Characterisation of a Novel Insect-Specific Virus Discovered in Rice Thrips, Haplothrips aculeatus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and RNA Extraction
2.2. Host Insect Identification
2.3. Transcriptomic and Small RNA (sRNA) Sequencing
2.4. Virus Discovery and Confirmation by Reverse Transcription-PCR (RT-PCR)
2.5. Determination of Viral Genome Termini and Transcript Abundance
2.6. Small RNA Analysis
2.7. Genome Annotation and Phylogenetic Analysis
3. Results
3.1. Discovery of RNA Virus-Related Sequences in H. aculeatus
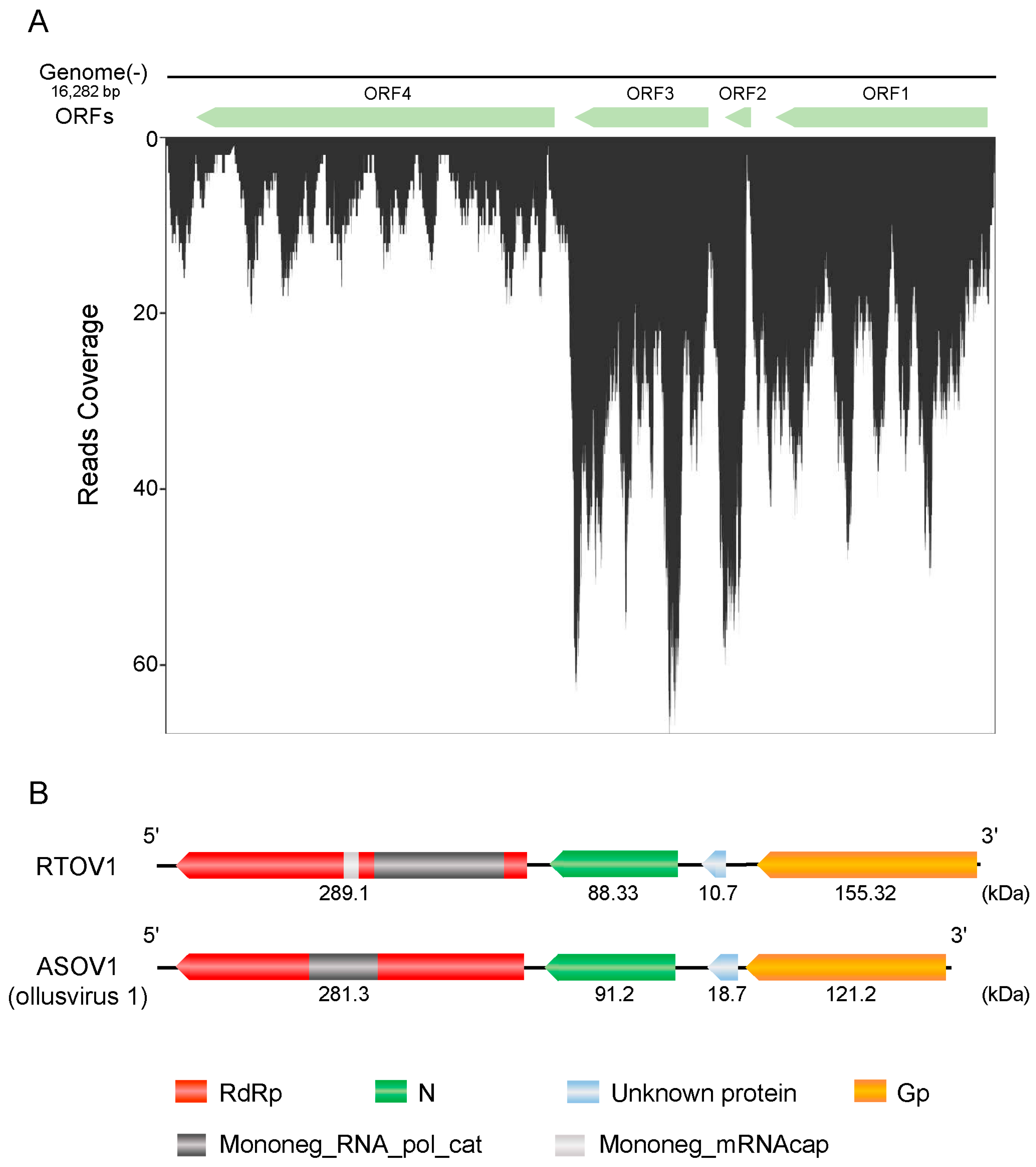
3.2. Characterisation of Rice Thrips Ollusvirus 1
3.3. Phylogenetic Analysis of RTOV1 and Related Jingchuvirals
3.4. Activation of Antiviral RNA Interference Pathway in H. aculeatus Responsive to ISVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, C.X.; Shi, M.; Tian, J.H.; Lin, X.D.; Kang, Y.J.; Chen, L.J.; Qin, X.C.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 2015, 4, 1–26. [Google Scholar] [CrossRef]
- Edgar, R.C.; Taylor, B.; Lin, V.; Altman, T.; Barbera, P.; Meleshko, D.; Lohr, D.; Novakovsky, G.; Buchfink, B.; Al-Shayeb, B.; et al. Petabase-scale sequence alignment catalyses viral discovery. Nature 2022, 602, 142–147. [Google Scholar] [CrossRef]
- Qi, Y.H.; Ye, Z.X.; Zhang, C.X.; Chen, J.P.; Li, J.M. Diversity of RNA viruses in agricultural insects. Comput. Struct. Biotechnol. J. 2023, 21, 4312–4321. [Google Scholar]
- de Almeida, J.P.; Aguiar, E.R.; Armache, J.N.; Olmo, R.P.; Marques, J.T. The virome of vector mosquitoes. Curr. Opin. Virol. 2021, 49, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M.; Dolja, V.V. The global virome: How much diversity and how many independent origins? Environ. Microbiol. 2023, 25, 40–44. [Google Scholar] [CrossRef]
- Bonning, B.C. The Insect Virome: Opportunities and Challenges. Curr. Issues Mol. Biol. 2020, 34, 1–12. [Google Scholar] [CrossRef]
- Carvalho, V.L.; Long, M.T. Insect-Specific Viruses: An overview and their relationship to arboviruses of concern to humans and animals. Virology 2021, 557, 34–43. [Google Scholar] [CrossRef]
- Kormelink, R.; Garcia, M.L.; Goodin, M.; Sasaya, T.; Haenni, A.L. Negative-strand RNA viruses: The plant-infecting counterparts. Virus Res. 2011, 162, 184–202. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.; Krupovic, M.; Zhang, Y.Z.; Maes, P.; Dolja, V.; Koonin, E.V.; Kuhn, J.H. Megataxonomy of Negative-Sense RNA Viruses; Proposal (Taxoprop). No. 2017.006M; International Committee for Taxonomy of Viruses: London, UK, 2017. [Google Scholar]
- Kuhn, J.H.; Dheilly, N.M.; Junglen, S.; Paraskevopoulou, S.; Shi, M.; Di Paola, N. ICTV Virus Taxonomy Profile: Jingchuvirales 2023. J. Gen Virol. 2023, 104, 001924. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Wu, H.M.; Pang, R.; Cheng, T.; Xue, L.; Zeng, H.Y.; Lei, T.; Chen, M.T.; Wu, S.; Ding, Y.; Zhang, J.M.; et al. Abundant and Diverse RNA Viruses in Insects Revealed by RNA-Seq Analysis: Ecological and Evolutionary Implications. mSystems 2020, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pakrashi, A.; Patidar, A.; Singha, D.; Kumar, V.; Tyagi, K. Comparative analysis of the two suborders of Thysanoptera and characterization of the complete mitochondrial genome of Thrips parvispinus. Arch. Insect Biochem. Physiol. 2023, 114, 1–15. [Google Scholar] [CrossRef]
- Mound, L.A. Thysanoptera: Diversity and interactions. Annu. Rev. Entomol. 2005, 50, 247–269. [Google Scholar] [CrossRef]
- Widana Gamage, S.M.K.; Rotenberg, D.; Schneweis, D.J.; Tsai, C.W.; Dietzgen, R.G. Transcriptome-wide responses of adult melon thrips (Thrips palmi) associated with capsicum chlorosis virus infection. PLoS ONE 2018, 13, e0208538. [Google Scholar] [CrossRef]
- Lu, G.; Ye, Z.X.; He, Y.J.; Zhang, Y.; Wang, X.; Huang, H.J.; Zhuo, J.C.; Sun, Z.T.; Yan, F.; Chen, J.P.; et al. Discovery of Two Novel Negeviruses in a Dungfly Collected from the Arctic. Viruses 2020, 12, 692. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Ren, P.P.; Ye, Z.X.; Wang, S.N.; Li, J.M.; Chen, J.P.; Zhang, C.X.; Lu, J.B. Complete genome analysis of a novel chuvirus from a southern green stink bug (Nezara viridula). Arch. Virol. 2022, 167, 2423–2427. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- He, Y.J.; Ye, Z.X.; Zhang, C.X.; Li, J.M.; Chen, J.P.; Lu, G. An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis. Insects 2022, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Takemae, H.; Nunomura, Y.; Yokota, T.; Oba, M.; Mizutani, T.; Hsu, W.L.; Sakamoto, Y. Novel ollusvirus detected in a solitary wild bee species (Osmia taurus) in Japan. Arch. Virol. 2023, 168, 183. [Google Scholar] [CrossRef]
- Bonning, B.C.; Saleh, M.C. The Interplay Between Viruses and RNAi Pathways in Insects. Annu. Rev. Entomol. 2021, 66, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Naganuma, M.; Tadakuma, H.; Tomari, Y. Single-molecule analysis of processive double-stranded RNA cleavage by Drosophila Dicer-2. Nat. Commun. 2021, 12, 4268. [Google Scholar] [CrossRef]
- Rotenberg, D.; Jacobson, A.L.; Schneweis, D.J.; Whitfield, A.E. Thrips transmission of tospoviruses. Curr. Opin. Virol. 2015, 15, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Muppudathi, S.P.; Natarajan, G.; Varagur Ganesan, M.; Sevugapperumal, N.; Subbarayalu, M.; John Samuel, K.; Perumal, R. Role of Thrips palmi and Parthenium hysterophorus pollen in active spread of tobacco streak virus in the cotton ecosystem. Virus Res. 2020, 284, 197979. [Google Scholar] [CrossRef] [PubMed]
- Mou, D.F.; Chen, W.T.; Li, W.H.; Chen, T.C.; Tseng, C.H.; Huang, L.H.; Peng, J.C.; Yeh, S.D.; Tsai, C.W. Transmission mode of watermelon silver mottle virus by Thrips palmi. PLoS ONE 2021, 16, e0247500. [Google Scholar] [CrossRef] [PubMed]
- Daimei, G.; Raina, H.S.; Devi, P.P.; Saurav, G.K.; Renukadevi, P.; Malathi, V.G.; Senthilraja, C.; Mandal, B.; Rajagopal, R. Influence of Groundnut bud necrosis virus on the Life History Traits and Feeding Preference of Its Vector, Thrips palmi. Phytopathology 2017, 107, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Messieha, M. Transmission of tobacco ringspot virus by thrips. Phytopathology 1969, 59, 943–945. [Google Scholar] [PubMed]
- Martin, R.R.; MacFarlane, S.; Sabanadzovic, S.; Quito, D.; Poudel, B.; Tzanetakis, I.E. Viruses and Virus Diseases of Rubus. Plant Dis. 2013, 97, 168–182. [Google Scholar] [CrossRef]
- Leach, A.; Fuchs, M.; Harding, R.; Nault, B.A. Iris Yellow Spot Virus Prolongs the Adult Lifespan of Its Primary Vector, Onion Thrips (Thrips tabaci) (Thysanoptera: Thripidae). J. Insect Sci. 2019, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Mound, L.A.; Wang, Z.; Lima, É.F.B.; Marullo, R. Problems with the Concept of “Pest” among the Diversity of Pestiferous Thrips. Insects 2022, 13, 61. [Google Scholar] [CrossRef]
- French, R.K.; Anderson, S.H.; Cain, K.E.; Greene, T.C.; Minor, M.; Miskelly, C.M.; Montoya, J.M.; Wille, M.; Muller, C.G.; Taylor, M.W.; et al. Host phylogeny shapes viral transmission networks in an island ecosystem. Nat. Ecol. Evol. 2023, 7, 1834–1843. [Google Scholar] [CrossRef]
- Luo, M.; Terrell, J.R.; Mcmanus, S.A. Nucleocapsid Structure of Negative Strand RNA Virus. Viruses 2020, 12, 835. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Shrestha, A.; Peters, J.R.; Carroll, B.J.; Srinivasan, R.; Pappu, H.R.; Mitter, N. The Tomato Spotted Wilt Virus Genome Is Processed Differentially in its Plant Host Arachis hypogaea and its Thrips Vector Frankliniella fusca. Front. Plant Sci. 2016, 7, 1349. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, H.; Ye, Z.; Lu, G.; Feng, K.; Zhang, M.; Sun, X.; Han, Z.; Jiang, S.; Wu, B.; Yin, X.; et al. Characterisation of a Novel Insect-Specific Virus Discovered in Rice Thrips, Haplothrips aculeatus. Insects 2024, 15, 303. https://doi.org/10.3390/insects15050303
Hong H, Ye Z, Lu G, Feng K, Zhang M, Sun X, Han Z, Jiang S, Wu B, Yin X, et al. Characterisation of a Novel Insect-Specific Virus Discovered in Rice Thrips, Haplothrips aculeatus. Insects. 2024; 15(5):303. https://doi.org/10.3390/insects15050303
Chicago/Turabian StyleHong, Hao, Zhuangxin Ye, Gang Lu, Kehui Feng, Mei Zhang, Xiaohui Sun, Zhilei Han, Shanshan Jiang, Bin Wu, Xiao Yin, and et al. 2024. "Characterisation of a Novel Insect-Specific Virus Discovered in Rice Thrips, Haplothrips aculeatus" Insects 15, no. 5: 303. https://doi.org/10.3390/insects15050303
APA StyleHong, H., Ye, Z., Lu, G., Feng, K., Zhang, M., Sun, X., Han, Z., Jiang, S., Wu, B., Yin, X., Xu, S., Li, J., & Xin, X. (2024). Characterisation of a Novel Insect-Specific Virus Discovered in Rice Thrips, Haplothrips aculeatus. Insects, 15(5), 303. https://doi.org/10.3390/insects15050303





