Wētā Aotearoa—Polyphyly of the New Zealand Anostostomatidae (Insecta: Orthoptera)
Abstract
Simple Summary
Abstract
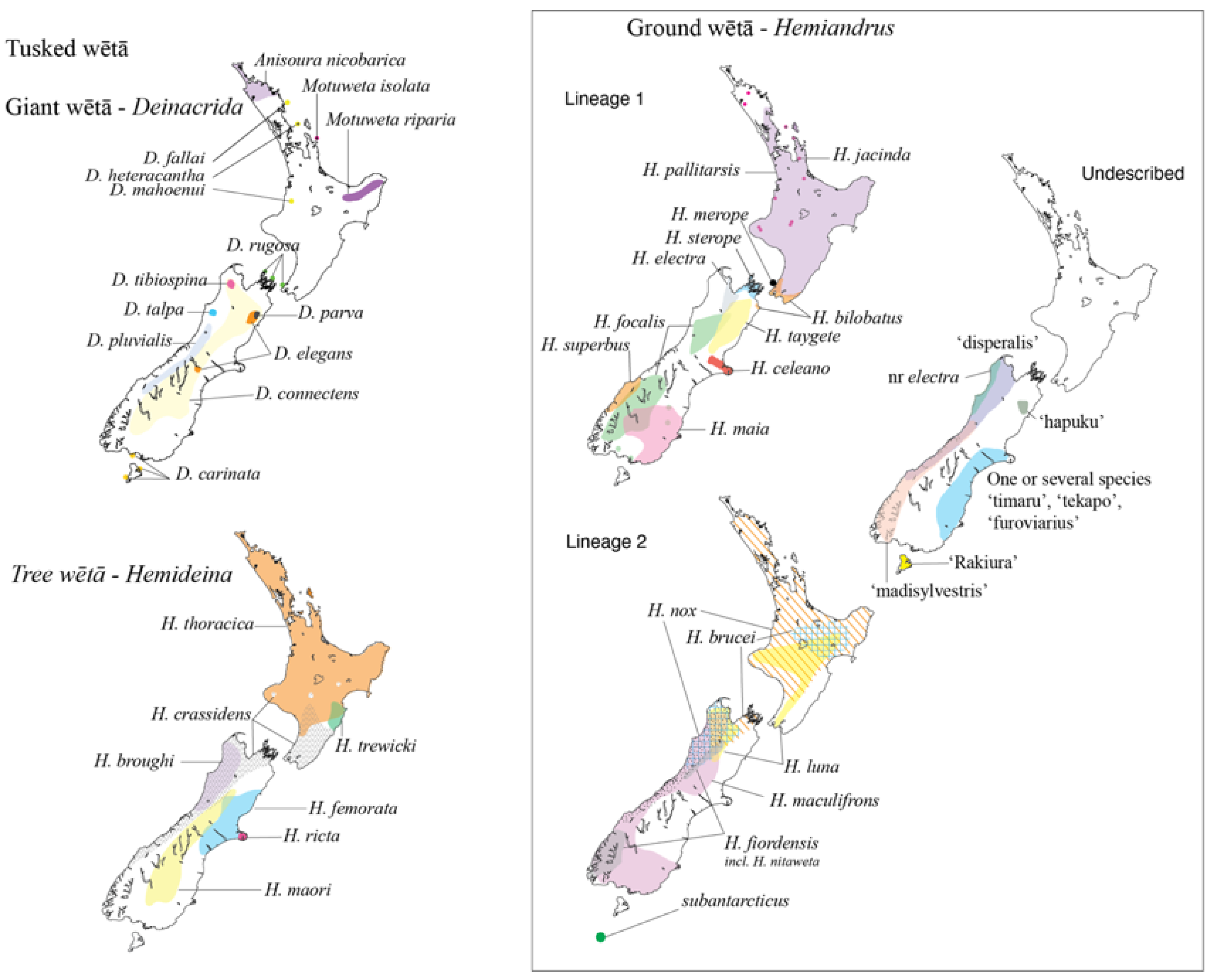
1. Introduction
2. Materials and Methods
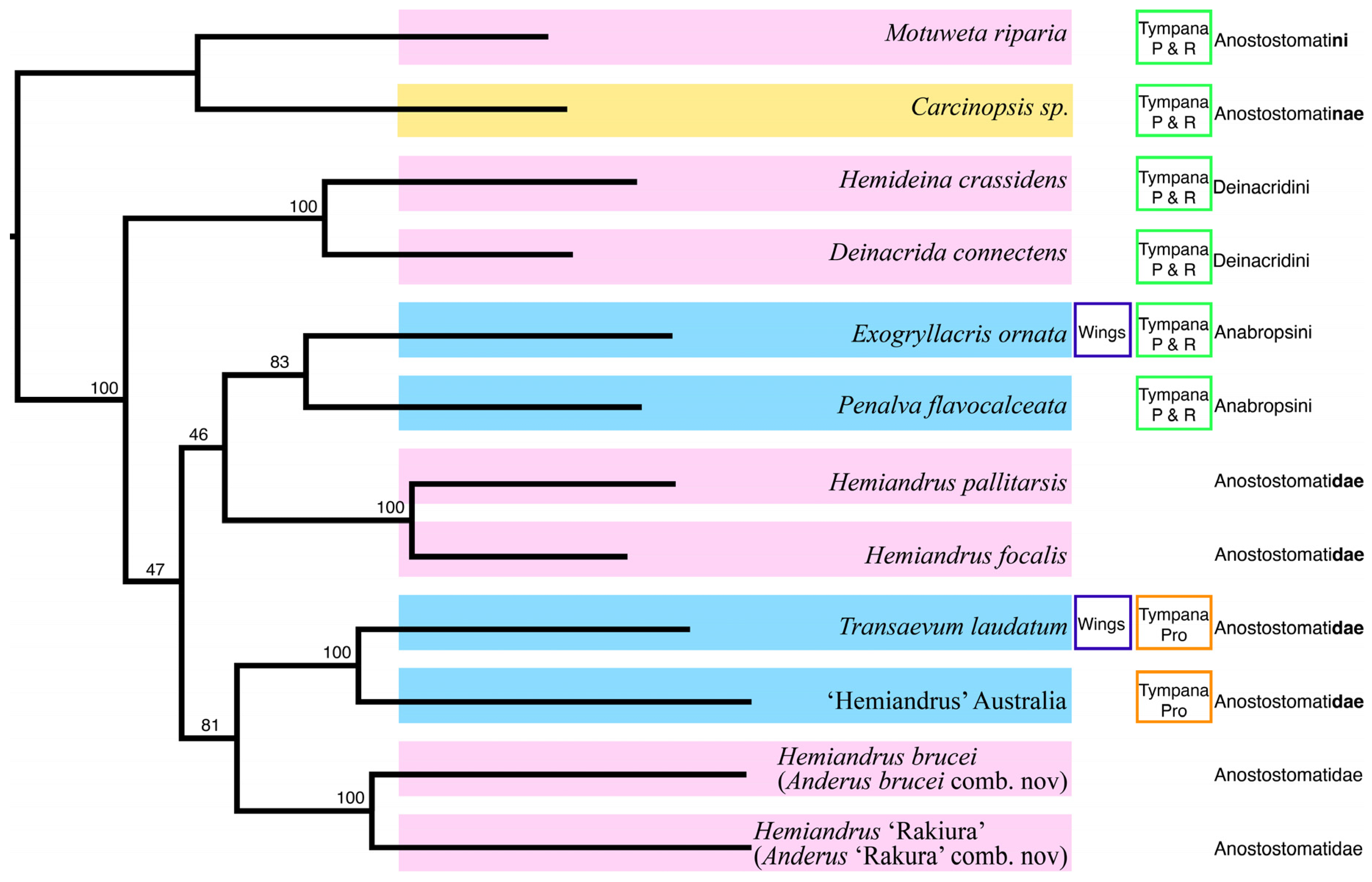
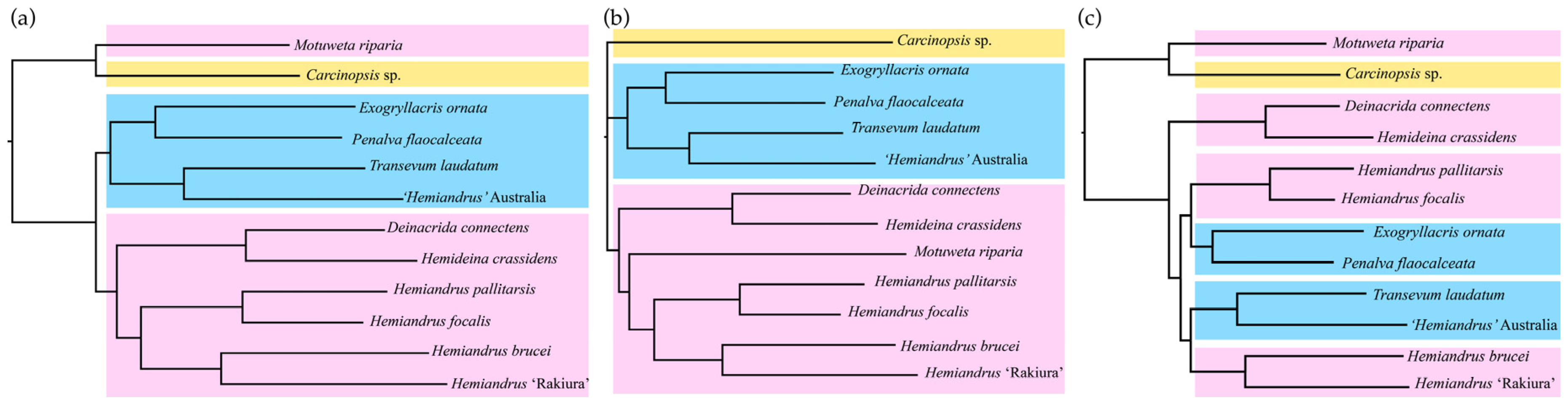
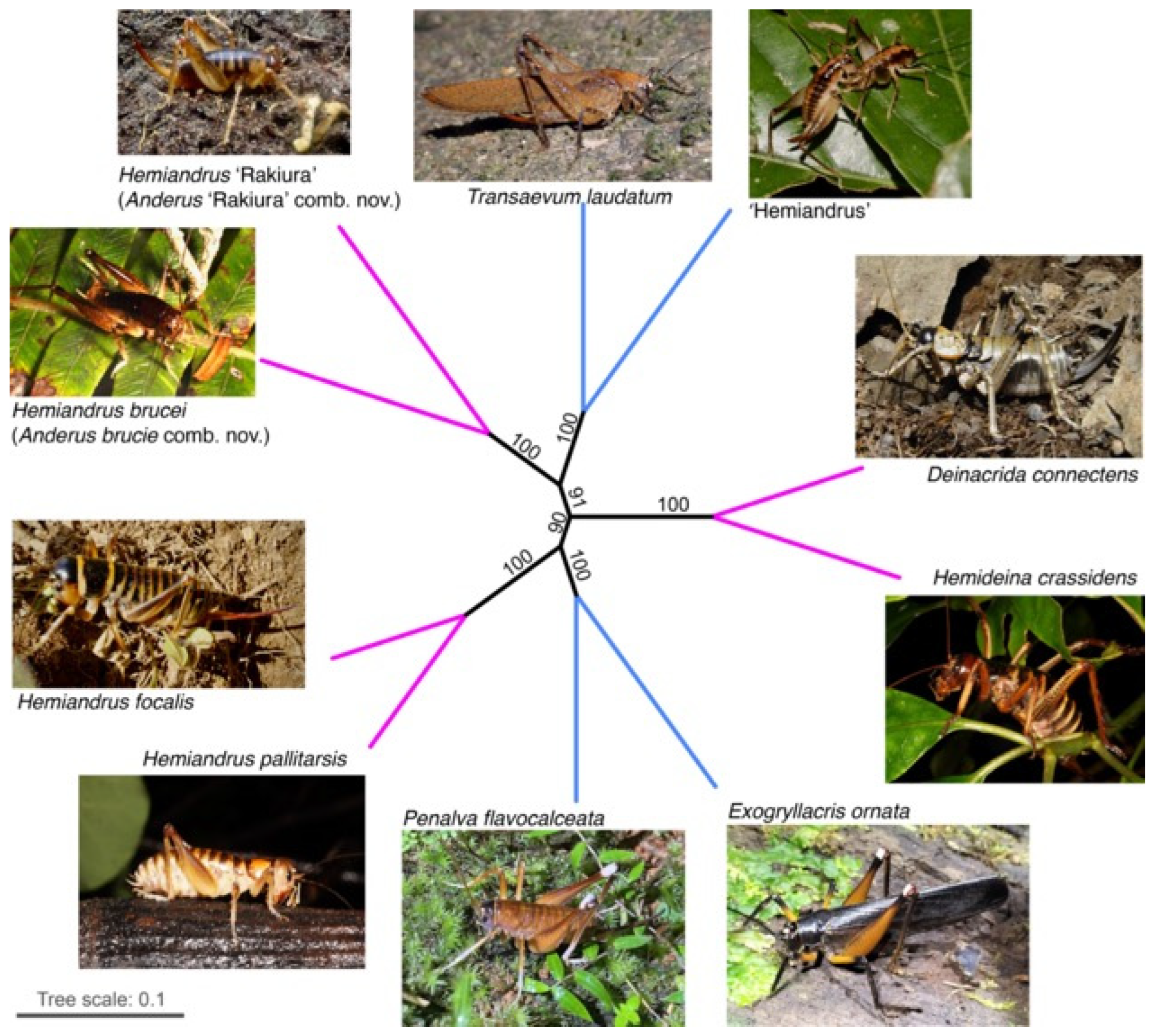
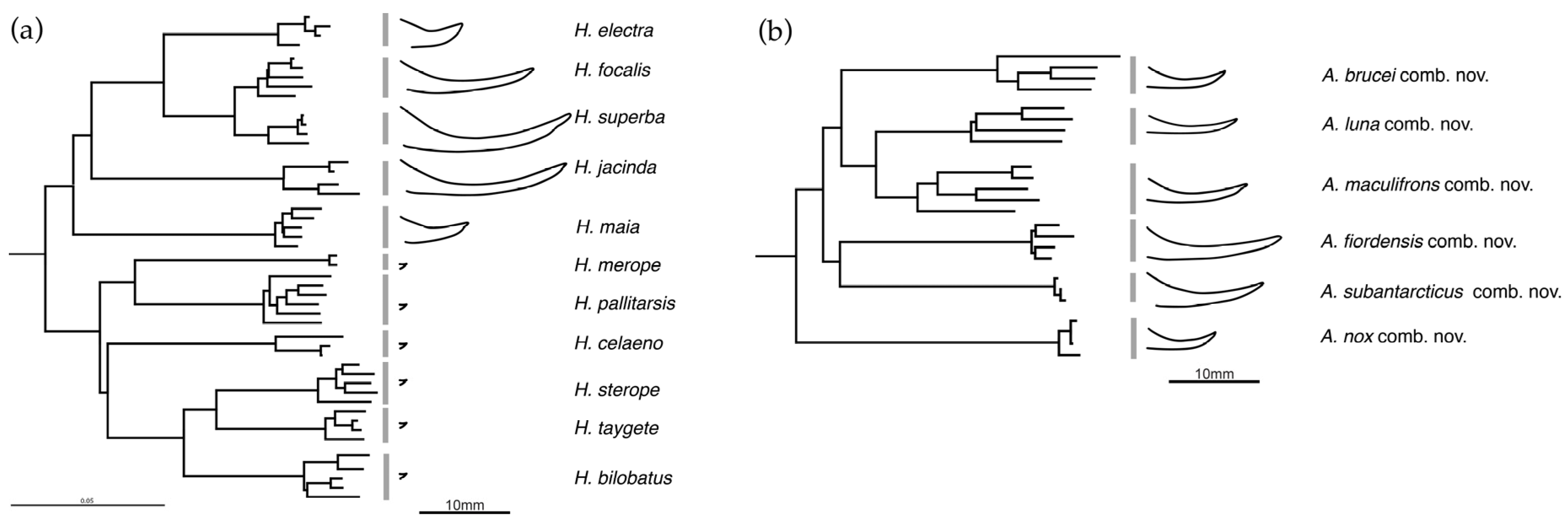
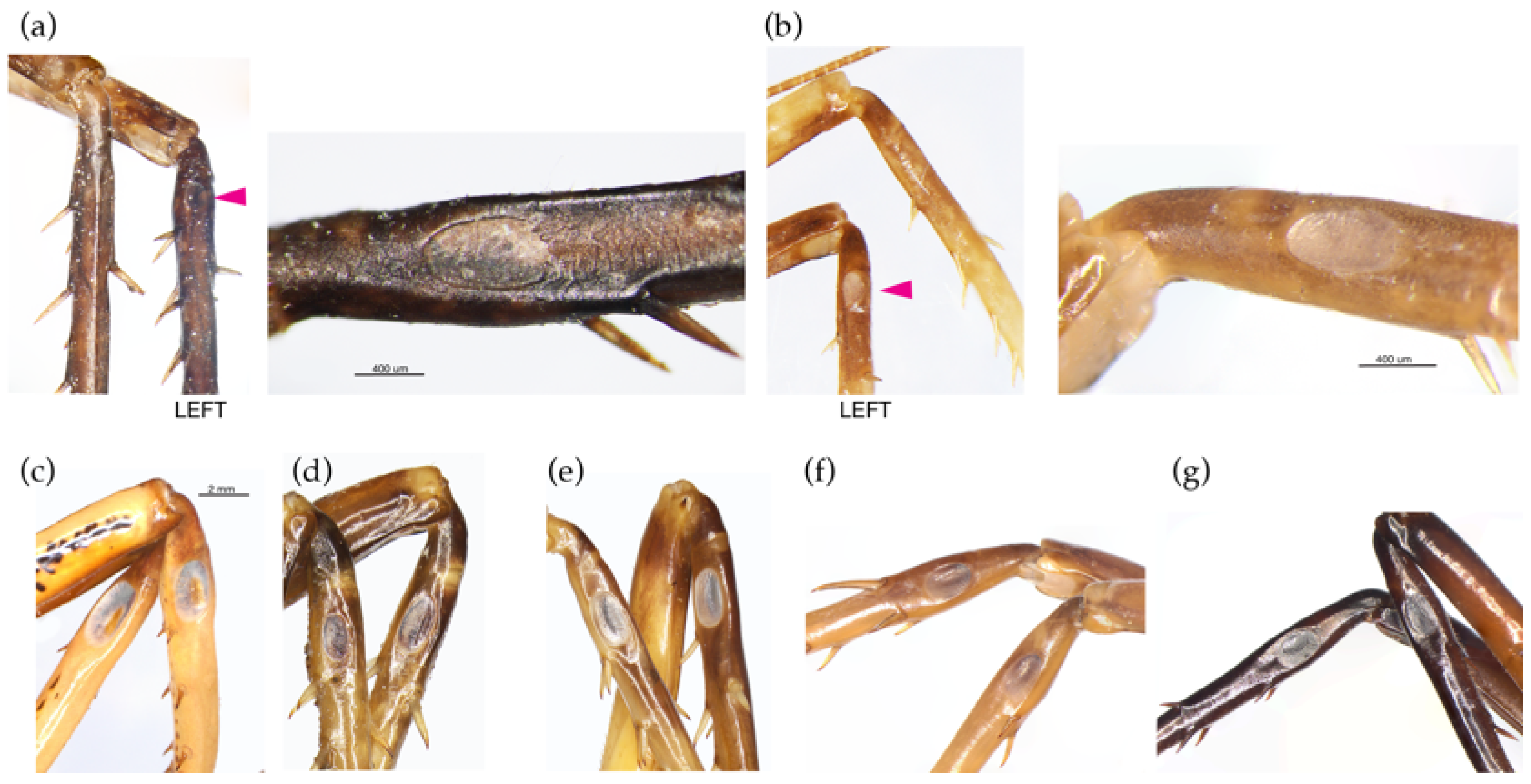
3. Results
Taxonomy
- Class Insecta
- Order Orthoptera
- Suborder Ensifera
- Superfamily Stenopelmatoidea
- Family Anostostomatidae Saussure (1859)
- Anderus gen. nov.
- zoobank.org:pub:65583178-0930-47F3-A9A8-C887572BC0A8
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johns, P.M. The Gondwanaland weta: Family Anostostomatidae (formerly Stenopelmatidae, Henicidae or Mimnermidae): Nomenclatural problems, world checklist, new genera and species. J. Orthoptera Res. 1997, 6, 125–138. [Google Scholar] [CrossRef]
- Cigliano, M.M.; Braun, H.; Eades, D.C.; Otte, D. Orthoptera Species File. Available online: http://orthoptera.speciesfile.org/ (accessed on 19 June 2024).
- Gorochov, A.V. The Families Stenopelmatidae and Anostostomatidae (Orthoptera). 1. Higher classification, new and littleknown taxa. Entomol. Rev. 2021, 100, 1106–1151. [Google Scholar] [CrossRef]
- Morgan-Richards, M.; Gibbs, G.W. A phylogenetic analysis of New Zealand giant and tree weta (Orthoptera: Anostostomatidae: Deinacrida and Hemideina) using morphological and genetic characters. Invertebr. Taxon. 2001, 15, 1–12. [Google Scholar] [CrossRef][Green Version]
- Trewick, S.A.; Morgan-Richards, M. Phylogenetics of New Zealand’s tree, giant and tusked weta (Orthoptera: Anostostomatidae): Evidence from mitochondrial DNA. J. Orthopteran Res. 2004, 13, 185–196. [Google Scholar] [CrossRef]
- Trewick, S.A.; Morgan-Richards, M. After the deluge: Mitochondrial DNA indicates Miocene radiation and Pliocene adaptation of tree and giant weta (Orthoptera: Anostostomatidae). J. Biogeog. 2005, 32, 295–309. [Google Scholar] [CrossRef]
- Twort, V.G.; Newcomb, R.D.; Buckley, T.R. New Zealand tree and giant wētā (Orthoptera) transcriptomics reveal divergent selection patterns in metabolic loci. Genome Biol. Evol. 2019, 11, 1293–1306. [Google Scholar] [CrossRef]
- Wyman, T.E.; Trewick, S.A.; Morgan-Richards, M.; Noble, A.D.L. Mutualism or opportunism? Tree fuchsia (Fuchsia excorticata) and tree weta (Hemideina) interactions. Austral Ecol. 2010, 36, 261–281. [Google Scholar] [CrossRef]
- Griffin, M.J.; Morgan-Richards, M.; Trewick, S.A. Is the tree weta Hemideina crassidens an obligate herbivore? N. Z. Nat. Sci. 2011, 36, 11–19. [Google Scholar]
- Monteith, G.B.; Field, L.H. Australian king crickets: Distribution, habitats and biology (Orthoptera: Anostostomatidae). In The Biology of Wetas, King Crickets and Their Allies; Field, L.H., Ed.; CABI Publishing: Wallingford, UK, 2001; pp. 79–94. [Google Scholar]
- Cary, P.R.L. Diet of the ground weta Zealandosandrus gracilis (Orthoptera: Semipalmated). N. Z. J. Zool. 1983, 10, 295–297. [Google Scholar] [CrossRef]
- Winks, C.J.; Fowler, S.V.; Ramsay, G.W. Captive-Rearing of the Middle Island Tusked Wētā; Science for Conservation 197; NZ Department of Conservation: Wellington, New Zealand, 2002. [Google Scholar]
- Gorochov, A.V. The higher classification, phylogeny and evolution of the superfamily Stenopelmatiodea. In The Biology of Wetas, King Crickets and Their Allies; Field, L.H., Ed.; CABI Publishing: Wallingford, UK, 2001; pp. 3–33. [Google Scholar]
- Gorochov, A.V.; Cadena-Castañeda, O.J. New and little known Stenopelmatoidea (Orthoptera: Ensifera) from America. Zoosystematica Ross. 2016, 25, 98–143. [Google Scholar] [CrossRef]
- Pratt, R.C.; Morgan-Richards, M.; Trewick, S.A. Diversification of New Zealand weta (Orthoptera: Ensifera: Anostostomatidae) and their relationships in Australasia. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3427–3438. [Google Scholar] [CrossRef] [PubMed]
- Vandergast, A.G.; Weissman, D.B.; Wood, D.A.; Rentz, D.C.F.; Bazelet, C.S.; Ueshima, N. Tackling an intractable problem: Can greater taxon sampling help resolve relationships within the Stenopelmatoidea (Orthoptera: Ensifera)? Zootaxa 2017, 4291, 1–33. [Google Scholar] [CrossRef]
- McKintyre, M. Ruakumara tusked weta: Field and captive observations. In Conservation Advisory Science Notes No. 219; Department of Conservation: Wellington, New Zealand, 1998. [Google Scholar]
- Stringer, I.A.N.; Mack, H.; Grant, E.A.; Winks, C.J. Growth and development of captive-reared Mercury Islands tusked weta, Motuweta isolata Johns (Orthoptera: Anostostomatidae). N. Z. Entomol. 2005, 29, 5–19. [Google Scholar] [CrossRef]
- Salmon, J.T. A revision of the New Zealand wetas—Anostostomatinae (Orthoptera: Stenopelmatidae). Dom. Mus. Rec. Entomol. 1950, 1, 121–177. [Google Scholar]
- Johns, P.M. Distribution and Conservation Status of Ground Weta, Hemiandrus Species (Orthoptera: Anostostomatidae); Science for Conservation, Department of Conservation: Wellington, New Zealand, 2001. [Google Scholar]
- Smith, B.L.T. Evolution and Diversity: Analysis of Species and Speciation in Hemiandrus Ground Wētā. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2015. [Google Scholar]
- Song, H.; Béthoux, O.; Shin, S.; Donath, A.; Letsch, H.; Liu, S.; McKenna, D.D.; Meng, G.; Misof, B.; Podsiadlowski, L.; et al. Phylogenomic analysis sheds light on the evolutionary pathways towards acoustic communication in Orthoptera. Nat. Commun. 2020, 11, 4939. [Google Scholar] [CrossRef]
- Brunner von Wattenwyl, C. Monographie der Stenopelmatiden und Gryllacriden. Verhandlungen Der Kais.-Königlichen Zool.-Bot. Ges. Wien 1888, 38, 247–394. [Google Scholar]
- Ander, K. Vergleichend-anatomische und phylogenetische Studien über die Ensifera (Saltatoria). In Opuscula Entomologica. Edidit Societas Entomologica Lundensis; Entomologiska Sallskapet: Lund, Sweden, 1963; p. 306. [Google Scholar]
- Willemse, C. A new genus and a new species of Orthoptera from Australia. Entomol. Ber. 1963, 23, 1–4. [Google Scholar]
- Taylor-Smith, B.L.; Trewick, S.A.; Morgan-Richards, M. Three new ground wētā and a redescription of Hemiandrus maculifrons. N. Z. J. Zool. 2016, 43, 363–383. [Google Scholar] [CrossRef]
- Hutton, F.W. The Stenopelmatidae of New Zealand. Trans. Proc. N. Z. Inst. 1897, 29, 208–242. [Google Scholar]
- Walker, F. Catalogue of the Specimens of Dermpatera Saltaroria in the Collection of the British Museum. Part 5; British Museum: London, UK, 1869; p. 108. [Google Scholar]
- Blanchard, E. Fauna Chilena. Insectos. Orden IV. Ortoperos. In Historiajsica y Politica de Chile; Zoologia Vol. 6 & Atlas Zoologico-Entomologia, Ortopteros; Gay, C., Ed.; Chilean Government: Paris, France, 1851; pp. 41–42. [Google Scholar]
- Gibbs, G.W. A new species of tusked weta from the Raukumara Range, North Island, New Zealand (Orthoptera: Anostostomatidae: Motuweta). N. Z. J. Zool. 2002, 29, 293–301. [Google Scholar] [CrossRef][Green Version]
- Karny, H.H. Revision der Gryllacriden des Naturhistorischen Museums in Wien einschließlich der Collection Brunner v. Wattenwyl. Ann. Naturhist. Mus. Wien 1929, 43, 3–186. [Google Scholar]
- Koot, E.M.; Morgan-Richards, M.; Trewick, S.A. An alpine grasshopper radiation older than the mountains, on Kā Tiritiri o te Moana (Southern Alps) of Aotearoa (New Zealand). Mol. Phylogenet. Evol. 2020, 147, 106783. [Google Scholar] [CrossRef] [PubMed]
- Vaux, F.; Hills, S.F.K.; Marshall, B.A.; Trewick, S.A.; Morgan-Richards, M. A phylogeny of Southern Hemisphere whelks (Gastropoda: Buccinulidae) and concordance with the fossil record. Mol. Phylogenet. Evol. 2017, 114, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.A.; Koot, E.M.; Morgan-Richards, M. Māwhitiwhiti Aotearoa: Phylogeny and synonymy of the silent alpine grasshopper radiation of New Zealand (Orthoptera: Acrididae). Zootaxa 2023, 5383, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Sunnucks, P.; Hale, D.F. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol. Biol. Evol. 1996, 13, 510–524. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Dowle, E.J.; Trewick, S.A.; Morgan-Richards, M. Fossil calibrated phylogenies of Southern cave wētā show dispersal and extinction confound biogeographic signal. R. Soc. Open Sci. 2024, 11, 231118. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2010, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. 2018 UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Kishino, H.; Miyata, T.; Hasegawa, M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol. 1990, 31, 151–160. [Google Scholar] [CrossRef]
- Kishino, H.; Hasegawa, M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989, 29, 170–179. [Google Scholar] [CrossRef]
- Shimodaira, H.; Hasegawa, M. Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Mol. Biol. Evol. 1999, 16, 1114. [Google Scholar] [CrossRef]
- Strimmer, K.; Rambaut, A. Inferring confidence sets of possibly misspecified gene trees. Proc. R. Soc. B 2002, 269, 137–142. [Google Scholar] [CrossRef]
- Shimodaira, H. An Approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef]
- Taylor Smith, B.L.; Morgan-Richards, M.; Trewick, S.A. New Zealand ground wētā (Anostostomatidae: Hemiandrus): Descriptions of two species with notes on their biology. N. Z. J. Zool. 2013, 40, 314–329. [Google Scholar] [CrossRef][Green Version]
- Trewick, S.A. A new species of large Hemiandrus ground wētā (Orthoptera: Anostostomatidae) from North Island, New Zealand. Zootaxa 2021, 4942, 207–218. [Google Scholar] [CrossRef]
- Trewick, S.A.; Taylor-Smith, B.L.; Morgan-Richards, M. Ecology and systematics of the wine wētā and allied species, with description of four new Hemiandrus species. N. Z. J. Zool. 2020, 48, 47–80. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Bechenhach, A.; Crespi, B.; Liue, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Bulgarella, M.; Trewick, S.A.; Minards, N.A.; Jacobson, M.J.; Morgan-Richards, M. Shifting ranges of two tree weta species (Hemideina spp.): Competitive exclusion and changing climate. J. Bioleography 2014, 41, 524–535. [Google Scholar] [CrossRef]
- Chappell, E.M.; Trewick, S.A.; Morgan-Richards, M. Shape and sound reveal genetic cohesion not speciation in the New Zealand orthopteran, Hemiandrus pallitarsis, despite high mtDNA divergence. Biol. J. Linn. Soc. 2012, 105, 169–186. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Burge, P.I. Repertoire of male-male agonistic behaviour in tusked weta (Motuweta riparia Gibbs) (Orthoptera: Anostostomatidae) compared to tree weta (Hemideina Walker) (Orthoptera: Anostostomatidae). N. Z. Entomol. 2005, 28, 15–27. [Google Scholar] [CrossRef]
- Griffini, A. Stenopelmatidae della Nuova-Caledonia. In Nova Caledonia A. Zoologie; C.W. Kreidel’s Verlag: Wiesbaden, Germany, 1914; Volume 1, pp. 283–311. [Google Scholar]
- Field, L.H.; Deans, N.A. Sexual selection and secondary sexual characters of wetas and king crickets. In The Biology of Wetas, King Crickets and Their Allies; Field, L.H., Ed.; CABI Publishing: Wallingford, UK, 2001; pp. 179–204. [Google Scholar]
- Ander, K. Diagnosen neuer Laubheuschrecken. Opusc. Entomol. 1938, 3, 50–56. [Google Scholar]
- Ingrisch, S. Review of the genus Pteranabropsis (Anostostomatidae: Anabropsinae) with description of six new species. J. Orthoptera Res. 2019, 28, 107–124. [Google Scholar] [CrossRef]
- Cadena-Castañeda, O.J.; Weissman, D.B.; Gonzalez-Castillo, N.S.; Castilo-Avilla, C.; Arias-Pineda, J.Y. A new unusual Anabropsis species from Mexico, with notes on the identity of A. rehni Griffini from Africa (Orthoptera: Anostostomatidae). Zootaxa 2020, 4860, 592–600. [Google Scholar] [CrossRef]
- Trewick, S.A. A new weta from the Chatham Islands (Orthoptera: Raphidophoridae). J. Royal Soc. N. Z. 1999, 29, 165–173. [Google Scholar] [CrossRef]
- Trewick, S.A. Flightless and phylogeny amongst endemic rails (Aves: Rallidae) of the New Zealand region. Phil. Trans. Royal Soc. 1997, 352, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Wright, N.A.; Steadman, D.W.; Witt, C.C. Predictable evolution toward flightlessness in volant island birds. Proc. Natl. Acad. Sci. USA 2016, 113, 4765–4770. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, J.; Gibb, G.C.; Trewick, S.A. Convergent morphological responses to loss of flight in rails (Aves: Rallidae). Ecol. Evol. 2020, 10, 6186–6207. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Gibb, G.C.; Crimp, E.A.; Penny, D. Tinamous and moa flock together: Mitochondrial genome sequence analysis reveals independent losses of flight among ratites. Syst. Biol. 2010, 59, 90–107. [Google Scholar] [CrossRef]
- Crisp, M.; Trewick, S.A.; Cook, L.G. Hypothesis testing in biogeography. Trends Ecol. Evol. 2011, 26, 66–72. [Google Scholar] [CrossRef]
- Grandcolas, P.; Nattier, R.; Trewick, S.A. Relict species: A relict concept? Trends Ecol. Evol. 2014, 29, 655–663. [Google Scholar] [CrossRef] [PubMed]


| Taxon | Classification | Location | MPN Code | GenBank mtDNA | GenBank Histone | Collector |
|---|---|---|---|---|---|---|
| Carcinopsis sp. Brunner 1888 [23] | Anostostomatinae | Col d’Amieu, New Caledonia | ORT388 | PQ442192 | PP965128, PP965140, PP965152, PP965164 | E.X.M. Trewick |
| Deinacrida connectens (Ander, 1939) [24] | Deinacridini | Mount Peel, South Island, New Zealand | Dco2011 | PQ442198 | PP965131, PP965143, PP965155, PP965167 | S.A. Trewick |
| Exogryllacris ornata Willemse, 1963 [25] | Anabropsini | Bartle Frere, Queensland, Australia | ORT193 | PQ442190 | PP965130, PP965142, PP965154, PP965166 | G. Monteith |
| Hemiandrus ‘Rakiura’ | Unplaced | Tin Range, Rakiura, New Zealand | GW1392 | PQ442194 | PP965139, PP965151, PP965163, PP965175 | D. Hegg |
| Hemiandrus brucei Taylor-Smith, 2016 [26] | Unplaced | Whanganui, North Island, New Zealand | Hbr2011 | PQ442197 | PP965138, PP965150, PP965162, PP965174 | B.L. Taylor-Smith |
| Hemiandrus focalis (Hutton, 1897) [27] | Unplaced | Lake Alta, South Island, New Zealand | GW262 | PQ442196 | PP965134, PP965146, PP965158, PP965170 | M. Morgan-Richards |
| Hemiandrus pallitarsis (Walker, 1869) [28] | Unplaced | Palmerston North, North Island, New Zealand | Hpa2012 | PQ442195 | PP965133, PP965145, PP965157, PP965169 | S.A. Trewick |
| ‘Hemiandrus’ sp. [1] | Unplaced | Bellenden Ker, Queensland, Australia | ORT190 | PQ442193 | PP965137, PP965149, PP965161, PP965173 | G. Monteith |
| Hemideina crassidens (Blanchard, 1851) [29] | Deinacridini | Maitai Valley, South Island, New Zealand | Hcr2011 | PQ452770 | PP965132, PP965144, PP965156, PP965168 | S.A. Trewick |
| Motuweta riparia (Gibbs, 2002) [30] | Anostostomatini | Raukumura Range, North Island, New Zealand | TW29 | PQ423746 | PP965129, PP965141, PP965153, PP965165 | E. Dowle |
| Penalva flavocalceata (Karny, 1929) [31] | Anabropsini | Bartle Frere, Queensland, Australia | ORT200 | PQ442191 | PP965136, PP965148, PP965160, PP965172 | G. Monteith |
| Transaevum laudatum (Johns, 1997) [1] | Unplaced | Mt Finnigan, Queensland, Australia | ORT178 | PQ442189 | PP965135, PP965147, PP965159, PP965171 | G. Monteith |
| Species | MPN Code | Location | Year | GenBank Accession |
|---|---|---|---|---|
| Hemiandrus brucei (Anderus brucei nov. comb.) | GW218 | Raurimu | 2007 | EU676796 |
| GW126 | Pureora Forest | 2005 | EU676793 | |
| GW49A | Puketi Forest | 1990 | EU676765 | |
| GW04 | Manganuku | 1998 | EU676798 | |
| GW93A | Pelorus Bridge | 2005 | EU676791 | |
| Hemiandrus luna (Anderus luna nov. comb.) | GW104 | Sky Farm | 2006 | EU676742 |
| GW143 | Lewis Pass | 2006 | EU676784 | |
| GW1385 | Tongariro | 2021 | PP34546 | |
| GW916B | Arthurs Pass | 2013 | PP34545 | |
| Hemiandrus fiordensis (Anderus fiordensis nov. comb.) | GW1481 | Stuart Mountains | 2022 | PP34550 |
| FD3(nitaweta) | Sinbad Valley | 2013 | PP34541 | |
| GW70 | Lake Roe | 2004 | PP34547 | |
| GW1516 | Shy Lake | 2022 | PP34553 | |
| Hemiandrus nox (Anderus nox nov. comb.) | GW834 | Hokitika | 2012 | PP34552 |
| GW76 | Awakiri Valley | 1997 | EU676766 | |
| GW896a | Denniston Plateau | 2012 | PP34549 | |
| GW899 | Denniston Plateau | 2013 | PP34548 | |
| GW1542 | St Arnaud | 2005 | PP34551 | |
| Hemiandrus maculifrons (Anderus maculifrons nov. comb.) | GW119 | Catlins Coast | 2006 | EU676770 |
| GW201 | Takitimu Mountains | 2006 | EU676772 | |
| GW68 | Mount Fyffe | 2004 | EU676787 | |
| GW217 | Kahurangi | 2007 | EU676776 | |
| GW150 | Franz Josef | 2006 | EU676786 | |
| Hemiandrus subantarcticus (Anderus subantarcticus nov. comb.) | GW988 | The Snares | 2010 | MW463359 |
| GW792 | The Snares | 2010 | PP34544 | |
| GW989 | The Snares | 2010 | MW463360 | |
| Hemiandrus electra | GW138 | St Arnaud | 2005 | EU676783 |
| GW1028 | Mount Richmond | 2013 | PP34562 | |
| GW1029 | Mount Richmond | 2013 | PP34561 | |
| Hemiandrus focalis | GW1212 | Takitimu Mountains | 2019 | PP34554 |
| GW1211 | Takitimu Mountains | 2019 | PP34555 | |
| FD7 | Eyre Mountains | 1999 | PP34542 | |
| GW206 | Obelisk | 2006 | EU676774 | |
| GW08 | Harris Saddle | 1999 | EU676773 | |
| Hemiandrus superbus | FD5 | Sinbad Valley | 2013 | PP34543 |
| FD10 | Sinbad Valley | 2010 | MW463358 | |
| FD8 | Sinbad Valley | 2011 | MW463357 | |
| GW1198 | Skippers Range | 2019 | PP34556 | |
| Hemiandrus jacinda | GW1376 | Pouakai | 2021 | PP34560 |
| GW1486 | Pukeiti | 2022 | PP34559 | |
| GW208 | Whareorino | 2006 | MW463354 | |
| GW1328 | Thames | 2020 | MW463352 | |
| GW62 | Moehau | 1990 | MW463353 | |
| Hemiandrus maia | GW125 | Portabello | 2006 | EU676744 |
| GW1067 | Mount Kyeburn | 2013 | PP34557 | |
| GW118 | Hampden | 2006 | EU676795 | |
| GW136 | Blue Mountains | 2006 | EU676780 | |
| Hemiandrus merope | GW674 | Kapiti Island | 2011 | MT6323126 |
| GW682 | Kapiti Island | 2011 | MT623127 | |
| Hemiandrus palllitarsis | GW227 | Hauturu | 2007 | JF895541 |
| GW226 | Hauturu | 2007 | JF895542 | |
| KA90 | Kauaeranga Valley | 2012 | JF895543 | |
| KA88 | Kauaeranga Valley | 2012 | F895547 | |
| MI98 | Middle Island | 2012 | JF895548 | |
| CO145 | Moehau | 2012 | JF895551 | |
| GW87 | Pohaninga Valley | 2004 | JF895554 | |
| Hemiandrus celaeno | GW120 | Banks Peninsular | 2005 | EU676771 |
| GW127 | Foggy Peak | 2006 | EU676778 | |
| GW129 | Foggy Peak | 2006 | EU676779 | |
| Hemiandrus sterope | GW1033 | Lewis Pass | 2014 | MT623110 |
| GW1047 | Cable Bay | 2014 | MT623103 | |
| GW717 | Manaroa | 2012 | MT623102 | |
| GW602 | Te Rua Bay | 2010 | MT623101 | |
| GW54 | Whites Bay | 1990 | EU676788 | |
| Hemiandrus taygete | GW491 | Kaikoura | 2009 | MT623112 |
| GW1031 | Mount Richmond | 2013 | MT623115 | |
| GW871 | Upper Clarence Valley | 2012 | MT623113 | |
| GW1231 | Youngman Stream | 2019 | PP34558 | |
| Hemiandrus bilobatus | GW586 | Awatere Valley | 2010 | MT623085 |
| GW25 | Wellington | 1999 | EU676794 | |
| GW240 | Wellington | 2007 | JF895562 | |
| GW657 | Mana Island | 2010 | MT623095 | |
| GW122 | Marfells Beach | 2000 | EU676777 | |
| GW55 | Marfells Beach | 1990 | EU676789 | |
| GW193 | Muritai | 2006 | JF895564 |
| Hypothesis | logL | deltaL | bp-RELL | p-KH | p-SH | c-ELW |
|---|---|---|---|---|---|---|
| 1 | −89,689.86177 | 679.86 | 0− | 0− | 0− | 3.53 × 10−231− |
| 2 | −90,220.76741 | 1210.8 | 0− | 0− | 0− | 0− |
| 3 | −89,010.00659 | 0 | 1+ | 1+ | 1+ | 1+ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trewick, S.A.; Taylor-Smith, B.L.; Morgan-Richards, M. Wētā Aotearoa—Polyphyly of the New Zealand Anostostomatidae (Insecta: Orthoptera). Insects 2024, 15, 787. https://doi.org/10.3390/insects15100787
Trewick SA, Taylor-Smith BL, Morgan-Richards M. Wētā Aotearoa—Polyphyly of the New Zealand Anostostomatidae (Insecta: Orthoptera). Insects. 2024; 15(10):787. https://doi.org/10.3390/insects15100787
Chicago/Turabian StyleTrewick, Steven A., Briar L. Taylor-Smith, and Mary Morgan-Richards. 2024. "Wētā Aotearoa—Polyphyly of the New Zealand Anostostomatidae (Insecta: Orthoptera)" Insects 15, no. 10: 787. https://doi.org/10.3390/insects15100787
APA StyleTrewick, S. A., Taylor-Smith, B. L., & Morgan-Richards, M. (2024). Wētā Aotearoa—Polyphyly of the New Zealand Anostostomatidae (Insecta: Orthoptera). Insects, 15(10), 787. https://doi.org/10.3390/insects15100787







