigra sp. nov. mitogenome, and 84.6% in the N. triangulella mitogenome; both genes show positive AT-skew (0.071 and 0.091) and GC-skew (0.368 and 0.363) values (Table S2). The two rRNA genes (rrnL and rrnS) are encoded on the N-strand and are separated by trnV (Figure 2). The rrnS gene (774 bp) and rrnL gene (1372 bp) in the N. seminigra sp. nov. mitogenome are slightly shorter than those in the N. triangulella mitogenome (781 bp and 1390 bp, respectively).
The A+T rich region is the longest noncoding region in both mitogenomes, with a length of 320 bp in N. seminigra sp. nov. mitogenome and 312 bp in N. triangulella mitogenome. Characterized by its high AT content, this region also displays a negative AT skew and GC skew.
3.3.4. Phylogenetic Analysis Based on Mitogenomes
A phylogenomic framework (Figure 3) was reconstructed using concatenated mitogenomes from two newly sequenced specimens and 57 publicly available sequences, analyzed through Maximum Likelihood (ML) and Bayesian Inference (BI) approaches. Phycitinae is confirmed as a robustly supported monophyletic lineage (BS = 100, PP = 1.00), which bifurcates into two major clades, with Clade A (BS = 93, PP = 1.00) exhibiting stronger nodal support than Clade B (BS = 74, PP = 1.00). The monophyly of each genus is well supported, although intergeneric relationships remain poorly resolved. The phylogenetic position of the new species is highly supported in both analyses (BS = 100, PP = 1.00), which is consistent with morphological evidence and confirms its classification within the genus Nyctegretis.
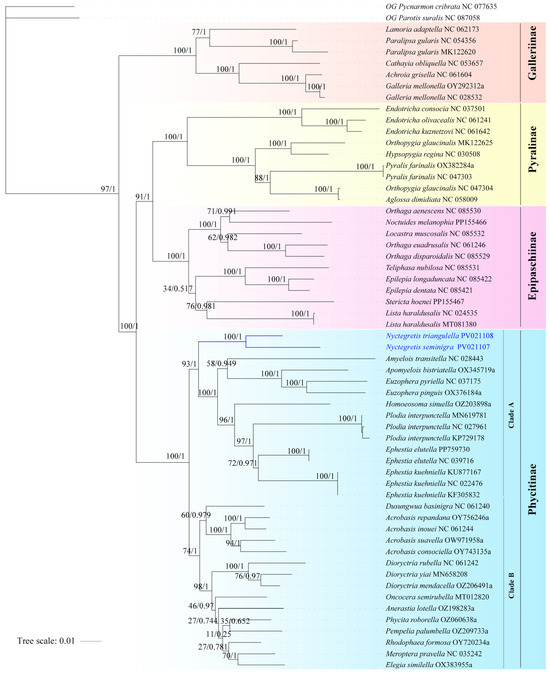
Figure 3.
Maximum likelihood (ML) and Bayesian inference (BI) phylogenetic tree of Pyralidae based on mitogenomes (PCG dataset). Numbers near branches refer to bootstrap support values and Bayesian posterior probabilities. Species names are followed by their corresponding GenBank accession numbers.
4. Discussion
The new species is classified within the genus Nyctegretis based on the following morphological characteristics: the forewing with stalked R3 + R4+5 and stalked M2 + M3, the hindwing with M2 and M3 fused and stalked with CuA1; the transtilla is characterized as a narrow band, and the broad valva has a rod-like costa that protrudes at its end in the male genitalia; the antrum is well-developed and the signum comprises a cluster of plate-like chitinous tubercles in the female genitalia. The results of the molecular phylogenetic analysis further corroborate this classification, demonstrating that mitochondrial genome data are valuable resources for species identification and taxonomic status determination.
It should be noted that some characteristics of Nyctegretis are also observed in other phycitines, and there are individual species within Nyctegretis that do not adhere to the genus characteristics (e.g., N. aenigmella lacks a signum; N. infractalis with stalked CuA1 + CuA2 in the forewing). This is common in Phycitinae. As indicated by Roe et al. [44], the considerable diversity within this subfamily, coupled with the high incidence of homoplasy (similar traits evolve independently in different lineages), poses great challenges for reconstructing phylogenetic relationships based solely on morphological features. In this case, the integration of molecular data analysis becomes particularly crucial. Roe et al. [44] provided the first molecular phylogeny of phycitine genera, resulting in the division of the subfamily into two clades. Similar results were obtained in later studies on the molecular phylogeny of Pyraloidea [9,45,46], including findings in this study. It is noteworthy, however, that these studies did not comprehensively sample most taxa of Phycitinae, focusing mainly on species from Phycitini. Only one species of Anerastiini (Peoria approximella) was included in the analysis and it is nested within Phycitini [44]. Additional representatives will need to be sampled to achieve a comprehensive molecular phylogenetic analysis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects16040413/s1, Figure S1: Amino acid frequency (values at the top of each bar) and relative synonymous codon usage of protein-coding genes in the mitogenomes of Nyctegretis seminigra sp. nov. and N. triangulella; Figure S2: Predicted secondary structure of tRNAs in Nyctegretis seminigra sp. nov. mitogenome; Figure S3: Predicted secondary structure of tRNAs in Nyctegretis triangulella mitogenome. Table S1: Organization of the mitochondrial genome for Nyctegretis seminigra sp. nov. (Ns) and N. triangulella (Nt); Table S2: Nucleotide composition and skewness of mitochondrial gemome for Nyctegretis seminigra sp. nov. (Ns) and N. triangulella (Nt); Table S3: Codon usage in mitochondrial PCGs of N. seminigra sp. nov. (NS) and N. triangulella (Nt).
Author Contributions
Conceptualization, L.Y. and Y.R.; methodology, L.Y.; software, L.Y. and Y.Z.; formal analysis, L.Y.; specimen collection, Y.R.; writing—original draft preparation, L.Y.; writing—review and editing, Y.R.; funding acquisition, L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Joint Fund of Henan Science and Technology R&D program (Advantageous Discipline Development), grant number 232301420119.
Data Availability Statement
DNA barcodes (PV156261–PV156270) and mitogenomes (PV021107–PV021108) generated in this study have been deposited in GenBank. Type specimens are deposited at HAASM and NKU. Publicly archived datasets used in this study were retrieved from the BOLD System (see materials and methods section for BIN numbers) and GenBank (see Figure 3 for accession numbers).
Acknowledgments
We’re grateful to Houhun Li (Nankai University, Tianjin) for providing the specimens used in our research, and to our colleagues who collected the specimens in the field. We also extend our gratitude to Tianyou Zhao (China Agricultural University, Beijing), and Ci Tang (Southwest University, Chongqing) for their suggestions on mitogenome assembly and annotation.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| HAASM | Insect Collection of the Institute of Plant Protection, Henan Academy of Agricultural Sciences, Zhengzhou, China |
| NKU | Insect Collection, College of Life Sciences, Nankai University, Tianjin, China |
| RSCU | relative synonymous codon usage |
| PCGs | protein-coding genes |
| Ns | Nyctegretis seminigra sp. nov. (in Tables S1–S3) |
| Nt | Nyctegretis triangulella (in Tables S1–S3) |
References
- Zeller, P.C. Die Gallerien und nackthornigen Phycideen. Isis Von Oken 1848, 8, 641–691. Available online: https://www.biodiversitylibrary.org/bibliography/13271 (accessed on 4 December 2024).
- Roesler, R.U. Phycitinae. Trifine Acrobasiina. In Microlepidoptera Palaearctica; Amsel, H.G., Gregor, F., Reisser, H., Eds.; Verlag Georg Fromme & Co.: Wien, Austria, 1973; Volume 4, pp. Part 1: i–xvi, 1–752; Part 2: 1–137, pls 1–170. [Google Scholar]
- Leraut, G.H.C. Contribution à la connaissance des Pyralidae Phycitinae de Madagascar, des Comores et des Mascareignes. I. Révision et inventaire des Phycitinae des Comores et description de 2 genres, 8 espèces et une sous-espèce. Antenor Études De Lépidoptérologie Trop. 2019, 6, 29–46. [Google Scholar]
- Leraut, G.H.C. A global comprehensive check-list of the Phycitinae (Lep.: Pyraloidea, Pyralidae). Rev. Française D’entomologie Générale 2021, 2 (Suppl. S5–S6), 1–371. [Google Scholar]
- Nuss, M.; Landry, B.; Mally, R.; Vegliante, F.; Tränkner, A.; Bauer, F.; Hayden, J.; Segerer, A.; Schouten, R.; Li, H.; et al. Global Information System on Pyraloidea. 2003–2024. Available online: http://www.pyraloidea.org (accessed on 28 January 2025).
- Du, Y.L.; Li, H.H.; Wang, S.X. A study on the genus Nyctegretis zeller from China (Lepidoptera: Pyralidae: Phycitinae). Acta Sci. Nat. Univ. Nankaiensis 2001, 34, 98–102. [Google Scholar]
- Timmermans, M.J.T.N.; Lees, D.C.; Simonsen, T.J. Towards a mitogenomic phylogeny of Lepidoptera. Mol. Phylogenetics Evol. 2014, 79, 169–178. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, L.; Liao, C.Q.; Wang, X.; Wang, M.; Huang, G.H. Comparative mitochondrial genome analysis and phylogenetic relationship among lepidopteran species. Gene 2022, 830, 146516. [Google Scholar] [CrossRef]
- Léger, T.; Mally, R.; Neinhuis, C.; Nuss, M. Refining the phylogeny of Crambidae with complete sampling of subfamilies (Lepidoptera, Pyraloidea). Zool. Scr. 2021, 50, 84–99. [Google Scholar] [CrossRef]
- Li, H.H. The Gelechiidae of China (I) (Lepidoptera: Gelechioidea); Nankai University Press: Tianjin, China, 2002; pp. 1–538. [Google Scholar]
- Slamka, F. Pyraloidea of Europe; Phycitinae Part 1; František Slamka: Bratislava, Slovakia, 2019; Volume 4, pp. 1–432. [Google Scholar]
- Yang, L.L.; Li, H.H. First report of the genus Pelecystola Meyrick (Lepidoptera, Tineidae) in China, with description of a new species. ZooKeys 2021, 1046, 189–206. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Meng, G.L.; Li, Y.Y.; Yang, C.T.; Liu, S.L. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, 63. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones Havas, S.; Cheung, M.; Sturrock, S. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Zakour, N.L.B.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genome 2011, 12, 402. [Google Scholar] [CrossRef]
- Hasenfuss, I. Die Larvalsystematik der Zünsler (Pyralidae). Abh. Zur Larvalsystematik Der Insekten 1960, 5, 1–263. [Google Scholar]
- Huertas-Dionisio, M.; Bernabé-Ruiz, P.M. Estados inmaturos de Lepidoptera (LXIII). Nyctegretis ruminella (La Harpe, 1860) en Huelva, España (Lepidoptera: Pyralidae, Phycitinae). SHILAP Rev. De Lepidopterol. 2023, 51, 123–131. [Google Scholar] [CrossRef]
- Gozmány, L. Notes on Hungarian Phycitidae (Lepidoptera). Ann. Hist. -Nat. Musei Natl. Hung. 1958, 50, 223–225. Available online: http://publication.nhmus.hu/pdf/annHNHM/Annals_HNHM_1958_Vol_50_223.pdf (accessed on 12 December 2024).
- Ragonot, E.L.; Hampson, G.F. Monographie des Phycitinae et des Galleriinae. In Mémoires sur les Lépidoptères; Romanoff, N.M., Ed.; Imprimerie de M.M. Stassuléwitch: Saint Petersburg, Russia, 1901; Volume 8, pp. i–xli, 1–602, pls 24–57. [Google Scholar] [CrossRef]
- Roesler, R.U. Das neue systematische Verzeichnis der deutschen Phycitinae (Lepidoptera, Pyralidae). Nachrichtenblatt Der Bayer. Entomol. 1968, 17, 25–28. [Google Scholar]
- Fletcher, D.S.; Nye, I.W.B. Pyraloidea. In The Generic Names of the Moths of the World; Natural History Museum Publications: London, UK, 1984; Volume 5, pp. i–xvi+1–185. [Google Scholar] [CrossRef]
- Turner, A.J. Studies in Australian Lepidoptera, Pyralidae. Proc. R. Soc. Qld. 1913, 24, 111–163. [Google Scholar] [CrossRef]
- Leraut, P.J.A. Contribution à l’étude des Phycitinae (Lepidoptera, Pyralidae). Nouv. Rev. D’entomologie 2002, 9, 141–177. [Google Scholar]
- Balinsky, B.I. New and inadequately described genera and species of Phycitinae (Pyralidae, Lepidoptera) from southern Africa. South Afr. J. Zool. 1991, 26, 93–114. [Google Scholar] [CrossRef]
- Ragonot, E.L. Nouveaux genres et espèces de Phycitidae & Galleriidae; Publié par l’auteur: Paris, France, 1888; pp. 1–52. [Google Scholar] [CrossRef]
- Balinsky, B.I. On genitalia of some southern African Phycitinae (Lepidoptera, Phycitinae). South Afr. J. Zool. 1991, 26, 11–35. [Google Scholar] [CrossRef]
- Leraut, G.H.C. Contribution à la connaissance des Pyralidae Phycitinae de Madagascar, des Comores et des Mascareignes (III)—Révision et inventaire des espèces présentes à Madagascar et dans les Mascareignes. Supplément à la Faune de Madagascar. Rev. Française D’entomologie Générale 2019, 1, 38–198. Available online: https://www.researchgate.net/publication/336346084 (accessed on 8 December 2024).
- Walker, F. Tineites. In List of the Specimens of Lepidopterous Insects in the Collection of the British Museum; Printed by order of the Trustees: London, UK, 1864; Volume 30, pp. 837–1096. [Google Scholar] [CrossRef]
- Roesler, R.U. Die Phycitinae von Sumatra (Lepidoptera: Pyralidae); Heterocera Sumatrana: Keltern, Germany, 1983; pp. 1–136, pls 1–69. [Google Scholar]
- Hampson, G.F. New genera and species of Phycitinae (Lepidoptera, Pyralidae). Ann. Mag. Nat. Hist. Incl. Zool. Bot. Geol. 1930, 5, 50–80. [Google Scholar] [CrossRef]
- Leraut, P.J.A. Moths of Europe, Pyralids 2; N.A.P. Editions: Verrières-le-Buisson, France, 2014; pp. 1–441, pls 1–69, text figs 1–190. [Google Scholar]
- Bae, Y.S.; Byun, B.K.; Paek, M.K. Pyralid Moths of Korea (Lepidoptera, Pyraloidea); Korea National Arboretum, SAMSUNGAD. COM: Seoul, Republic of Korea, 2008; pp. 1–426. [Google Scholar]
- Hirowatari, T.; Nasu, Y.; Sakamaki, Y.; Kishida, Y. The Standard of Moths in Japan; Gakken Education Publishing: Tokyo, Japan, 2013; Volume 3, pp. 1–359. [Google Scholar]
- Sinev, S.Y.; Streltzov, A.N.; Trofimova, T.A. Pyralidae. In Catalogue of the Lepidoptera of Russia, 2nd ed.; Sinev, S.Y., Ed.; Zoological Institute RAS: Saint Petersburg, Russia, 2019; pp. 165–178. [Google Scholar]
- Roesler, R.U. Neue Taxa für die Phycitinen-Fauna von Madagaskar. Phycitinen-Studien XX (Lepidoptera, Pyralidae). Bull. Du Muséum Natl. D’histoire Nat. 1982, 3, 855–891. [Google Scholar] [CrossRef]
- La Harpe, J.-J.C. Contributions a la faune de la Sicile. Lépidoptères. Bull. De La Société Vaudoise Des Sci. Nat. 1860, 6, 386–418. [Google Scholar]
- Kasy, F. Korrekturen und Bemerkungen zur Bearbeitung der Gattung Nyctegretis Zeller in Microlepidoptera Palearctica, Bd. 4 (Lepidopt.; Pyralidae, Phycitinae). Z. Der Arbeitsgemeinschaft Osterr. Entomol. 1975, 26, 51–60. Available online: https://www.zobodat.at/pdf/ZAOE_26_0051-0060.pdf (accessed on 8 January 2025).
- Chen, S.; Li, F.H.; Lan, X.E.; You, P. Complete mitochondrial genomes of three Spilomelinae species and a preliminary phylogenetic analysis of the Pyraloidea (Insecta: Lepidotera). Chin. J. Appl. Entomol. 2017, 54, 22–34. [Google Scholar] [CrossRef]
- Wang, R.H.; Lei, Z.Y.; Kuang, M.Z.; Liu, Y.; Wang, Y.; Rong, H. Sequencing and analysis of the complete mitochondrial genome of Pycnarmon cribrata (Fabricius, 1794) (Crambidae: Spilomelinae). J. South. Agric. 2024, 55, 1355–1365. [Google Scholar] [CrossRef]
- Roe, A.D.; Simonsen, T.J.; Scholtens, B.; Sperling, F.A.H.; Weller, S.J. Phycitinae Phylogeny Based on Two Genes, with Implications for Morphological Trait Evolution and Heinrich’s Tribal Classification (Lepidoptera: Pyralidae). J. Lepid. Soc. 2015, 69, 157–172. [Google Scholar] [CrossRef]
- Qi, M.; Zhao, H.; Yu, F.; Zhang, A.; Li, H. The first mitogenomes of the subfamily Odontiinae (Lepidoptera, Crambidae) and phylogenetic analysis of Pyraloidea. Insects 2021, 12, 486. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qi, M.; Xu, H.; Wu, Z.; Hu, L.; Yang, M.; Li, H. Nine Mitochondrial Genomes of the Pyraloidea and Their Phylogenetic Implications (Lepidoptera). Insects 2021, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).