Chitin Deacetylase Gene Family Positively Regulates the Accumulation of Rice Stripe Virus in Laodelphax striatellus Fallén (Hemiptera: Delphacidae) Ovaries
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RSV, Plants and Insects
2.2. Identification and Phylogenetic Analysis of CDAs
2.3. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.4. Cloning and Characterization of LsCDA1
2.5. Yeast Two-Hybrid Assays
2.6. GST Pull-Down Assays
2.7. RNA Interference
2.8. Western Blotting
2.9. Immunofluorescence Microscopy
2.10. Evaluation of the Number of Offsprings and Egg Morphology
2.11. Statistical Analysis
3. Results
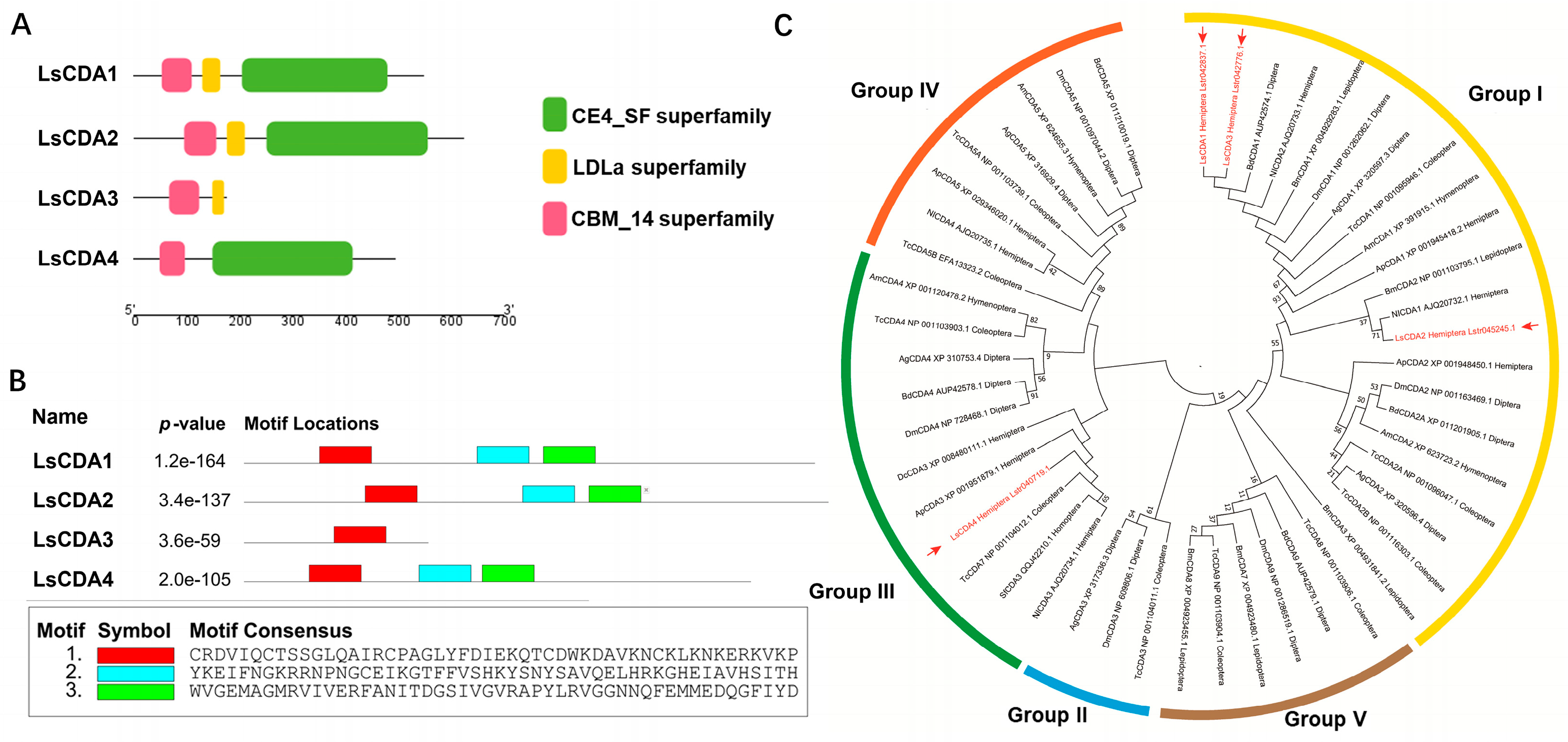
3.1. Identification of CDA Genes in SBPH
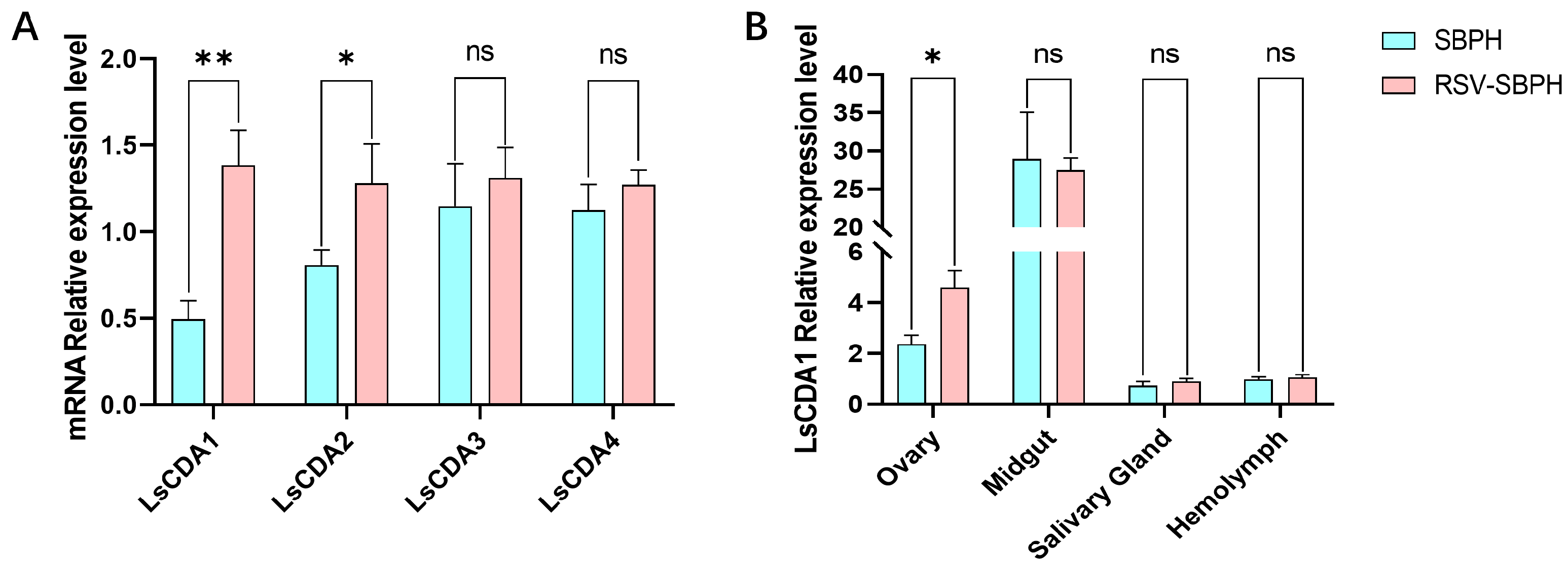
3.2. RSV Infection Induces LsCDA1 Expression in SBPH
3.3. RSV NP and NS2 Interact with LsCDA1 In Vitro
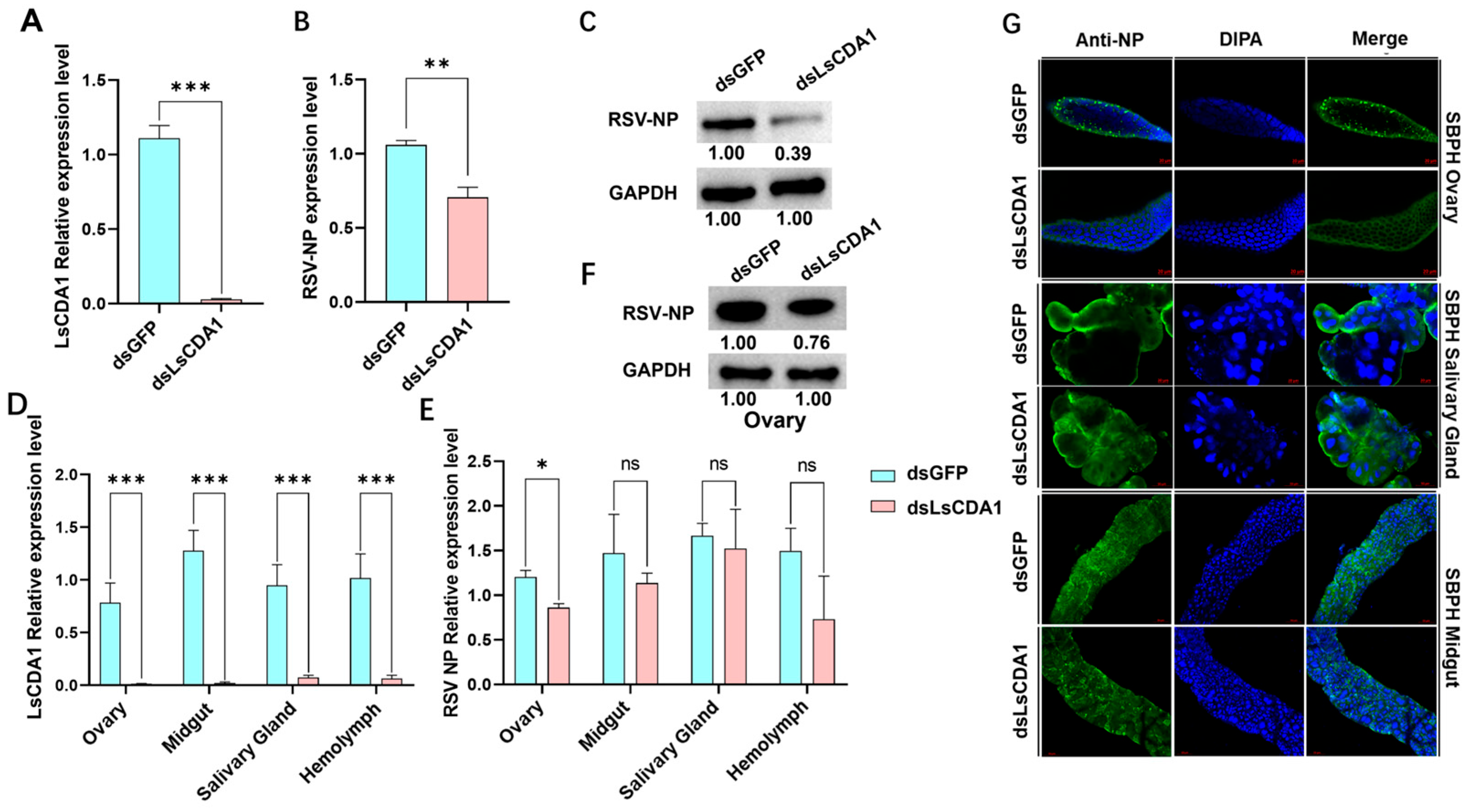
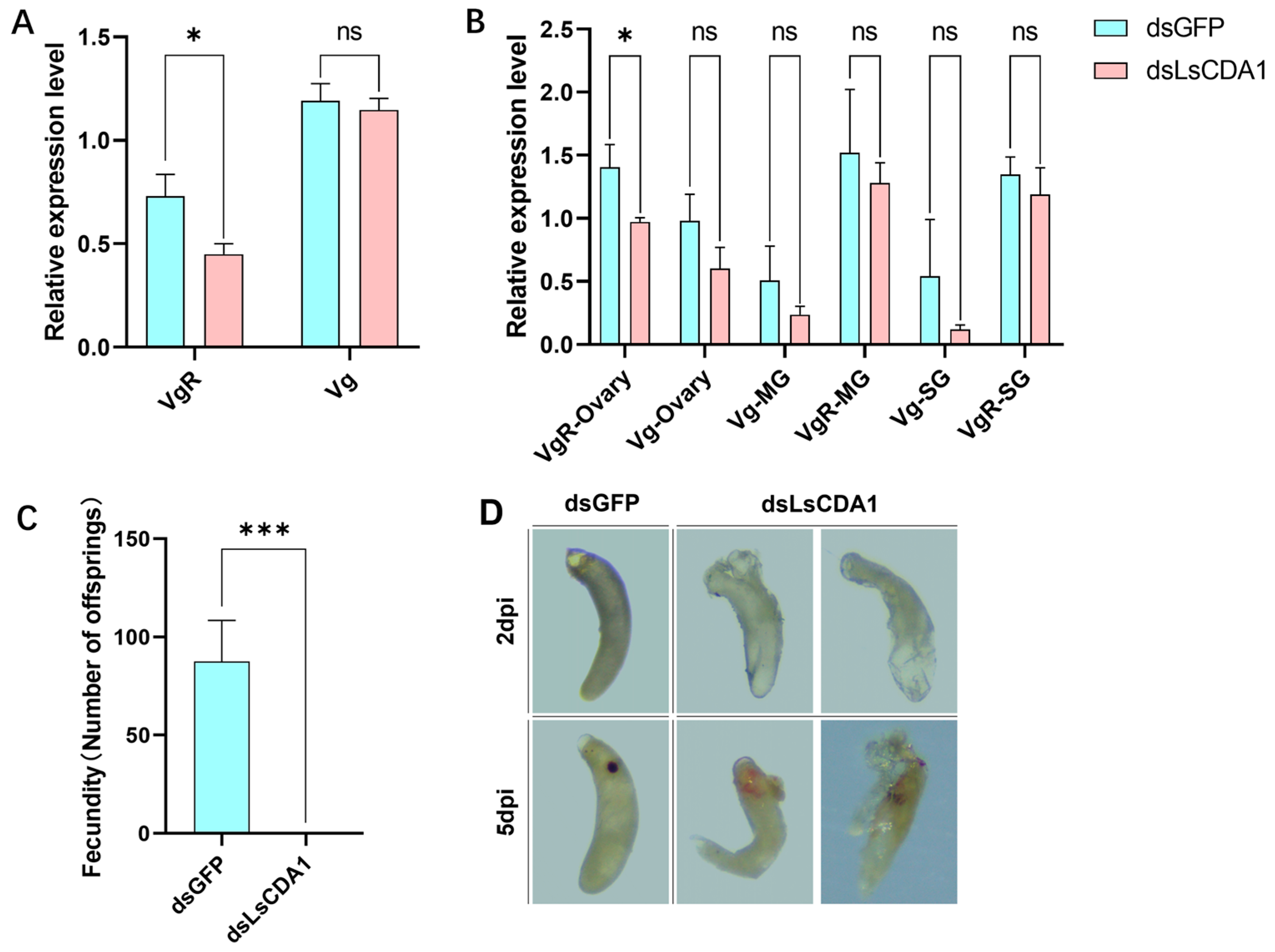
3.4. LsCDA1 Facilitates RSV Accumulation in Ovaries and Promotes SBPH Fecundity by Inducing VgR Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, D.M.; Qiao, J.H.; Gao, Q.; Zhang, J.; Zang, Y.; Xie, L.; Zhang, Y.; Wang, Y.; Fu, J.; Zhang, H.; et al. A plant cytorhabdovirus modulates locomotor activity of insect vectors to enhance virus transmission. Nat. Commun. 2023, 14, 5754. [Google Scholar] [CrossRef]
- Tatineni, S.; Hein, G.L. Plant viruses of agricultural importance: Current and future perspectives of virus disease management strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479–480, 278–289. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant virus-insect vector interactions: Current and potential future research directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Ammar, E.D.; Whitfield, A.E.; Redinbaugh, M.G. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, D.; Hu, J.; Zhang, K.; Kang, L.; Chen, Y.; Huang, L.; Zhang, L.; Xiang, Y.; Song, Q.; et al. The alpha-tubulin of Laodelphax striatellus mediates the passage of rice stripe virus (RSV) and enhances horizontal transmission. PLoS Pathog. 2020, 16. [Google Scholar] [CrossRef]
- Wei, J.; He, Y.Z.; Guo, Q.; Guo, T.; Liu, Y.Q.; Zhou, X.P.; Liu, S.S.; Wang, X.W. Vector development and vitellogenin determine the transovarial transmission of begomoviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 6746–6751. [Google Scholar] [CrossRef]
- Brasset, E.; Taddei, A.R.; Arnaud, F.; Faye, B.; Fausto, A.M.; Mazzini, M.; Giorgi, F.; Vaury, C. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology 2006, 3, 25. [Google Scholar] [CrossRef]
- Jia, D.; Mao, Q.; Chen, Y.; Liu, Y.; Chen, Q.; Wu, W.; Zhang, X.; Chen, H.; Li, Y.; Wei, T. Insect symbiotic bacteria harbour viral pathogens for transovarial transmission. Nat. Microbiol. 2017, 2, 17025. [Google Scholar] [CrossRef]
- Muthukrishnan, S.; Arakane, Y.; Noh, M.Y.; Mun, S.; Merzendorfer, H.; Boehringer, C.; Wellmeyer, B.; Yang, Q.; Qu, M.; Liu, L. Chitin in insect cuticle. Adv. Insect. Phys. 2022, 62, 1–110. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, L.; Wan, N.; Ji, X.; Zhang, H.; Jiang, J. Transcriptomic analysis of the interactions between the Spodoptera exigua midgut and nucleopolyhedrovirus. Pestic. Biochem. Physiol. 2020, 163, 241–253. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; Caccia, S.; Gordon, K.H.; Ferre, J.; Herrero, S. Downregulation of a chitin deacetylase-like protein in response to baculovirus infection and its application for improving baculovirus infectivity. J. Virol. 2010, 84, 2547–2555. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, S.; Tao, X.; Zhou, X. Rice stripe virus: Exploring molecular weapons in the arsenal of a negative-sense RNA virus. Annu. Rev. Phytopathol. 2021, 59, 351–371. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avsic-Zupanc, T.; Ayllon, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef] [PubMed]
- Kakutani, T.; Hayano, Y.; Hayashi, T.; Minobe, Y. Ambisense segment 4 of rice stripe virus: Possible evolutionary relationship with phleboviruses and uukuviruses (Bunyaviridae). J. Gen. Virol. 1990, 71, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Kakutani, T.; Hayano, Y.; Hayashi, T.; Minobe, Y. Ambisense segment 3 of rice stripe virus: The first instance of a virus containing two ambisense segments. J. Gen. Virol. 1991, 72, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.A.; Toriyama, S.H.; Hamamatsu, C.H.; Ishihama, A.K. Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J. Gen. Virol. 1993, 74, 769–773. [Google Scholar] [CrossRef]
- Toriyama, S.; Takahashi, M.; Sano, Y.; Shimizu, T.; Ishihama, A. Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 1994, 75, 3569–3579. [Google Scholar] [CrossRef]
- Huo, Y.; Liu, W.; Zhang, F.; Chen, X.; Li, L.; Liu, Q.; Zhou, Y.; Wei, T.; Fang, R.; Wang, X. Transovarial transmission of a plant virus is mediated by vitellogenin of its insect vector. PLoS Pathog. 2014, 10, e1003949. [Google Scholar] [CrossRef]
- Huo, Y.; Yu, Y.; Chen, L.; Li, Q.; Zhang, M.; Song, Z.; Chen, X.; Fang, R.; Zhang, L. Insect tissue-specific vitellogenin facilitates transmission of plant virus. PLoS Pathog. 2018, 14, e1006909. [Google Scholar] [CrossRef]
- Huo, Y.; Yu, Y.; Liu, Q.; Liu, D.; Zhang, M.; Liang, J.; Chen, X.; Zhang, L.; Fang, R. Rice stripe virus hitchhikes the vector insect vitellogenin ligand-receptor pathway for ovary entry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180312. [Google Scholar] [CrossRef]
- He, K.; Lin, K.; Ding, S.; Wang, G.; Li, F. The vitellogenin receptor has an essential role in vertical transmission of rice stripe virus during oogenesis in the small brown planthopper. Pest Manag. Sci. 2019, 75, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-X.; Seo, E.-Y.; Cho, I.-S.; Kim, J.-K.; Song, Z.; Kim, K.-H.; Eom, W.-S.; Jung, S.-H.; Hammond, J.; Lim, H.-S. Reassortment of infectious clones of radish mosaic virus shows that systemic necrosis in Nicotiana benthamiana is determined by RNA1. Phytopathology® 2022, 112, 1361–1372. [Google Scholar] [CrossRef]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef]
- Wu, H.; Zhao, D.; Guo, X.C.; Liu, Z.R.; Li, R.J.; Lu, X.J.; Guo, W. Group V chitin deacetylases influence the structure and composition of the midgut of beet armyworm, Spodoptera exigua. Int. J. Mol. Sci. 2023, 24, 3076. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, K.P.; Panes, V.A.; Santos, M.D. Molecular cloning and expression of chitin deacetylase 1 gene from the gills of Penaeus monodon (black tiger shrimp). Fish Shellfish Immunol. 2016, 55, 484–489. [Google Scholar] [CrossRef]
- Liu, Q.; Meng, X.; Song, Z.; Shao, Y.; Zhao, Y.; Fang, R.; Huo, Y.; Zhang, L. Insect-transmitted plant virus balances its vertical transmission through regulating Rab1-mediated receptor localization. Cell Rep. 2024, 43, 114571. [Google Scholar] [CrossRef]
- Zhang, X.; Ji, Y.; Moussian, B.; Yang, S.; Zhang, J.; Zhang, T.; Zhang, M. Serpentine and vermiform are produced autonomously to fulfill their function in Drosophila wings. Insects 2023, 14, 406. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Yu, P.H.; Xia, X.; Zhang, R.; Wang, L.H.; Fang, J.C.; Hoffmann, A.A.; Luo, G.H. Identification of a fatty acid synthase gene (FAS1) from Laodelphax striatellus planthoppers contributing to fecundity. Insect. Sci. 2023, 30, 599–610. [Google Scholar] [CrossRef]
- Jing, Y.P.; Wen, X.; Li, L.; Zhang, S.; Zhang, C.; Zhou, S. The vitellogenin receptor functionality of the migratory locust depends on its phosphorylation by juvenile hormone. Proc. Natl. Acad. Sci. USA 2021, 118, e2106908118. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, S.; Batz, T.; Armbruster, K.; Krasnow, M.A. Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006, 16, 186–194. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; You, A.; Zhang, J.; Li, Y.; Zuo, S.; Liu, F.; Zhang, L. Chitin Deacetylase Gene Family Positively Regulates the Accumulation of Rice Stripe Virus in Laodelphax striatellus Fallén (Hemiptera: Delphacidae) Ovaries. Insects 2025, 16, 334. https://doi.org/10.3390/insects16040334
Hu W, You A, Zhang J, Li Y, Zuo S, Liu F, Zhang L. Chitin Deacetylase Gene Family Positively Regulates the Accumulation of Rice Stripe Virus in Laodelphax striatellus Fallén (Hemiptera: Delphacidae) Ovaries. Insects. 2025; 16(4):334. https://doi.org/10.3390/insects16040334
Chicago/Turabian StyleHu, Wenxing, Ao You, Jiao Zhang, Yao Li, Shimin Zuo, Fang Liu, and Lu Zhang. 2025. "Chitin Deacetylase Gene Family Positively Regulates the Accumulation of Rice Stripe Virus in Laodelphax striatellus Fallén (Hemiptera: Delphacidae) Ovaries" Insects 16, no. 4: 334. https://doi.org/10.3390/insects16040334
APA StyleHu, W., You, A., Zhang, J., Li, Y., Zuo, S., Liu, F., & Zhang, L. (2025). Chitin Deacetylase Gene Family Positively Regulates the Accumulation of Rice Stripe Virus in Laodelphax striatellus Fallén (Hemiptera: Delphacidae) Ovaries. Insects, 16(4), 334. https://doi.org/10.3390/insects16040334




