Basic Structures of Gut Bacterial Communities in Eusocial Insects
Abstract
Simple Summary
Abstract
1. Introduction
2. General Ecology of Social Insects
3. Eusocial Bee Gut Microbiota
3.1. Apis mellifera and Other Honey Bees
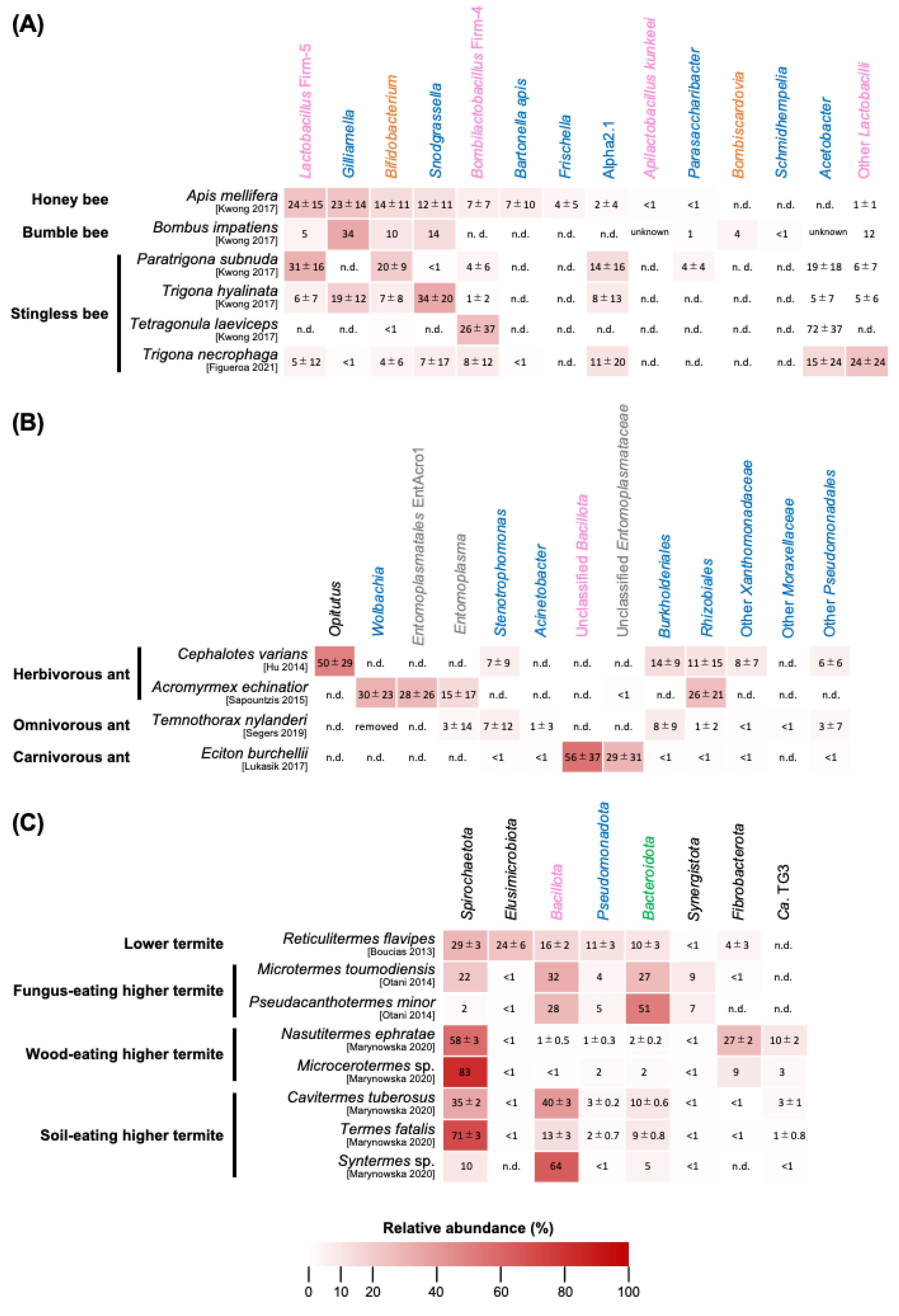

3.2. Bumble Bees
3.3. Stingless Bees
3.4. Summary of Bee Gut Microbiota
4. Ant Gut Microbiota
4.1. Herbivorous Ants
4.2. Omnivorous Ants
4.3. Carnivorous Ants
4.4. Summary of Ant Gut Microbiota
5. Termite Gut Microbiota
5.1. Lower Termites
5.2. Fungus-Eating Higher Termites
5.3. Wood/Soil-Eating Higher Termites
5.4. Conclusions of Termite Gut Microbiota
6. Gut Microbiota in Other Social Insects
7. Concluding Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sommer, F.; Backhed, F. The gut microbiota-masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Moeller, A.H.; Foerster, S.; Wilson, M.L.; Pusey, A.E.; Hahn, B.H.; Ochman, H. Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2016, 2, e1500997. [Google Scholar] [CrossRef] [PubMed]
- Raulo, A.; Ruokolainen, L.; Lane, A.; Amato, K.; Knight, R.; Leigh, S.; Stumpf, R.; White, B.; Nelson, K.E.; Baden, A.L.; et al. Social behaviour and gut microbiota in red-bellied lemurs (Eulemur rubriventer): In search of the role of immunity in the evolution of sociality. J. Anim. Ecol. 2018, 87, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Pukall, R.; Abt, B.; Foesel, B.U.; Meier-Kolthoff, J.P.; Kumar, N.; Bresciani, A.; Martinez, I.; Just, S.; Ziegler, C.; et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 2016, 1, 16131. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Bisch, G.; Neuvonen, M.M.; Pierce, N.E.; Russell, J.A.; Koga, R.; Sanders, J.G.; Lukasik, P.; Andersson, S.G.E. Genome Evolution of Bartonellaceae Symbionts of Ants at the Opposite Ends of the Trophic Scale. Genome Biol. Evol. 2018, 10, 1687–1704. [Google Scholar] [CrossRef]
- Hu, Y.; Sanders, J.G.; Lukasik, P.; D’Amelio, C.L.; Millar, J.S.; Vann, D.R.; Lan, Y.; Newton, J.A.; Schotanus, M.; Kronauer, D.J.C.; et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018, 9, 964. [Google Scholar] [CrossRef]
- Kesnerova, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef]
- Hammer, T.J.; Moran, N.A. Links between metamorphosis and symbiosis in holometabolous insects. Philos. Trans. R Soc. Lond. B Biol. Sci. 2019, 374, 20190068. [Google Scholar] [CrossRef]
- Moll, R.M.; Romoser, W.S.; Modrzakowski, M.C.; Moncayo, A.C.; Lerdthusnee, K. Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J. Med. Entomol. 2001, 38, 29–32. [Google Scholar] [CrossRef]
- Bright, M.; Bulgheresi, S. A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 2010, 8, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Kikuchi, Y.; Fukatsu, T. How many symbionts are provided by mothers, acquired by offspring, and needed for successful vertical transmission in an obligate insect-bacterium mutualism? Mol. Ecol. 2007, 16, 5316–5325. [Google Scholar] [CrossRef]
- Hongoh, Y.; Deevong, P.; Inoue, T.; Moriya, S.; Trakulnaleamsai, S.; Ohkuma, M.; Vongkaluang, C.; Noparatnaraporn, N.; Kudo, T. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 2005, 71, 6590–6599. [Google Scholar] [CrossRef] [PubMed]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of Acquisition of the Gut Microbiota of the Honey Bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Shapira, M. Gut Microbiotas and Host Evolution: Scaling Up Symbiosis. Trends Ecol. Evol. 2016, 31, 539–549. [Google Scholar] [CrossRef]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Hammer, T.J.; Le, E.; Martin, A.N.; Moran, N.A. The gut microbiota of bumblebees. Insectes Soc. 2021, 68, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Hornett, E.A.; Kageyama, D.; Hurst, G.D.D. Sex determination systems as the interface between male-killing bacteria and their hosts. Proc. Biol. Sci. 2022, 289, 20212781. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Shropshire, J.D.; Cross, K.L.; Leigh, B.; Mansueto, A.J.; Stewart, V.; Bordenstein, S.R.; Bordenstein, S.R. Living in the endosymbiotic world of Wolbachia: A centennial review. Cell Host. Microbe. 2021, 29, 879–893. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; Boyd, B.M.; Dale, C. The Life of an Insect Endosymbiont from the Cradle to the Grave. Curr. Biol. 2019, 29, R485–R495. [Google Scholar] [CrossRef]
- Leonhardt, S.D.; Menzel, F.; Nehring, V.; Schmitt, T. Ecology and Evolution of Communication in Social Insects. Cell 2016, 164, 1277–1287. [Google Scholar] [CrossRef]
- Holldobler, B.; Wilson, E.O. The Ants; Springer: Berlin/Heidelberg, Germany, 1990. [Google Scholar]
- Goulson, D.; Peat, J.; Stout, J.C.; Tucker, J.; Darvill, B.; Derwent, L.C.; Hughes, W.O.H. Can alloethism in workers of the bumblebee, Bombus terrestris, be explained in terms of foraging efficiency? Anim. Behav. 2002, 64, 123–130. [Google Scholar] [CrossRef]
- Gruter, C.; Menezes, C.; Imperatriz-Fonseca, V.L.; Ratnieks, F.L. A morphologically specialized soldier caste improves colony defense in a neotropical eusocial bee. Proc. Natl. Acad. Sci. USA 2012, 109, 1182–1186. [Google Scholar] [CrossRef]
- Koto, A.; Motoyama, N.; Tahara, H.; McGregor, S.; Moriyama, M.; Okabe, T.; Miura, M.; Keller, L. Oxytocin/vasopressin-like peptide inotocin regulates cuticular hydrocarbon synthesis and water balancing in ants. Proc. Natl. Acad. Sci. USA 2019, 116, 5597–5606. [Google Scholar] [CrossRef]
- Seid, M.A.; Traniello, J.F.A. Age-related repertoire expansion and division of labor in Pheidole dentata (Hymenoptera: Formicidae): A new perspective on temporal polyethism and behavioral plasticity in ants. Behav. Ecol. Sociobiol. 2006, 60, 631–644. [Google Scholar] [CrossRef]
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Gunkel, S.; Meusemann, K.; Kozlov, A.; Podsiadlowski, L.; Petersen, M.; Lanfear, R.; et al. Evolutionary History of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef]
- Mateus, S.; Ferreira-Caliman, M.J.; Menezes, C.; Grüter, C. Beyond temporal-polyethism: Division of labor in the eusocial bee Melipona marginata. Insectes Sociaux 2019, 66, 317–328. [Google Scholar] [CrossRef]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella apicola. MBio 2016, 7, e01326-16. [Google Scholar] [CrossRef] [PubMed]
- Holtof, M.; Lenaerts, C.; Cullen, D.; Vanden Broeck, J. Extracellular nutrient digestion and absorption in the insect gut. Cell Tissue Res. 2019, 377, 397–414. [Google Scholar] [CrossRef]
- Han, F.; Wallberg, A.; Webster, M.T. From where did the Western honeybee (Apis mellifera) originate? Ecol. Evol. 2012, 2, 1949–1957. [Google Scholar] [CrossRef]
- Raffiudin, R.; Crozier, R.H. Phylogenetic analysis of honey bee behavioral evolution. Mol. Phylogenet. Evol. 2007, 43, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, MA, USA; London, UK, 1991. [Google Scholar]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.J.Y.; Ascher, J.S.; Jaffe, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Cannone, J.J.; Gutell, R.R.; Kellner, K.; Plowes, R.M.; Mueller, U.G. Specificity between lactobacilli and hymenopteran hosts is the exception rather than the rule. Appl. Environ. Microbiol. 2013, 79, 1803–1812. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 2019, 10, 446. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Fruciano, C.; Marchant, J.; Hildebrand, F.; Forslund, S.; Bork, P.; Engel, P.; Hughes, W.O.H. The gut microbiome is associated with behavioural task in honey bees. Insectes Soc. 2018, 65, 419–429. [Google Scholar] [CrossRef]
- Anderson, K.E.; Ricigliano, V.A.; Mott, B.M.; Copeland, D.C.; Floyd, A.S.; Maes, P. The queen’s gut refines with age: Longevity phenotypes in a social insect model. Microbiome 2018, 6, 108. [Google Scholar] [CrossRef]
- Boucias, D.G.; Cai, Y.; Sun, Y.; Lietze, V.U.; Sen, R.; Raychoudhury, R.; Scharf, M.E. The hindgut lumen prokaryotic microbiota of the termite Reticulitermes flavipes and its responses to dietary lignocellulose composition. Mol. Ecol. 2013, 22, 1836–1853. [Google Scholar] [CrossRef]
- Figueroa, L.L.; Maccaro, J.J.; Krichilsky, E.; Yanega, D.; McFrederick, Q.S. Why Did the Bee Eat the Chicken? Symbiont Gain, Loss, and Retention in the Vulture Bee Microbiome. mBio 2021, 12, e0231721. [Google Scholar] [CrossRef]
- Hu, Y.; Lukasik, P.; Moreau, C.S.; Russell, J.A. Correlates of gut community composition across an ant species (Cephalotes varians) elucidate causes and consequences of symbiotic variability. Mol. Ecol. 2014, 23, 1284–1300. [Google Scholar] [CrossRef]
- Lukasik, P.; Newton, J.A.; Sanders, J.G.; Hu, Y.; Moreau, C.S.; Kronauer, D.J.C.; O’Donnell, S.; Koga, R.; Russell, J.A. The structured diversity of specialized gut symbionts of the New World army ants. Mol. Ecol. 2017, 26, 3808–3825. [Google Scholar] [CrossRef]
- Marynowska, M.; Goux, X.; Sillam-Dusses, D.; Rouland-Lefevre, C.; Halder, R.; Wilmes, P.; Gawron, P.; Roisin, Y.; Delfosse, P.; Calusinska, M. Compositional and functional characterisation of biomass-degrading microbial communities in guts of plant fibre- and soil-feeding higher termites. Microbiome 2020, 8, 96. [Google Scholar] [CrossRef]
- Otani, S.; Mikaelyan, A.; Nobre, T.; Hansen, L.H.; Kone, N.A.; Sorensen, S.J.; Aanen, D.K.; Boomsma, J.J.; Brune, A.; Poulsen, M. Identifying the core microbial community in the gut of fungus-growing termites. Mol. Ecol. 2014, 23, 4631–4644. [Google Scholar] [CrossRef] [PubMed]
- Sapountzis, P.; Zhukova, M.; Hansen, L.H.; Sorensen, S.J.; Schiott, M.; Boomsma, J.J. Acromyrmex Leaf-Cutting Ants Have Simple Gut Microbiota with Nitrogen-Fixing Potential. Appl. Environ. Microbiol. 2015, 81, 5527–5537. [Google Scholar] [CrossRef] [PubMed]
- Segers, F.; Kaltenpoth, M.; Foitzik, S. Abdominal microbial communities in ants depend on colony membership rather than caste and are linked to colony productivity. Ecol. Evol. 2019, 9, 13450–13467. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Suenami, S.; Miyazaki, R.; Engel, P. Vast Differences in Strain-Level Diversity in the Gut Microbiota of Two Closely Related Honey Bee Species. Curr. Biol. 2020, 30, 2520–2531.e2527. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef] [PubMed]
- Kesnerova, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Otani, S.; Zhukova, M.; Kone, N.A.; da Costa, R.R.; Mikaelyan, A.; Sapountzis, P.; Poulsen, M. Gut microbial compositions mirror caste-specific diets in a major lineage of social insects. Environ. Microbiol. Rep. 2019, 11, 196–205. [Google Scholar] [CrossRef]
- Belsky, J.E.; Camp, A.A.; Lehmann, D.M. The Importance of Males to Bumble Bee (Bombus Species) Nest Development and Colony Viability. Insects 2020, 11, 506. [Google Scholar] [CrossRef]
- Sadd, B.M.; Barribeau, S.M.; Bloch, G.; de Graaf, D.C.; Dearden, P.; Elsik, C.G.; Gadau, J.; Grimmelikhuijzen, C.J.; Hasselmann, M.; Lozier, J.D.; et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015, 16, 76. [Google Scholar] [CrossRef]
- Powell, E.; Ratnayeke, N.; Moran, N.A. Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol. Ecol. 2016, 25, 4461–4471. [Google Scholar] [CrossRef]
- Martinson, V.G.; Magoc, T.; Koch, H.; Salzberg, S.L.; Moran, N.A. Genomic features of a bumble bee symbiont reflect its host environment. Appl. Environ. Microbiol. 2014, 80, 3793–3803. [Google Scholar] [CrossRef] [PubMed]
- Killer, J.; Kopecny, J.; Mrazek, J.; Havlik, J.; Koppova, I.; Benada, O.; Rada, V.; Kofronova, O. Bombiscardovia coagulans gen. nov., sp. nov., a new member of the family Bifidobacteriaceae isolated from the digestive tract of bumblebees. Syst. Appl. Microbiol. 2010, 33, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef] [PubMed]
- Leger, L.; McFrederick, Q.S. The Gut-Brain-Microbiome Axis in Bumble Bees. Insects 2020, 11, 517. [Google Scholar] [CrossRef]
- Li, L.; Solvi, C.; Zhang, F.; Qi, Z.; Chittka, L.; Zhao, W. Gut microbiome drives individual memory variation in bumblebees. Nat. Commun. 2021, 12, 6588. [Google Scholar] [CrossRef]
- Li, J.; Powell, J.E.; Guo, J.; Evans, J.D.; Wu, J.; Williams, P.; Lin, Q.; Moran, N.A.; Zhang, Z. Two gut community enterotypes recur in diverse bumblebee species. Curr. Biol. 2015, 25, R652–R653. [Google Scholar] [CrossRef]
- Parmentier, A.; Meeus, I.; Van Nieuwerburgh, F.; Deforce, D.; Vandamme, P.; Smagghe, G. A different gut microbial community between larvae and adults of a wild bumblebee nest (Bombus pascuorum). Insect Sci. 2018, 25, 66–74. [Google Scholar] [CrossRef]
- Wang, L.; Wu, J.; Li, K.; Sadd, B.M.; Guo, Y.; Zhuang, D.; Zhang, Z.; Chen, Y.; Evans, J.D.; Guo, J.; et al. Dynamic Changes of Gut Microbial Communities of Bumble Bee Queens through Important Life Stages. mSystems 2019, 4, e00631-19. [Google Scholar] [CrossRef]
- Bosmans, L.; Pozo, M.I.; Verreth, C.; Crauwels, S.; Wackers, F.; Jacquemyn, H.; Lievens, B. Hibernation Leads to Altered Gut Communities in Bumblebee Queens (Bombus terrestris). Insects 2018, 9, 188. [Google Scholar] [CrossRef]
- Rasmussen, C.; Cameron, S.A. Global stingless bee phylogeny supports ancient divergence, vicariance, and long distance dispersal. Biol. J. Linn. Soc. 2009, 99, 206–232. [Google Scholar] [CrossRef]
- Hartfelder, K.; Makert, G.R.; Judice, C.C.; Pereira, G.A.G.; Santana, W.C.; Dallacqua, R.; Bitondi, M.M.G. Physiological and genetic mechanisms underlying caste development, reproduction and division of labor in stingless bees. Apidologie 2006, 37, 144–163. [Google Scholar] [CrossRef]
- Gruter, C.; Segers, F.H.; Menezes, C.; Vollet-Neto, A.; Falcon, T.; von Zuben, L.; Bitondi, M.M.; Nascimento, F.S.; Almeida, E.A. Repeated evolution of soldier sub-castes suggests parasitism drives social complexity in stingless bees. Nat. Commun. 2017, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Sarton-Loheac, G.; Nunes da Silva, C.G.; Mazel, F.; Baud, G.; de Bakker, V.; Das, S.; El Chazli, Y.; Ellegaard, K.; Garcia-Garcera, M.; Glover, N.; et al. Deep Divergence and Genomic Diversification of Gut Symbionts of Neotropical Stingless Bees. mBio 2023, 14, e0353822. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, A.E.S.; Hammer, T.J.; Moran, N.A.; Santana, W.C.; Kasuya, M.C.M.; da Silva, C.C. Extinction of anciently associated gut bacterial symbionts in a clade of stingless bees. ISME J. 2021, 15, 2813–2816. [Google Scholar] [CrossRef] [PubMed]
- Roubik, D.W. Obligate necrophagy in a social bee. Science 1982, 217, 1059–1060. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A.; Brettell, L.E.; Liu, H.; Nacko, S.; Spooner-Hart, R.; Riegler, M.; Cook, J.M. Temporal changes in the microbiome of stingless bee foragers following colony relocation. FEMS Microbiol. Ecol. 2020, 97, fiaa236. [Google Scholar] [CrossRef]
- Moreau, C.S. Symbioses among ants and microbes. Curr. Opin. Insect Sci. 2020, 39, 1–5. [Google Scholar] [CrossRef]
- Cook, S.C.; Davidson, D.W. Nutritional and functional biology of exudate-feeding ants. Entomologia Experimentalis Applicata 2006, 118, 1–10. [Google Scholar] [CrossRef]
- Roche, R.K.; Wheeler, D.E. Morphological specializations of the digestive tract ofZacryptocerus rohweri (Hymenoptera: Formicidae). J. Morphol. 1997, 234, 253–262. [Google Scholar] [CrossRef]
- Sanders, J.G.; Lukasik, P.; Frederickson, M.E.; Russell, J.A.; Koga, R.; Knight, R.; Pierce, N.E. Dramatic Differences in Gut Bacterial Densities Correlate with Diet and Habitat in Rainforest Ants. Integr. Comp. Biol. 2017, 57, 705–722. [Google Scholar] [CrossRef]
- Anderson, K.E.; Russell, J.A.; Moreau, C.S.; Kautz, S.; Sullam, K.E.; Hu, Y.; Basinger, U.; Mott, B.M.; Buck, N.; Wheeler, D.E. Highly similar microbial communities are shared among related and trophically similar ant species. Mol. Ecol. 2012, 21, 2282–2296. [Google Scholar] [CrossRef] [PubMed]
- Urbani, C.B.; de Andrade, M.L. Pollen Eating, Storing, and Spitting by Ants. Naturwissenschaften 1997, 84, 256–258. [Google Scholar] [CrossRef]
- Russell, J.A.; Moreau, C.S.; Goldman-Huertas, B.; Fujiwara, M.; Lohman, D.J.; Pierce, N.E. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl. Acad. Sci. USA 2009, 106, 21236–21241. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.B.; Hansen, L.H.; Sapountzis, P.; Sorensen, S.J.; Boomsma, J.J. Specificity and stability of the Acromyrmex-Pseudonocardia symbiosis. Mol. Ecol. 2013, 22, 4307–4321. [Google Scholar] [CrossRef]
- Zhukova, M.; Sapountzis, P.; Schiott, M.; Boomsma, J.J. Diversity and Transmission of Gut Bacteria in Atta and Acromyrmex Leaf-Cutting Ants during Development. Front. Microbiol. 2017, 8, 1942. [Google Scholar] [CrossRef]
- Hammer, T.J.; Sanders, J.G.; Fierer, N. Not all animals need a microbiome. FEMS Microbiol. Lett. 2019, 366, fnz117. [Google Scholar] [CrossRef]
- Russell, J.A.; Sanders, J.G.; Moreau, C.S. Hotspots for symbiosis: Function, evolution, and specificity of ant-microbe associations from trunk to tips of the ant phylogeny (Hymenoptera: Formicidae). Myrmecol. News. 2017, 24, 43–69. [Google Scholar] [CrossRef]
- Brown, B.P.; Wernegreen, J.J. Deep divergence and rapid evolutionary rates in gut-associated Acetobacteraceae of ants. BMC Microbiol. 2016, 16, 140. [Google Scholar] [CrossRef]
- Koto, A.; Nobu, M.K.; Miyazaki, R. Deep Sequencing Uncovers Caste-Associated Diversity of Symbionts in the Social Ant Camponotus japonicus. mBio 2020, 11, e00408-20. [Google Scholar] [CrossRef]
- Moreau, C.S.; Rubin, B.E.R. Diversity and Persistence of the Gut Microbiome of the Giant Neotropical Bullet Ant. Integr. Comp. Biol. 2017, 57, 682–689. [Google Scholar] [CrossRef]
- Chouvenc, T.; Sobotnik, J.; Engel, M.S.; Bourguignon, T. Termite evolution: Mutualistic associations, key innovations, and the rise of Termitidae. Cell Mol. Life Sci. 2021, 78, 2749–2769. [Google Scholar] [CrossRef] [PubMed]
- Bucek, A.; Sobotnik, J.; He, S.; Shi, M.; McMahon, D.P.; Holmes, E.C.; Roisin, Y.; Lo, N.; Bourguignon, T. Evolution of Termite Symbiosis Informed by Transcriptome-Based Phylogenies. Curr. Biol. 2019, 29, 3728–3734.e3724. [Google Scholar] [CrossRef] [PubMed]
- Mikaelyan, A.; Dietrich, C.; Kohler, T.; Poulsen, M.; Sillam-Dusses, D.; Brune, A. Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol. Ecol. 2015, 24, 5284–5295. [Google Scholar] [CrossRef]
- Mikaelyan, A.; Meuser, K.; Brune, A. Microenvironmental heterogeneity of gut compartments drives bacterial community structure in wood- and humus-feeding higher termites. FEMS Microbiol. Ecol. 2017, 93, fiw210. [Google Scholar] [CrossRef]
- Donovan, S.E.; Jones, D.T.; Sands, W.A.; Eggleton, P. Morphological phylogenetics of termites (Isoptera). Biol. J. Linn. Soc. 2000, 70, 467–513. [Google Scholar] [CrossRef]
- Ohkuma, M. Symbioses of flagellates and prokaryotes in the gut of lower termites. Trends Microbiol. 2008, 16, 345–352. [Google Scholar] [CrossRef]
- Berlanga, M.; Paster, B.J.; Grandcolas, P.; Guerrero, R. Comparison of the gut microbiota from soldier and worker castes of the termite Reticulitermes grassei. Int. Microbiol. 2011, 14, 83–93. [Google Scholar] [CrossRef]
- Cameron, S.L.; Lo, N.; Bourguignon, T.; Svenson, G.J.; Evans, T.A. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): Robust support for interfamilial relationships and molecular synapomorphies define major clades. Mol. Phylogenet. Evol. 2012, 65, 163–173. [Google Scholar] [CrossRef]
- Reid, N.M.; Addison, S.L.; West, M.A.; Lloyd-Jones, G. The bacterial microbiota of Stolotermes ruficeps (Stolotermitidae), a phylogenetically basal termite endemic to New Zealand. FEMS Microbiol. Ecol. 2014, 90, 678–688. [Google Scholar] [CrossRef]
- Dietrich, C.; Kohler, T.; Brune, A. The cockroach origin of the termite gut microbiota: Patterns in bacterial community structure reflect major evolutionary events. Appl. Environ. Microbiol. 2014, 80, 2261–2269. [Google Scholar] [CrossRef]
- Hyodo, F.; Tayasu, I.; Inoue, T.; Azuma, J.I.; Kudo, T.; Abe, T. Differential role of symbiotic fungi in lignin degradation and food provision for fungus-growing termites (Macrotermitinae: Isoptera). Funct. Ecol. 2003, 17, 186–193. [Google Scholar] [CrossRef]
- Su, L.; Yang, L.; Huang, S.; Su, X.; Li, Y.; Wang, F.; Wang, E.; Kang, N.; Xu, J.; Song, A. Comparative Gut Microbiomes of Four Species Representing the Higher and the Lower Termites. J. Insect Sci. 2016, 16, 97. [Google Scholar] [CrossRef]
- Mikaelyan, A.; Kohler, T.; Lampert, N.; Rohland, J.; Boga, H.; Meuser, K.; Brune, A. Classifying the bacterial gut microbiota of termites and cockroaches: A curated phylogenetic reference database (DictDb). Syst. Appl. Microbiol. 2015, 38, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G.; Mikaelyan, A.; Fukui, C.; Matsuura, Y.; Watanabe, H.; Fujishima, M.; Brune, A. Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci. USA 2018, 115, E11996–E12004. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, T.; Lo, N.; Dietrich, C.; Sobotnik, J.; Sidek, S.; Roisin, Y.; Brune, A.; Evans, T.A. Rampant Host Switching Shaped the Termite Gut Microbiome. Curr. Biol. 2018, 28, 649–654 e642. [Google Scholar] [CrossRef]
- Suenami, S.; Konishi Nobu, M.; Miyazaki, R. Community analysis of gut microbiota in hornets, the largest eusocial wasps, Vespa mandarinia and V. simillima. Sci. Rep. 2019, 9, 9830. [Google Scholar] [CrossRef]
- Cini, A.; Meriggi, N.; Bacci, G.; Cappa, F.; Vitali, F.; Cavalieri, D.; Cervo, R. Gut microbial composition in different castes and developmental stages of the invasive hornet Vespa velutina nigrithorax. Sci. Total Environ. 2020, 745, 140873. [Google Scholar] [CrossRef]
- Crespi, B.J. Eusociality in Australian gall thrips. Nature 1992, 359, 724–726. [Google Scholar] [CrossRef]
- Kranz, B.D.; Schwarz, M.P.; Mound, L.A.; Crespi, B.J. Social biology and sex ratios of the eusocial gall-inducing thripsKladothrips hamiltoni. Ecol. Entomol. 1999, 24, 432–442. [Google Scholar] [CrossRef]
- Shibao, H.; Kutsukake, M.; Fukatsu, T. Temporal division of labor in an aphid social system. Sci. Rep. 2021, 11, 1183. [Google Scholar] [CrossRef]
- Xu, T.T.; Chen, J.; Jiang, L.Y.; Qiao, G.X. Diversity of bacteria associated with Hormaphidinae aphids (Hemiptera: Aphididae). Insect Sci. 2021, 28, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.T.; Jiang, L.Y.; Chen, J.; Qiao, G.X. Host Plants Influence the Symbiont Diversity of Eriosomatinae (Hemiptera: Aphididae). Insects 2020, 11, 217. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Yorimoto, S. Aphid hologenomics: Current status and future challenges. Curr. Opin. Insect Sci. 2022, 50, 100882. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suenami, S.; Koto, A.; Miyazaki, R. Basic Structures of Gut Bacterial Communities in Eusocial Insects. Insects 2023, 14, 444. https://doi.org/10.3390/insects14050444
Suenami S, Koto A, Miyazaki R. Basic Structures of Gut Bacterial Communities in Eusocial Insects. Insects. 2023; 14(5):444. https://doi.org/10.3390/insects14050444
Chicago/Turabian StyleSuenami, Shota, Akiko Koto, and Ryo Miyazaki. 2023. "Basic Structures of Gut Bacterial Communities in Eusocial Insects" Insects 14, no. 5: 444. https://doi.org/10.3390/insects14050444
APA StyleSuenami, S., Koto, A., & Miyazaki, R. (2023). Basic Structures of Gut Bacterial Communities in Eusocial Insects. Insects, 14(5), 444. https://doi.org/10.3390/insects14050444




