Parasitization of Aphis gossypii Glover by Binodoxys communis Gahan Causes Shifts in the Ovarian Bacterial Microbiota
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Collection
2.2. Sample Collection and Processing
2.3. DNA Extraction, PCR Amplification, Library Preparation, and Sequencing
2.4. Sequence Data Processing and Analysis
2.5. Quantification of Bacterial Communities
3. Results
3.1. Analysis of the 16S rDNA Sequencing Results
3.2. Community Diversity Analyses
3.3. Analysis of the Microbial Community Composition in th Ovaries
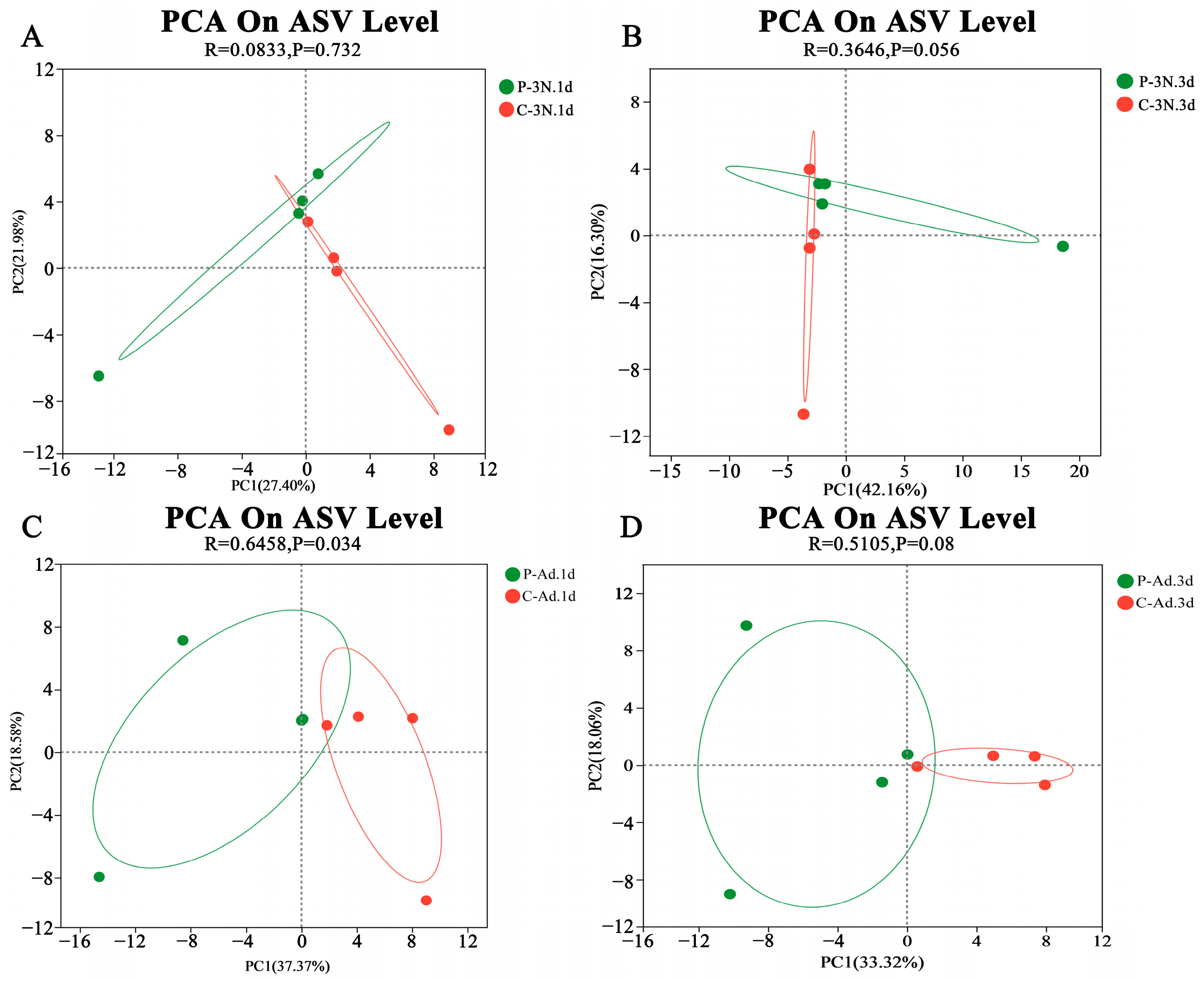
3.4. Microbial Community Alterations Due to Parasitism
3.5. Functional Prediction
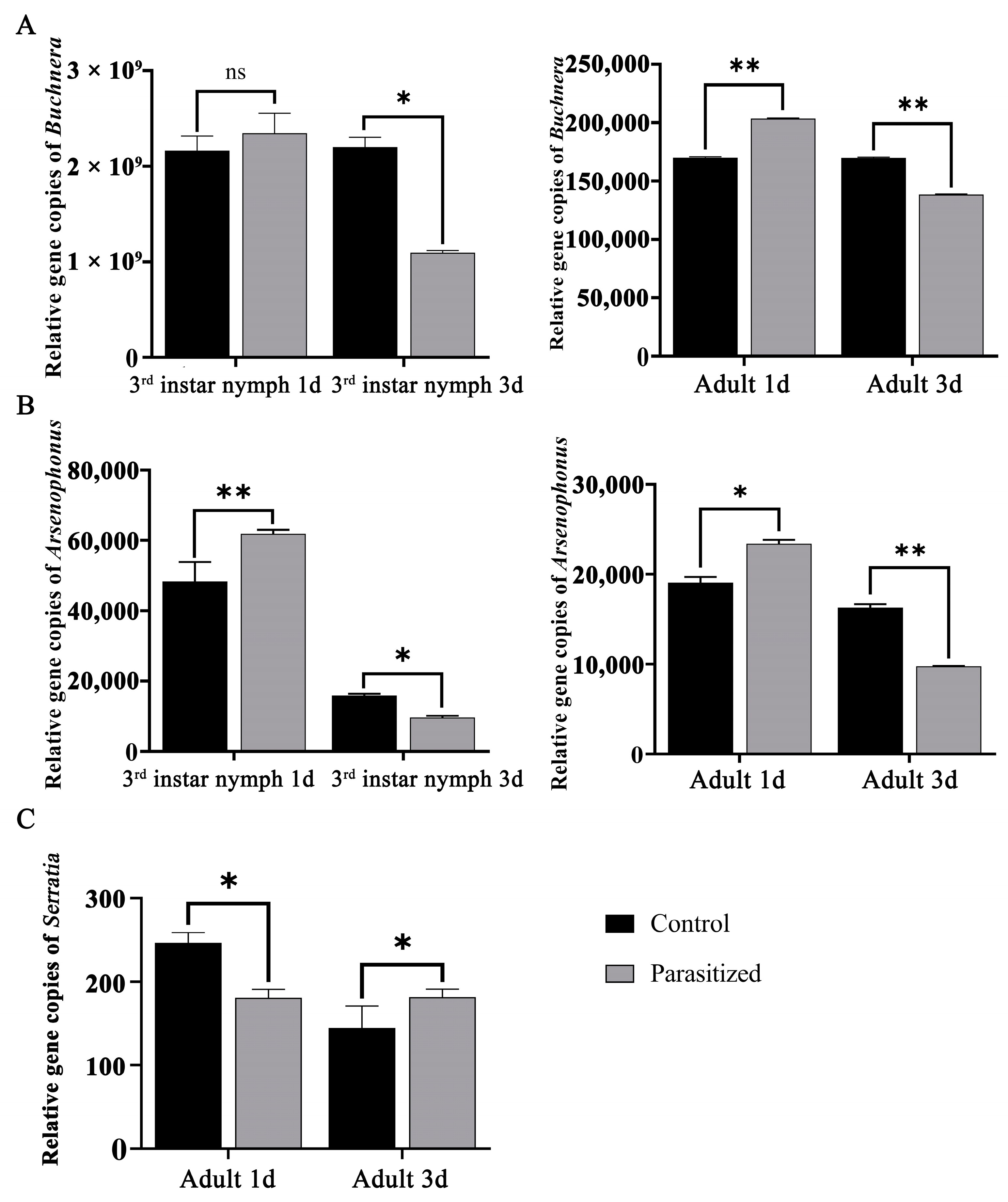
3.6. RT-qPCR of Core Symbiotic Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Koga, R.; Tsuchida, T.; Fukatsu, T. Changing partners in an obligate symbiosis: A facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. B Biol. Sci. 2004, 270, 2543–2550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, H.; Zeng, L.; Yu, Y.; Lin, X.; Huang, X. Coexistence of Three Dominant Bacterial Symbionts in a Social Aphid and Implications for Ecological Adaptation. Insects 2021, 12, 416. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Russell, J.A.; Moran, N.A.; Hunter, M.S. Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 2003, 100, 1803–1807. [Google Scholar] [CrossRef]
- Łukasik, P.; van Asch, M.; Guo, H.; Ferrari, J.; Godfray, H.C. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 2013, 16, 214–218. [Google Scholar] [CrossRef]
- Tsuchida, T.; Koga, R.; Horikawa, M.; Tsunoda, T.; Maoka, T.; Matsumoto, S.; Simon, J.C.; Fukatsu, T. Symbiotic Bacterium Modifies Aphid Body Color. Science 2010, 330, 1102–1104. [Google Scholar] [CrossRef]
- Burke, G.; Fiehn, O.; Moran, N. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. Isme J. 2010, 4, 242–252. [Google Scholar] [CrossRef]
- Simon, J.C.; Boutin, S.; Tsuchida, T.; Koga, R.; Gallic, J.; Frantz, A.; Outreman, Y.; Fukatsu, T. Facultative Symbiont Infections Affect Aphid Reproduction. PLoS ONE 2011, 6, e21831. [Google Scholar] [CrossRef]
- Brownlie, J.C.; Johnson, K.N. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009, 17, 348–354. [Google Scholar] [CrossRef]
- Kaltenpoth, M.; Gttler, W.; Herzner, G.; Strohm, E. Symbiotic Bacteria Protect Wasp Larvae from Fungal Infestation—ScienceDirect. Curr. Biol. 2005, 15, 882. [Google Scholar] [CrossRef]
- Scarborough, C.L.; Ferrari, J.; Godfray, H.C. Aphid protected from pathogen by endosymbiont. Science 2005, 310, 1781. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702. [Google Scholar] [CrossRef] [PubMed]
- Kellner, R.; Dettner, K. Differential efficacy of toxic pederin in deterring potential arthropod predators of Paederus (Coleoptera: Staphylinidae) offspring. Oecologia 1996, 107, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Vorburger, C.; Sandrock, C.; Gouskov, A.; Ferrari, L.E.C.E. Genotypic variation and the role of defensive endosymbionts in an all-parthenogenetic host-parasitoid interaction. Evolution 2009, 63, 1439–1450. [Google Scholar] [CrossRef]
- Vorburger, C.; Gehrer, L.; Rodriguez, P. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol. Lett. 2010, 6, 109–111. [Google Scholar] [CrossRef]
- Jaenike, J.; Unckless, R.; Cockburn, R.N.; Boelio, R.M.; Perlman, R.J. Adaptation via Symbiosis: Recent Spread of a Drosophila Defensive Symbiont. Science 2010, 329, 212–215. [Google Scholar] [CrossRef]
- Douglas, A.E. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 1998, 43, 17–37. [Google Scholar] [CrossRef]
- Russell, J.A.; Oliver, K.M.; Hansen, A.K. Band-aids for Buchnera and B vitamins for all. Mol. Ecol. 2017, 26, 2199–2203. [Google Scholar] [CrossRef]
- Javed, K.; Javed, H.; Qiu, D. Biocontrol Potential of Purified Elicitor Protein PeBL1 Extracted from Brevibacillus laterosporus Strain A60 and Its Capacity in the Induction of Defense Process against Cucumber Aphid (Myzus persicae) in Cucumber (Cucumis sativus). Biology 2020, 9, 179. [Google Scholar] [CrossRef]
- Russell, J.A.; Moran, N.A. Costs and benefits of symbiont infection in aphids: Variation among symbionts and across temperatures. Proc. Biol. Sci. 2006, 273, 603–610. [Google Scholar] [CrossRef]
- Oliver, K.; Moran, N.; Hunter, M. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. USA 2005, 102, 12795–12800. [Google Scholar] [CrossRef] [PubMed]
- Leonardo, T.E.; Mondor, E.B. Symbiont modifies host life-history traits that affect gene flow. Proc. R. Soc. B Biol. Sci. 2006, 273, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.S.; Moraiet, M.; Ahmad, S. Insecticides: Impact on the Environment and Human Health. In Environmental Deterioration and Human Health; Springer: Dordrecht, The Netherlands, 2014; pp. 99–123. [Google Scholar]
- Sunil, J.; Rabindra, R.J.; Rajendran, T.P. Biological control of aphids. J. Biol. Control 2010, 24, 185–202. [Google Scholar]
- Wyckhuys, K.A.G.; Stone, L.; Desneux, N.; Hoelmer, K.A.; Hopper, K.R.; Heimpel, G.E. Parasitism of the soybean aphid, Aphis glycines by Binodoxys communis: The role of aphid defensive behaviour and parasitoid reproductive performance. Bull. Entomol. Res. 2008, 98, 361–370. [Google Scholar] [CrossRef]
- Wan, B.; Goguet, E.; Ravallec, M.; Pierre, O.; Lemauf, S.; Volkoff, A.-N.; Gatti, J.-L.; Poirié, M. Venom Atypical Extracellular Vesicles as Interspecies Vehicles of Virulence Factors Involved in Host Specificity: The Case of a Drosophila Parasitoid Wasp. Front. Immunol. 2019, 10, 1688. [Google Scholar] [CrossRef] [PubMed]
- Charnov, E.L. Parasitoids and Darwinian Theory:Parasitoids: Behavioral and Evolutionary Ecology. H. C. J. Godfray. Q. Rev. Biol. 1994, 69, 73–76. [Google Scholar] [CrossRef]
- Gao, X.; Xue, H.; Luo, J.; Ji, J.; Cui, J. Molecular Evidence that Lysiphlebia japonica Regulates the Development and Physiological Metabolism of Aphis gossypii. Int. J. Mol. Sci. 2020, 21, 4610. [Google Scholar] [CrossRef]
- Gao, X.; Niu, R.; Zhu, X.; Wang, L.; Ji, J.; Niu, L.; Wu, C.; Zhang, S.; Luo, J.; Cui, J. Characterization and comparison of the bacterial microbiota of Lysiphlebia japonica parasitioid wasps and their aphid host Aphis gosypii. Pest Manag. Sci. 2021, 77, 2710–2718. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Norman, J.A. Tentacles of venom: Toxic protein convergence in the Kingdom Animalia. J. Mol. Evol. 2009, 68, 311–321. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R.; Somerfield, P.J.; Chapman, M.G. On resemblance measures for ecological studies, including taxonomic dissimilarities and a zero-adjusted Bray–Curtis coefficient for denuded assemblages. J. Exp. Mar. Biol. Ecol. 2006, 330, 55–80. [Google Scholar] [CrossRef]
- Carletto, J.; Gueguen, G.; Fleury, F.; Vanlerberghe-Masutti, F. Screening the bacterial endosymbiotic community of sap-feeding insects by terminal-restriction fragment length polymorphism analysis. Entomol. Exp. Et Appl. 2008, 129, 228–234. [Google Scholar] [CrossRef]
- Augustinos, A.A.; Santos-Garcia, D.; Dionyssopoulou, E.; Moreira, M.; Bourtzis, K. Detection and Characterization of Wolbachia Infections in Natural Populations of Aphids: Is the Hidden Diversity Fully Unraveled? PLoS ONE 2011, 6, e28695. [Google Scholar] [CrossRef]
- Jousselin, E.; D’Acier, A.C.; Vanlerberghe-Masutti, F.; Duron, O. Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol. Ecol. 2013, 22, 260–270. [Google Scholar] [CrossRef]
- Manjula, T.R.; Kannan, G.S.; Sivasubramanian, P. Field efficacy of Pseudomonas fluorescens against the cotton aphid, Aphis gossypii Glover (Hemiptera: Aphididae) in Bt and non Bt cotton. Agric. Update 2017, 12, 720–728. [Google Scholar] [CrossRef]
- Li, Q.; Fan, J.; Sun, J.X.; Wang, M.Q.; Chen, J.L. Plant-Mediated Horizontal Transmission of Hamiltonella defensa in the Wheat Aphid Sitobion miscanthi. J. Agric. Food Chem. 2018, 66, 13367–13377. [Google Scholar] [CrossRef]
- Desneux, N.; Barta, R.J.; Hoelmer, K.A.; Hopper, K.R.; Heimpel, G.E. Multifaceted determinants of host specificity in an aphid parasitoid. Oecologia 2009, 160, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Colella, S.; Parisot, N.; Simonet, P.; Gaget, K.; Duport, G.; Baa-Puyoulet, P.; Rahbé, Y.; Charles, H.; Febvay, G.; Callaerts, P.; et al. Bacteriocyte Reprogramming to Cope With Nutritional Stress in a Phloem Sap Feeding Hemipteran, the Pea Aphid Acyrthosiphon pisum. Front. Physiol. 2018, 9, 1498. [Google Scholar] [CrossRef] [PubMed]
- Koga, R.; Meng, X.Y.; Tsuchida, T.; Fukatsu, T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc. Natl. Acad. Sci. USA 2012, 109, E1230–E1237. [Google Scholar] [CrossRef] [PubMed]
- Pers, D.; Hansen, A.K. The boom and bust of the aphid’s essential amino acid metabolism across nymphal development. G3 2021, 11, jkab115. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Minhas, B.F.; Li-Byarlay, H.; Hansen, A.K. Key Transport and Ammonia Recycling Genes Involved in Aphid Symbiosis Respond to Host-Plant Specialization. G3 2018, 8, 2433–2443. [Google Scholar] [CrossRef]
- Tian, P.P.; Chang, C.Y.; Miao, N.H.; Li, M.Y.; Liu, X.D. Infections with Arsenophonus Facultative Endosymbionts Alter Performance of Aphids (Aphis gossypii) on an Amino-Acid-Deficient Diet. Appl. Env. Microbiol. 2019, 85, e01407-19. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Higashi, C.H. Variations on a protective theme: Hamiltonella defensa infections in aphids variably impact parasitoid success. Curr. Opin. Insect Sci. 2019, 32, 1–7. [Google Scholar] [CrossRef]
- Boyd, B.M.; Chevignon, G.; Patel, V.; Oliver, K.M.; Strand, M.R. Evolutionary genomics of APSE: A tailed phage that lysogenically converts the bacterium Hamiltonella defensa into a heritable protective symbiont of aphids. Virol. J. 2021, 18, 219. [Google Scholar] [CrossRef]
- Zhang, S.; Su, H.; Jiang, W.; Hu, D.; Ali, I.; Jin, T.; Yang, Y.; Ma, X. Symbiotic microbial studies in diverse populations of Aphis gossypii, existing on altered host plants in different localities during different times. Ecol. Evol. 2021, 11, 13948–13960. [Google Scholar] [CrossRef]
- Zhou, X.; Ling, X.; Guo, H.; Zhu-Salzman, K.; Ge, F.; Sun, Y. Serratia symbiotica Enhances Fatty Acid Metabolism of Pea Aphid to Promote Host Development. Int. J. Mol. Sci. 2021, 22, 5951. [Google Scholar] [CrossRef]
- Sakurai, M.; Koga, R.; Tsuchida, T.; Meng, X.Y.; Fukatsu, T. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: Novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 2005, 71, 4069–4075. [Google Scholar] [CrossRef] [PubMed]
- Attia, S.; Renoz, F.; Pons, I.; Louâpre, P.; Foray, V.; Piedra, J.M.; Sanané, I.; Le Goff, G.; Lognay, G.; Hance, T. The aphid facultative symbiont Serratia symbiotica influences the foraging behaviors and the life-history traits of the parasitoid Aphidius ervi. Entomol. Gen. 2022, 42, 21–33. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; An, Z.; Luo, J.; Zhu, X.; Wang, L.; Zhang, K.; Li, D.; Ji, J.; Niu, L.; Gao, X.; et al. Parasitization of Aphis gossypii Glover by Binodoxys communis Gahan Causes Shifts in the Ovarian Bacterial Microbiota. Insects 2023, 14, 314. https://doi.org/10.3390/insects14040314
Li J, An Z, Luo J, Zhu X, Wang L, Zhang K, Li D, Ji J, Niu L, Gao X, et al. Parasitization of Aphis gossypii Glover by Binodoxys communis Gahan Causes Shifts in the Ovarian Bacterial Microbiota. Insects. 2023; 14(4):314. https://doi.org/10.3390/insects14040314
Chicago/Turabian StyleLi, Jinming, Zhe An, Junyu Luo, Xiangzhen Zhu, Li Wang, Kaixin Zhang, Dongyang Li, Jichao Ji, Lin Niu, Xueke Gao, and et al. 2023. "Parasitization of Aphis gossypii Glover by Binodoxys communis Gahan Causes Shifts in the Ovarian Bacterial Microbiota" Insects 14, no. 4: 314. https://doi.org/10.3390/insects14040314
APA StyleLi, J., An, Z., Luo, J., Zhu, X., Wang, L., Zhang, K., Li, D., Ji, J., Niu, L., Gao, X., & Cui, J. (2023). Parasitization of Aphis gossypii Glover by Binodoxys communis Gahan Causes Shifts in the Ovarian Bacterial Microbiota. Insects, 14(4), 314. https://doi.org/10.3390/insects14040314




