Characterization of CrufCSP1 and Its Potential Involvement in Host Location by Cotesia ruficrus (Hymenoptera: Braconidae), an Indigenous Parasitoid of Spodoptera frugiperda (Lepidoptera: Noctuidae) in China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. RNA Extraction, cDNA Synthesis, and Gene Cloning
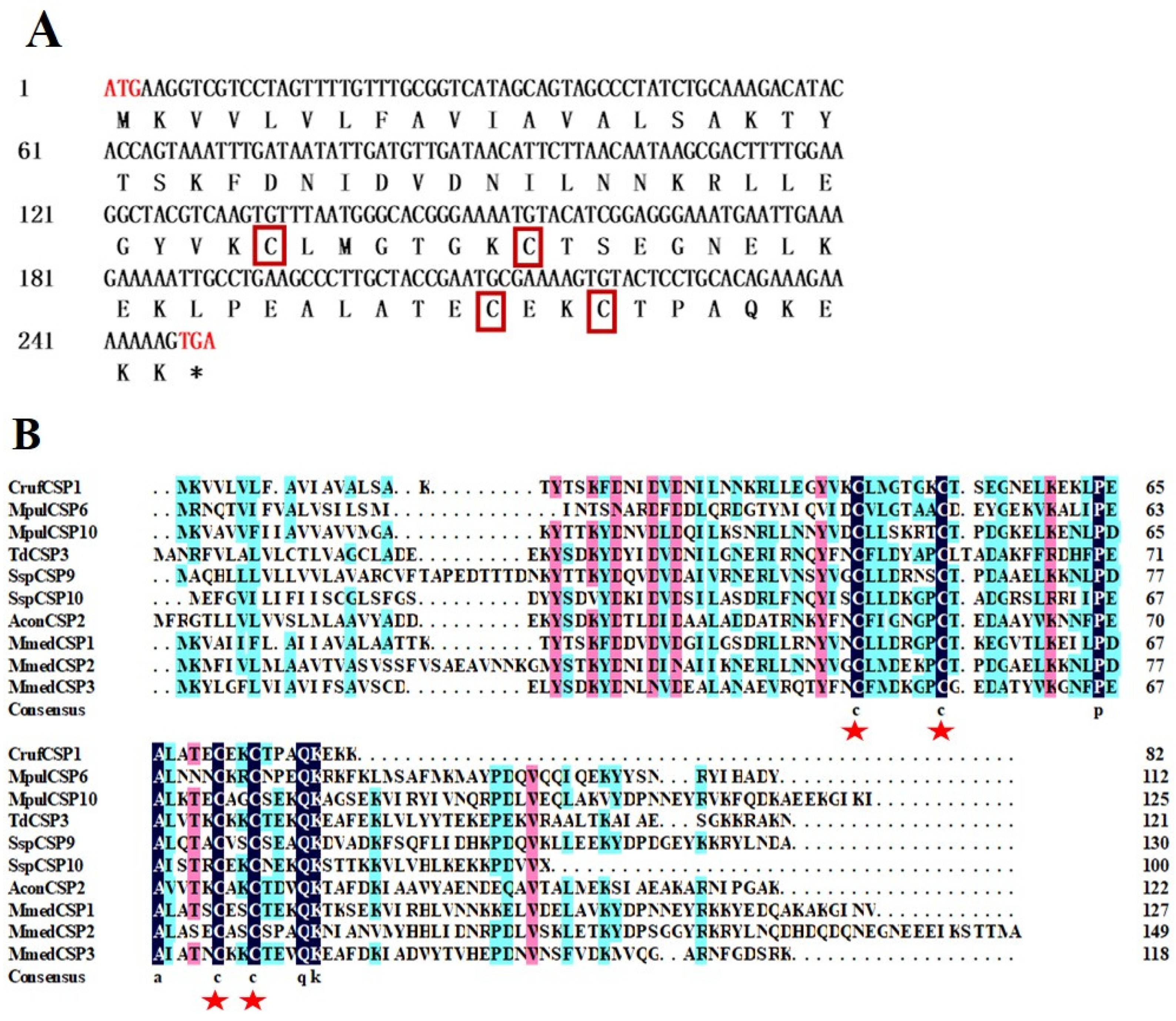
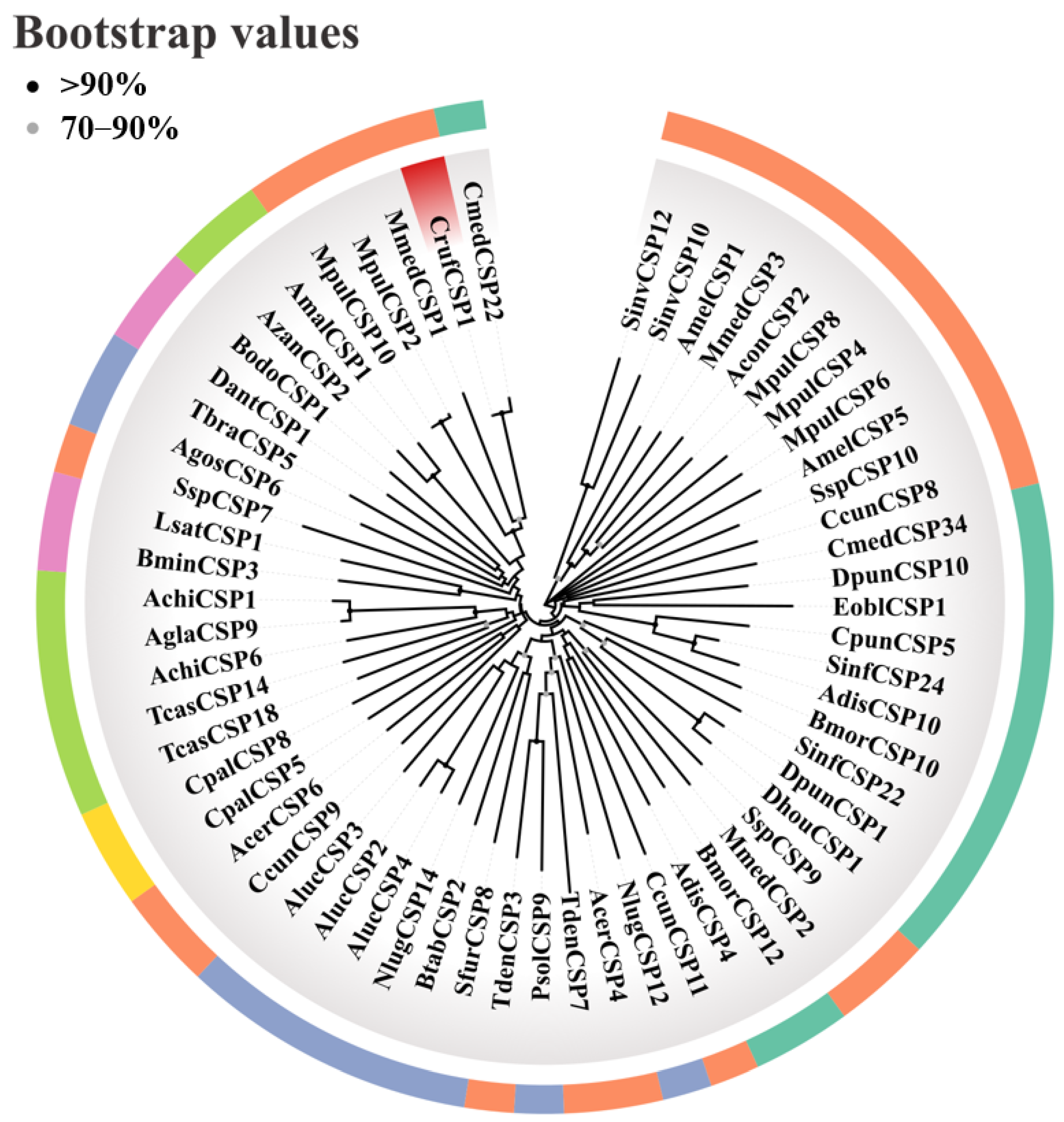
2.3. Identification, Sequence Analysis, and Phylogenetic Tree Construction of CrufCSP1
2.4. Expression of CrufCSP1 Gene
2.5. Cloning and Construction of Recombinant Plasmids
2.6. Prokaryotic Expression and Purification of CrufCSP1
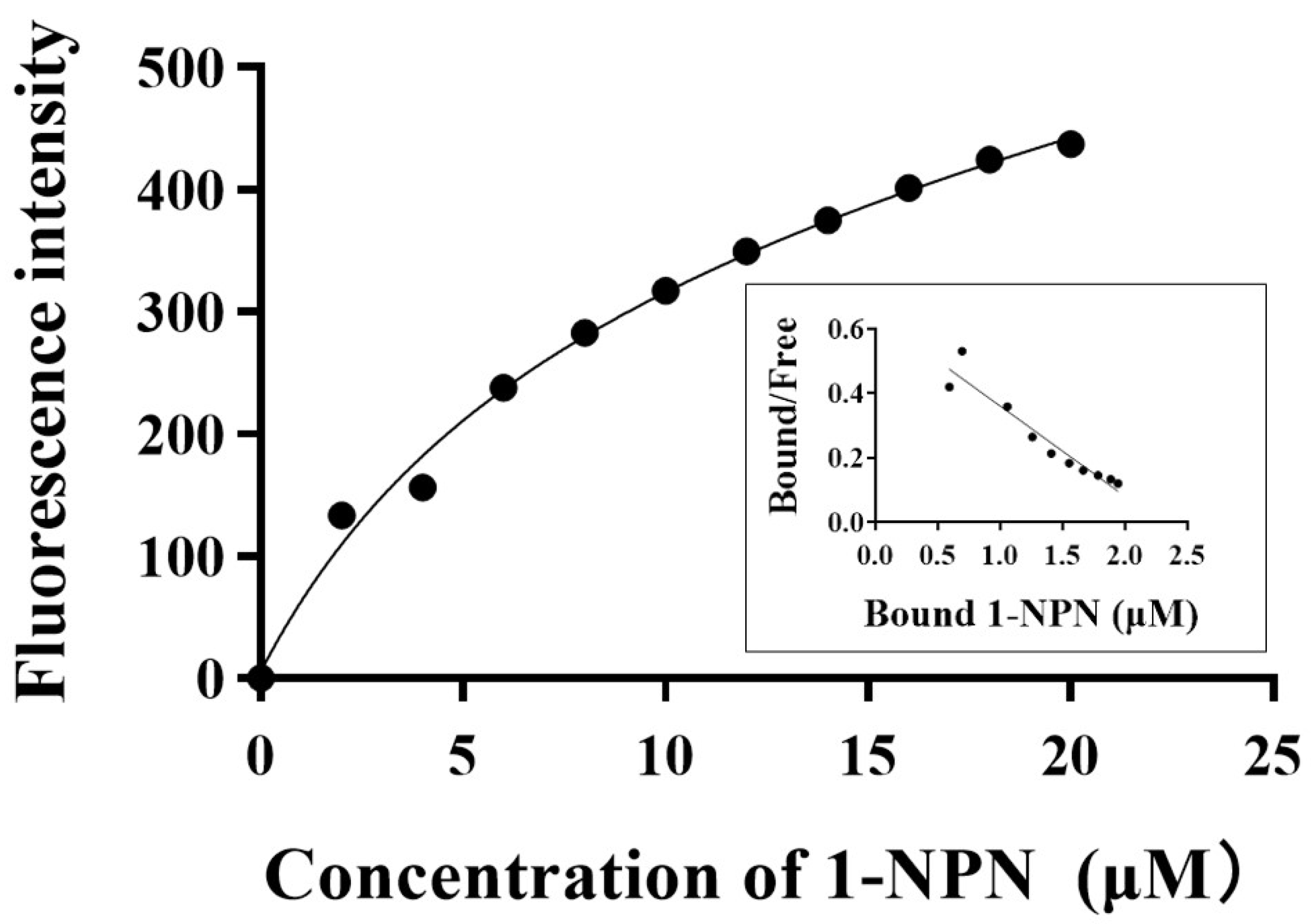
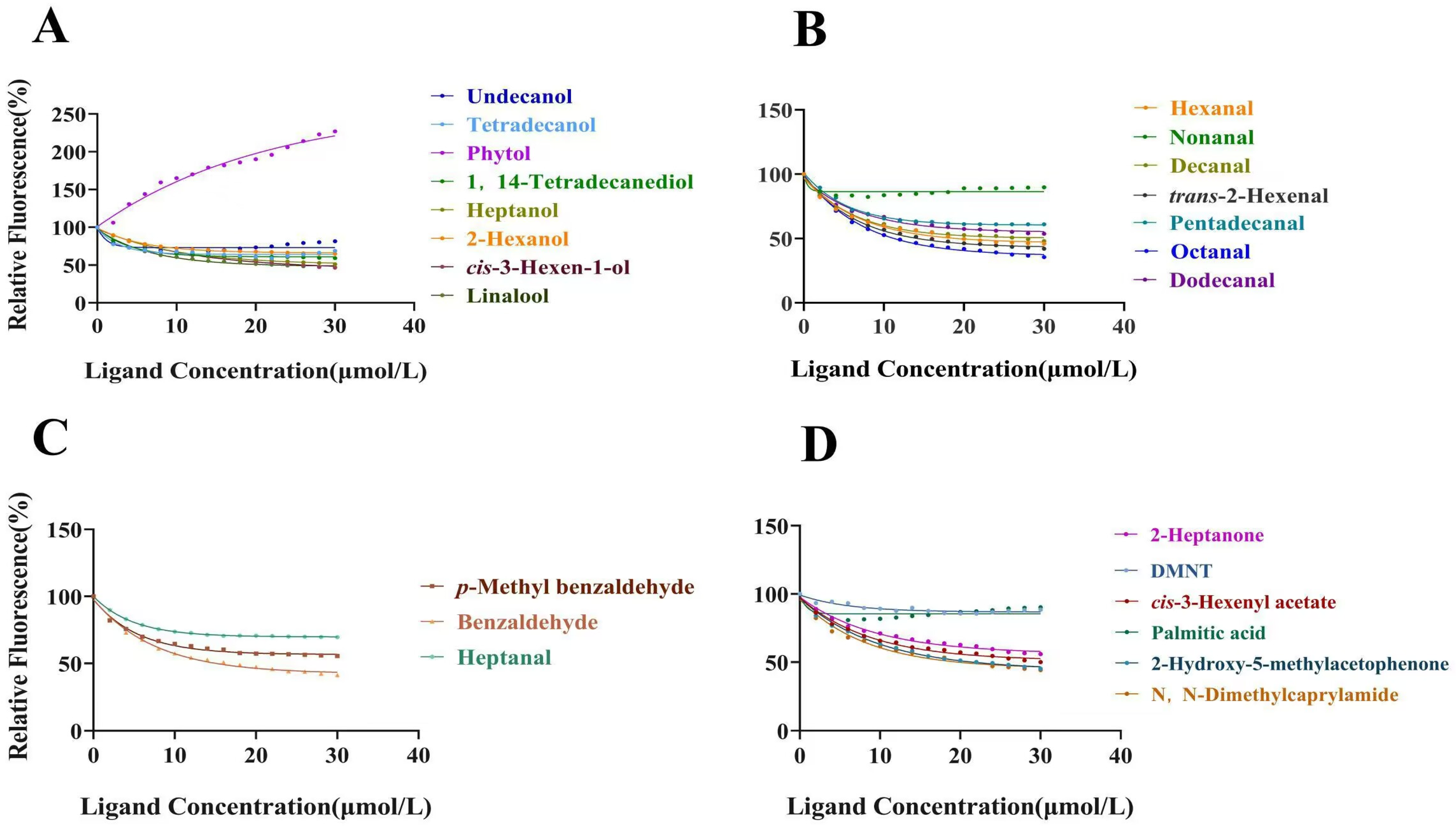
2.7. Fluorescence Binding Assays
2.8. Three-Dimensional Structure and Molecular Docking of CrufCSP1
3. Results
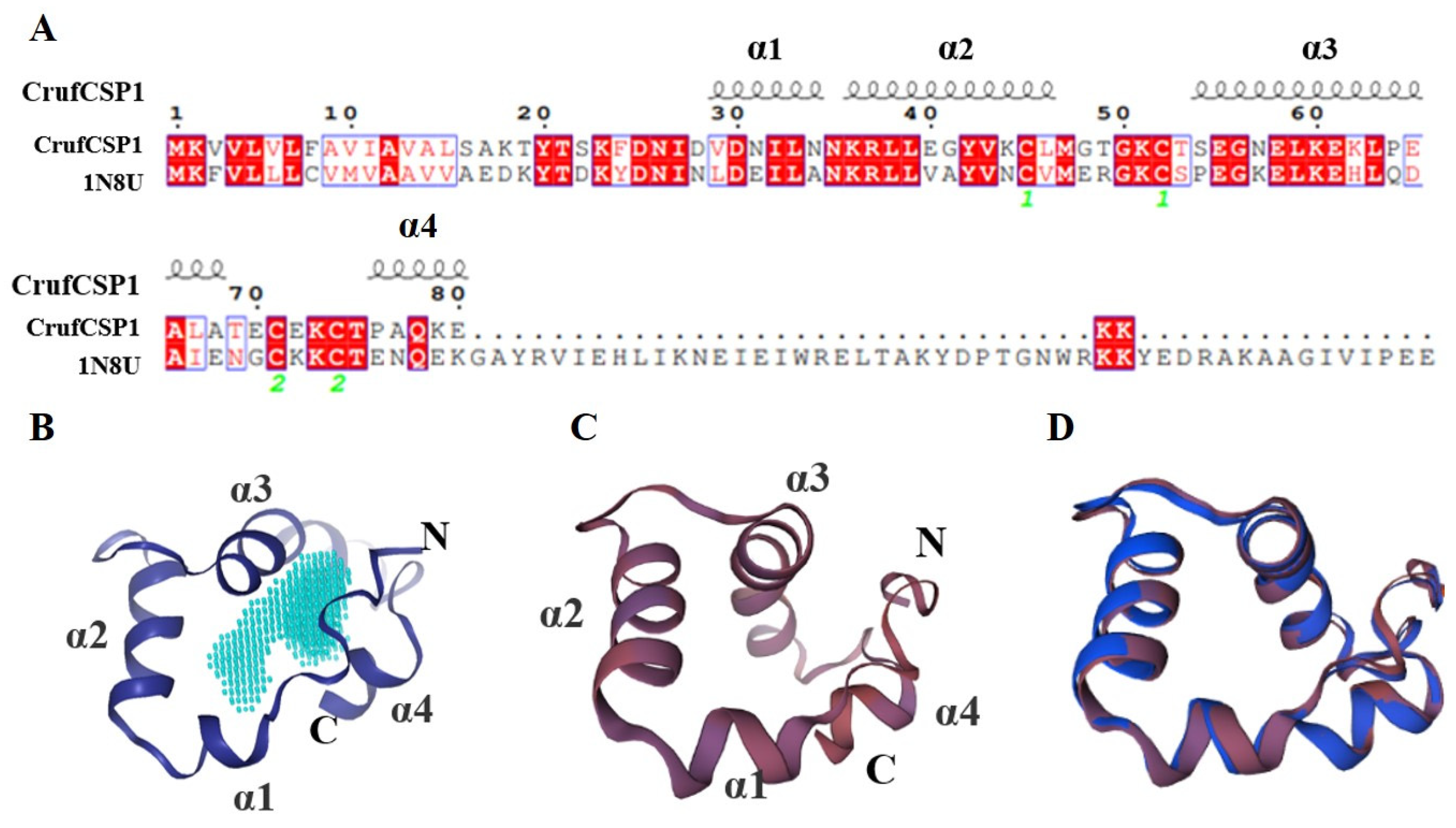
3.1. Identification and Phylogenetic Analysis of CrufCSP1 in C. ruficrus
3.2. Expression Profile Analysis of CrufCSP1
3.3. Recombinant Protein Expression and Purification of CrufCSP1
3.4. Fluorescence Binding Assays with CrufCSP1
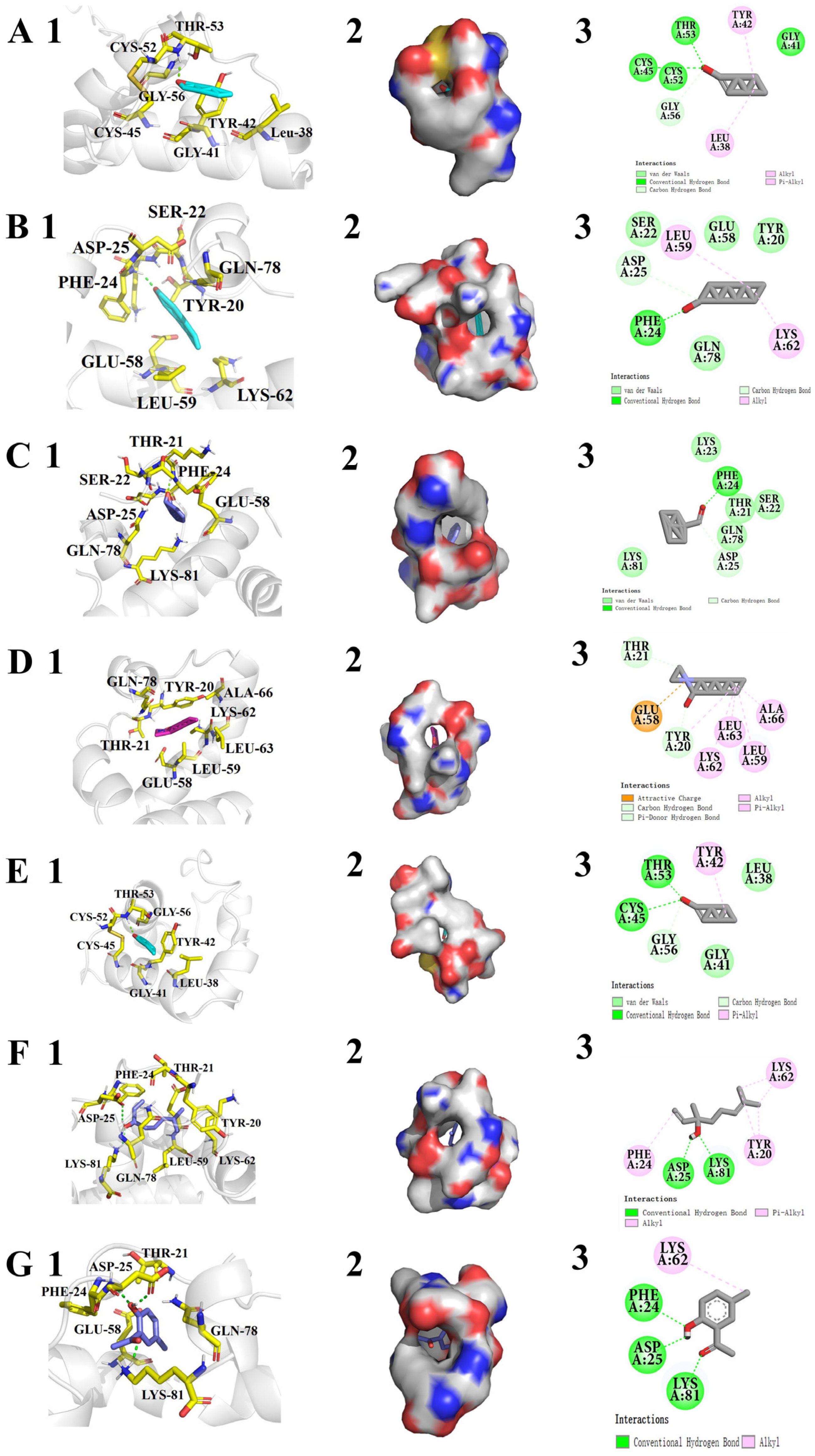
3.5. Three-Dimensional Model Structuring and Molecular Docking of CrufCSP1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science 2001, 291, 2141–2144. [Google Scholar] [CrossRef]
- Fatouros, N.E.; Van Loon, J.J.A.; Hordijk, K.A.; Smid, H.M.; Dicke, M. Herbivore-induced plant volatiles mediate in-flight host discrimination by parasitoids. J. Chem. Ecol. 2005, 31, 2033–2047. [Google Scholar] [CrossRef]
- Magalhaes, D.M.; Da-Silva, I.T.F.A.; Borges, M.; Laumann, R.A.; Blassioli-Moraes, M.C. Anthonomus grandis aggregation pheromone induces cotton indirect defence and attracts the parasitic wasp Bracon vulgaris. J. Exp. Bot. 2019, 70, 1891–1901. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Oliveira, D.S.; Brito, N.F.; Franco, T.A.; Moreira, M.F.; Leal, W.S.; Melo, A.C.A. Functional characterization of odorant binding protein 27 (RproOBP27) from Rhodnius prolixus antennae. Front. Physiol. 2018, 9, 1175. [Google Scholar] [CrossRef]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbash, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- Stocker, R.F. The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 1994, 275, 3–26. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Benton, R.; Sachse, S.; Michnick, S.W.; Vosshall, L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006, 4, 240–257. [Google Scholar] [CrossRef]
- Gadenne, C.; Barrozo, R.B.; Anton, S. Plasticity in Insect olfaction: To smell or not to smell? Annu. Rev. Entomol. 2016, 61, 317–333. [Google Scholar] [CrossRef]
- Li, X.W.; Cheng, J.H.; Chen, L.M.; Huang, J.; Zhang, Z.J.; Zhang, J.M.; Ren, X.Y.; Hafeez, M.; Zhou, S.X.; Dong, W.Y. Comparison and functional analysis of odorant-binding proteins and chemosensory proteins in two closely related thrips species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) based on antennal transcriptome analysis. Int. J. Mol. Sci. 2022, 23, 13900. [Google Scholar] [CrossRef]
- Calvello, M.; Brandazza, A.; Navarrini, A.; Dani, F.R.; Turillazzi, S.; Felicioli, A.; Pelosi, P. Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera. Insect Biochem. Mol. Biol. 2005, 35, 297–307. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Ze, S.Z.; Yang, B. Identification and expression profiling of six chemosensory protein genes in the beet armyworm, Spodoptera exigua. J. Asia-Pac. Entomol. 2015, 18, 61–66. [Google Scholar] [CrossRef]
- Cui, H.H.; Gu, S.H.; Zhu, X.Q.; Wei, Y.; Liu, H.W.; Khalid, H.D.; Guo, Y.Y.; Zhang, Y.J. Odorant-binding and chemosensory proteins identified in the antennal transcriptome of Adelphocoris suturalis Jakovlev. Comp. Biochem. Phys. D 2017, 45, 163–174. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, S.N.; Li, K.M.; Liu, J.T.; Zheng, Y.; Shan, S.; Yang, Y.Q.; Li, R.J.; Zhang, Y.J.; Guo, Y.Y. Identification of odorant binding proteins and chemosensory proteins in Microplitis mediator as well as functional characterization of chemosensory protein 3. PLoS ONE 2017, 12, e0180775. [Google Scholar] [CrossRef]
- Li, Z.Q.; Zhang, S.; Luo, J.Y.; Zhu, J.; Cui, J.J.; Dong, S.L. Expression analysis and binding assays in the chemosensory protein gene family indicate multiple roles in Helicoverpa armigera. J. Chem. Ecol. 2015, 41, 473–485. [Google Scholar] [CrossRef]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.L.; Bulet, P.; Imler, J.L. Pherokine-2 and-3, two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 2003, 270, 3398–3407. [Google Scholar] [CrossRef]
- Kitabayashi, A.N.; Arai, T.; Kubo, T.; Natori, S. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem. Mol. Biol. 1998, 28, 785–790. [Google Scholar] [CrossRef]
- Bautista, M.A.M.; Bhandary, B.; Wijeratne, A.J.; Michel, A.P.; Hoy, C.W.; Mittapalli, O. Evidence for trade-offs in detoxification and chemosensation gene signatures in Plutella xylostella. Pest Manag. Sci. 2015, 71, 423–432. [Google Scholar] [CrossRef]
- Montezano, D.G.; Specht, A.; Sosa-Gómez, D.R.; Roque-Specht, V.F.; Sousa-Silva, J.C.; Paula Moraes, S.V.D.; Peterson, J.A.; Hunt, T. Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Wang, W.W.; He, P.Y.; Zhang, Y.Y.; Liu, T.X.; Jing, X.F.; Zhang, S.Z. The population growth of Spodoptera frugiperda on six cash crop species and implications for its occurrence and damage potential in China. Insects 2020, 11, 639. [Google Scholar] [CrossRef]
- Guo, J.Y.; Zhang, Y.J.; Wang, Z.Y. Research progress in managing the invasive fall armyworm, Spodoptera frugiperda, in China. Plant Prot. 2022, 48, 79–87. (In Chinese) [Google Scholar]
- Sobhy, I.S.; Tamiru, A.; Morales, X.C.; Nyagol, D.; Cheruiyot, D.; Chidawanyika, F.; Subramanian, S.; Midega, C.A.; Bruce, T.J.A.; Khan, Z.R. Bioactive volatiles from push-pull companion crops repel fall armyworm and attract its parasitoids. Front. Ecol. Evol. 2022, 10, e883020. [Google Scholar] [CrossRef]
- Ornella, B.B.; John, L.; Raúl, O.R.S.; Fernando, B.; del-Val, E. Differential biocontrol efficacy of the parasitoid wasp Campoletis sonorensis on the fall armyworm Spodoptera frugiperda feeding on landrace and hybrid maize. Biocontrol Sci. Technol. 2021, 31, 713–724. [Google Scholar]
- Sokame, B.M.; Obonyo, J.; Sammy, E.M.; Mohamed, S.A.; Subramanian, S.; Kilalo, D.C.; Juma, G.; Calatayud, P.A. Impact of the exotic fall armyworm on larval parasitoids associated with the lepidopteran maize stemborers in Kenya. Biocontrol 2021, 66, 205. [Google Scholar]
- Mccutcheon, G.S.; Salley, W.Z.; Turnipseed, S.G. Biology of Apanteles ruficrus, an imported parasitoid of Pseudoplusia includens, Trichoplusia ni, and Spodoptera frugiperda (Lepidoptera: Noctuidae). Environ. Entomol. 1983, 12, 1055–1058. [Google Scholar] [CrossRef]
- Li, Z.M.; Dai, L.L.; Chu, H.L.; Fu, D.Y.; Sun, Y.Y.; Chen, H. Identification, expression patterns, and functional characterization of chemosensory proteins in Dendroctonus armandi (Coleoptera: Curculionidae: Scolytinae). Front. Physiol. 2018, 9, 291. [Google Scholar] [CrossRef]
- He, P.Y.; Li, X.; Liu, T.X.; Zhang, S.Z. Ontogeny of the Cotesia ruficrus, a parasitoid of Spodoptera frugiperda. Chin. J. Biol. Control 2023, 1–10. (In Chinese) [Google Scholar] [CrossRef]
- He, P.Y.; Li, X.; Liu, T.X.; Zhang, S.Z. Effects of oviposition times and host larva instars on the biological characters of Cotesia ruficrus (Hymenoptera: Braconidae), an indigenous parasitiod of Spodoptera frugiperda (Lepidoptera: Noctuidae). Acta Entomol. Sin. 2023, 66, 1095–1104. (In Chinese) [Google Scholar]
- Saraiva, N.B.; Prezoto, F.; Fonseca, M.G.; Blassioli-Moraes, M.C.; Borges, M.; Laumann, R.A.; Auad, A.M. The social wasp Polybia fastidiosuscula Saussure (Hymenoptera: Vespidae) uses herbivore-induced maize plant volatiles to locate its prey. J. Appl. Entomol. 2017, 141, 620–629. [Google Scholar] [CrossRef]
- Chen, P.; Liu, D.D.; Xiu, X.J.; Yu, J.; Hou, M.L. Corn and rice volatile components eliciting olfactory responses in Spodoptera frugiperda. Plant Prot. 2022, 48, 90–97. (In Chinese) [Google Scholar]
- Cui, X.; Liu, D.; Sun, K.; He, Y.; Shi, X. Expression profiles and functional characterization of two odorant-binding proteins from the apple buprestid beetle Agrilus mali (Coleoptera: Buprestidae). J. Econ. Entomol. 2018, 111, 1420–1432. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechsel-berger, R.; Sergio, A.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 10606–10613. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Liu, Y.Q.; Shi, M.; Huang, J.H.; Chen, X.X. Parasitoid wasps as effective biological control agents. J. Integr. Agric. 2019, 18, 705–715. [Google Scholar] [CrossRef]
- Lu, D.G.; Li, X.R.; Liu, X.X.; Zhang, Q.W. Identification and molecular cloning of putative odorant-binding proteins and chemosensory protein from the bethylid wasp, Scleroderma guani Xiao et Wu. J. Chem. Ecol. 2007, 33, 1359–1375. [Google Scholar] [CrossRef]
- Nishimura, O.; Brillada, C.; Yazawa, S.; Maffei, M.E.; Arimura, G. Transcriptome pyrosequencing of the parasitoid wasp Cotesia vestalis: Genes involved in the antennal odorant-sensory system. PLoS ONE 2012, 7, e50664. [Google Scholar] [CrossRef]
- Wang, K.; He, Y.Y.; Zhang, Y.J.; Guo, Z.J.; Xie, W.; Wu, Q.J.; Wang, S.L. Characterization of the chemosensory protein EforCSP3 and its potential involvement in host location by Encarsia formosa. J. Integr. Agric. 2023, 22, 514–525. [Google Scholar] [CrossRef]
- Ayasse, M.; Paxton, R.J.; Tengo, J. Mating behavior and chemical communication in the order Hymenoptera. Annu. Rev. Entomol. 2001, 46, 31–78. [Google Scholar] [CrossRef]
- Turlings, T.C.J.; Erb, M. Tritrophic interactions mediated by herbivore-induced plant volatiles: Mechanisms, ecological relevance, and application potential. Annu. Rev. Entomol. 2018, 63, 433–452. [Google Scholar] [CrossRef]
- Ruther, J.; Stahl, L.M.; Steiner, S.; Garbe, L.A.; Tolasch, T. A male sex pheromone in a parasitic wasp and control of the behavioral response by the female’s mating status. J. Exp. Biol. 2007, 210, 2163–2169. [Google Scholar] [CrossRef]
- Benelli, G.; Canale, A. Do tephritid-induced fruit volatiles attract males of the fruit flies parasitoid Psyttalia concolor (Sz’epligeti) (Hymenoptera: Braconidae)? Chemoecology 2013, 23, 191–199. [Google Scholar] [CrossRef]
- Xu, H.; Turlings, T.C.J. Plant volatiles as mate-finding cues for insects. Trends Plant Sci. 2018, 23, 100–111. [Google Scholar] [CrossRef]
- Guo, H.; Mo, B.T.; Li, G.C.; Li, Z.L.; Huang, L.Q.; Sun, Y.L.; Dong, J.F.; Smith, D.P.; Wang, C.Z. Sex pheromone communication in an insect parasitoid, Campoletis chlorideae Uchida. Proc. Natl. Acad. Sci. USA 2022, 119, e2215442119. [Google Scholar] [CrossRef]
- Xu, H.; Desurmont, G.; Degen, T.; Zhou, G.X.; Laplanche, D.; Henryk, L.; Turlings, T.C.J. Combined use of herbivore-induced plant volatiles and sex pheromones for mate location in braconid parasitoids. Plant Cell Environ. 2017, 40, 330–339. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, B.; Zhou, T.; Yu, X.; Hu, M.; Dai, W. Involvement of chemosensory protein BodoCSP1 in perception of host plant volatiles in Bradysia odoriphaga. J. Agric. Food Chem. 2021, 69, 10797–10806. [Google Scholar] [CrossRef]
- Li, Z.X.; Liu, L.; Zong, S.X.; Tao, J. Molecular characterization and expression profiling of chemosensory proteins in male Eogystia hippophaecolus (Lepidoptera: Cossidae). J. Entomol. Sci. 2021, 56, 217–234. [Google Scholar]
- Li, H.L.; Ni, C.X.; Tan, J.; Zhang, L.Y.; Hu, F.L. Chemosensory proteins of the eastern honeybee, Apis cerana: Identification, tissue distribution and olfactory related functional characterization. Comp. Biochem. Physiol. B 2016, 19, 11–19. [Google Scholar] [CrossRef]
- Zhu, J.; Iovinella, I.; Dani, F.R.; Liu, Y.L.; Huang, L.Q.; Liu, Y.; Wang, C.Z.; Pelosi, P.; Wang, G. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. Int. J. Biol. Sci. 2016, 12, 1394–1404. [Google Scholar] [CrossRef]
- Magalhães, D.M.; Borges, M.; Laumann, R.A.; Woodcock, C.M.; Pickett, J.A.; Birkett, M.A.; Blassioli-Moraes, M.C. Influence of two acyclic homoterpenes (Tetranorterpenes) on the foraging behavior of Anthonomus grandis Boh. J. Chem. Ecol. 2016, 42, 305–313. [Google Scholar] [CrossRef]
- Gurr, F.M.; Liu, J.; Pickett, J.A.; Stevenson, P.C. Review of the chemical ecology of homoterpenes in arthropod plant interactions. Austral Entomol. 2023, 61, 3–14. [Google Scholar] [CrossRef]
- Liu, J.; Sun, L.; Fu, D.; Zhu, J.; Liu, M.; Xiao, F.; Xiao, R. Herbivore-induced rice volatiles attract and affect the predation ability of the wolf spiders, Pirata subpiraticus and Pardosa pseudoannulata. Insects 2022, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.Y.; Liu, Y.P.; Wang, G.R.; Zhou, Q. Identification and functional analysis of a chemosensory protein from Bactrocera minax (Diptera: Tephritidae). Pest Manag. Sci. 2022, 78, 3479–3488. [Google Scholar] [CrossRef] [PubMed]
- Younas, A.; Waris, M.I.; Shaaban, M.; Qamar, M.T.; Wang, M.Q. Appraisal of MsepCSP14 for chemosensory functions in Mythimna separata. Insect Sci. 2022, 29, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Pan, Y.F.; Ma, Y.F.; Wang, J.; He, M.; He, P. Binding affinity characterization of an antennae-enriched chemosensory protein from the white-backed planthopper, Sogatella furcifera (Horvath), with host plant volatiles. Pestic. Biochem. Phys. 2018, 152, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.T.; Hua, D.K.; Zhu, J.Q.; Wang, F.; Zhang, Y.J. Chemosensory protein 4 is required for Bradysia odoriphaga to be olfactory attracted to sulfur compounds released from Chinese chives. Front. Physiol. 2022, 13, e989601. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, X.; Liu, J.; Hu, M.; Zhong, G.; Geng, P.; Yi, X. Molecular cloning, expression and molecular modeling of chemosensory protein from Spodoptera litura and its binding properties with Rhodojaponin III. PLoS ONE 2012, 7, e47611. [Google Scholar] [CrossRef]
- Zhang, T.T.; Wang, W.X.; Zhang, Z.D.; Zhang, Y.J.; Guo, Y.Y. Functional characteristics of a novel chemosensory protein in the cotton bollworm Helicoverpa armigera (Hubner). J. Integr. Agric. 2013, 12, 853–861. [Google Scholar] [CrossRef]
- Li, G.W.; Chen, X.L.; Chen, L.H.; Wang, W.Q.; Wu, J.X. Functional analysis of the chemosensory protein GmolCSP8 from the oriental fruit moth, Grapholita molesta (Busck) (Lepidoptera: Tortricidae). Front. Physiol. 2019, 10, 552. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Muhammad, T.U.Q.; Muhammad, S.; Prager, S.M.; Wang, M.Q. Functional analysis of the chemosensory protein MsepCSP8 from the oriental armyworm Mythimna separata. Front. Physiol. 2018, 9, 872. [Google Scholar] [CrossRef]

| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| For cloning CSP1 open reading frames | |
| CrufCSP1-Sense | ATGAAGGTCGTCCTAGTTTTG |
| CrufCSP1-Anti-sense | TTAAACATCAATTCCTTTTGC |
| For RT-qPCR of CSP1 | |
| CrufCSP1-Sense | TTTTGTTTGCGGTCATAGC |
| CrufCSP1-Anti-sense | ATTTTCCCGTGCCCATT |
| β-actin-Sense | GCTCCATCAACCATGAAG |
| β-actin-Anti-sense | TGGAAGGTGGACAGAGAAG |
| Heterologous expression of CSP1 | |
| CrufCSP1-Sense | CGCGGATCCATGAAGGTCGTCCTAGTT |
| CrufCSP1-Anti-sense | CCTCGAGGGCTTTTTTTCTTTCTGTGC |
| Ligand | Formula | CAS | Purity | Source | IC50 (μM) | Ki (μM) |
|---|---|---|---|---|---|---|
| Body volatiles | ||||||
| Undecanol | C11H24O | 112-42-5 | 98% | Macklin | >50 | >50 |
| Tetradecanol | C14H30O | 112-72-1 | 99.5% | Macklin | >50 | >50 |
| 1,14-Tetradecanediol | C14H30O2 | 19812-64-7 | 98% | Macklin | >50 | >50 |
| 2-Hexanol | C6H14O | 626-93-7 | 98% | Energy Chemical | >50 | >50 |
| Phytol | C20H40O | 150-86-7 | 90% | Macklin | >50 | >50 |
| Dodecanal | C12H24O | 112-54-9 | 95% | Macklin | 41.06 | 35.96 |
| Pentadecanal | C15H30O | 629-62-9 | 98% | Aladdin | 44.48 | 38.96 |
| p-Methyl benzaldehyde | C8H8O | 104-87-0 | 97% | Macklin | 45.96 | 40.26 |
| Palmitic acid | C16H32O2 | 57-10-3 | 97% | TCL | >50 | >50 |
| 2-Hydroxy-5-methylacetophenone | C9H10O2 | 1450-72-2 | 98% | Macklin | 22.79 | 19.97 |
| N,N-Dimethylcaprylamide | C10H21NO | 1118-92-9 | 95% | Macklin | 22.02 | 19.29 |
| Corn volatiles | ||||||
| Hexanal | C6H12O | 66-25-1 | 99% | Sigma | 22.10 | 19.36 |
| Heptanal | C7H14O | 111-71-7 | 99% | Macklin | >50 | >50 |
| Octanal | C8H16O | 124-13-0 | 99.5% | Macklin | 17.17 | 15.04 |
| Nonanal | C9H18O | 124-19-6 | 99% | Macklin | >50 | >50 |
| Decanal | C10H20O | 112-31-2 | 99% | Aladdin | 24.90 | 21.81 |
| Benzaldehyde | C7H6O | 100-52-7 | 98% | Sigma | 17.39 | 15.24 |
| trans-2-Hexenal | C6H10O | 6728-26-3 | 90% | TCL | 16.54 | 14.49 |
| Heptanol | C7H16O | 111-70-6 | 98% | Sigma | 31.56 | 27.65 |
| cis-3-Hexen-1-ol | C6H12O | 928-96-1 | 97% | Sigma | 28.23 | 24.73 |
| Linalool | C10H18O | 78-70-6 | 98% | Sigma | 22.60 | 19.80 |
| DMNT | C11H18 | 19945-61-0 | 97% | Yuanye bio-technology | >50 | >50 |
| cis-3-Hexenyl acetate | C8H14O2 | 3681-71-8 | 98% | Sigma | 33.36 | 29.22 |
| 2-Heptanone | C7H14O | 110-43-0 | 95% | Sigma | >50 | >50 |
| Ligand | Binding Energy (Kcal/mol) | Residues Involved in Hydrogen Bonds | Close-Contact Interacting Residues |
|---|---|---|---|
| trans-2-Hexenal | −3.68 | Thr53 (1.9 Å) | Leu38, Gly41, Tyr42, Cys45, Cys52, and Gly56 |
| Octanal | −3.77 | Phe24 (1.9 Å) | Tyr20, Ser22, Asp25, Glu58, Leu59, Lys62, and Gln78 |
| Benzaldehyde | −4.08 | Phe24 (1.9 Å) | Thr21, Ser22, Lys23, Asp25, Glu58, Gln78, and Lys81 |
| N,N-Dimethylcaprylamide | −4.65 | - | Tyr20, Thr21, Glu58, Leu59, Lys62, Leu63, Ala66, and Gln78 |
| Hexanal | −3.52 | Thr53 (2.0 Å) | Leu38, Gly41, Tyr42, Cys45, Cys52, and Gly56 |
| Linalool | −5.01 | ASP25 (2.2 Å) LYS81 (2.4 Å) | Tyr20, Thr21, Phe24, Glu58, Leu59, Lys62, and Gln78 |
| 2-Hydroxy-5-methylacetophenone | −5.24 | Phe 24 (2.5 Å) ASP25 (2.0 Å) LYS81 (1.9 Å) | Thr21, Glu58, and Gln78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.-R.; Wang, W.-W.; Yang, W.-Q.; Li, X.; Liu, T.-X.; Zhang, S.-Z. Characterization of CrufCSP1 and Its Potential Involvement in Host Location by Cotesia ruficrus (Hymenoptera: Braconidae), an Indigenous Parasitoid of Spodoptera frugiperda (Lepidoptera: Noctuidae) in China. Insects 2023, 14, 920. https://doi.org/10.3390/insects14120920
Han K-R, Wang W-W, Yang W-Q, Li X, Liu T-X, Zhang S-Z. Characterization of CrufCSP1 and Its Potential Involvement in Host Location by Cotesia ruficrus (Hymenoptera: Braconidae), an Indigenous Parasitoid of Spodoptera frugiperda (Lepidoptera: Noctuidae) in China. Insects. 2023; 14(12):920. https://doi.org/10.3390/insects14120920
Chicago/Turabian StyleHan, Kai-Ru, Wen-Wen Wang, Wen-Qin Yang, Xian Li, Tong-Xian Liu, and Shi-Ze Zhang. 2023. "Characterization of CrufCSP1 and Its Potential Involvement in Host Location by Cotesia ruficrus (Hymenoptera: Braconidae), an Indigenous Parasitoid of Spodoptera frugiperda (Lepidoptera: Noctuidae) in China" Insects 14, no. 12: 920. https://doi.org/10.3390/insects14120920
APA StyleHan, K.-R., Wang, W.-W., Yang, W.-Q., Li, X., Liu, T.-X., & Zhang, S.-Z. (2023). Characterization of CrufCSP1 and Its Potential Involvement in Host Location by Cotesia ruficrus (Hymenoptera: Braconidae), an Indigenous Parasitoid of Spodoptera frugiperda (Lepidoptera: Noctuidae) in China. Insects, 14(12), 920. https://doi.org/10.3390/insects14120920








