Simple Summary
The Aleurocanthus camelliae cryptic species complex, which includes a number of morphospecies and/or haplotypes, is one of the growing biological issues, the underlying mechanism of which is still unknown. It is well-known that Wolbachia infection can produce significant mitochondrial divergence in insects, which may eventually result in cryptic speciation. Therefore, the diversity and phenotypic characteristics of Wolbachia natural infections in the A. camelliae cryptic species complex were investigated. Two morphospecies were found to have distinct infection statuses. A. spiniferus morphospecies was the uninfected population, while A. camelliae morphospecies was fixed for infections. The oscillation hypothesis is discussed in light of the current discovery of novel cryptic species of A. camelliae. This idea may offer insights into cryptic speciation, specifically on how specialization and host expansion have been observed among these species. Additionally, this research discovered a parasitoid wasp from the genus Eretmocerus in A. camelliae for the first time in Japan.
Abstract
Wolbachia, an alphaproteobacterial reproductive parasite, can cause profound mitochondrial divergence in insects, which might eventually be a part of cryptic speciation. Aleurocanthus camelliae is a cryptic species complex consisting of several morphospecies and/or haplotypes that are genetically different but morphologically indistinctive. However, little is known about the Wolbachia infection status in these tea and Citrus pests. Thus, this study aimed to profile the diversity and phenotypic characteristics of Wolbachia natural infections in the A. camelliae cryptic species complex. A monophyletic strain of Wolbachia that infected the A. camelliae cryptic species complex (wAlec) with different patterns was discovered. Whiteflies that are morphologically identical to Aleurocanthus spiniferus (Aleurocanthus cf. A. spiniferus in Eurya japonica and A. spiniferus in Citrus) were grouped into uninfected populations, whereas the fixed infection was detected in A. camelliae B1 from Theaceae. The rapid evolution of wAlec was also found to occur through a high recombination event, which produced subgroups A and B in wAlec. It may also be associated with the non-cytoplasmic incompatibility (CI) phenotype of wAlec due to undetectable CI-related genes from phage WO (WOAlec). The current discovery of a novel cryptic species of A. camelliae led to a discussion about the oscillation hypothesis, which may provide insights on cryptic speciation, particularly on how specialization and host expansion have been recorded among these species. This study also identified a parasitoid wasp belonging to the genus Eretmocerus in A. camelliae, for the first time in Japan.
1. Introduction
Wolbachia is a well-known reproductive parasite that is one of the most common facultative symbiotic bacteria (secondary symbionts) of insects [1,2] and a speciation agent [3]. Wolbachia has a wide range of relationships with the host, from facultative parasitic to obligate mutualist [4]. Fixed infections (obligate mutualist) and phenotypic strain diversity (facultative parasitic) are important characteristics of Wolbachia infections associated with their significant roles in the induction of parthenogenesis and cytoplasmic incompatibility (CI), respectively [3]. Wolbachia, in its more extreme role as a speciation agent, Wolbachia may reduce gene flow between geographically distant and genetically distinct populations that overlap before the reproductive barrier mechanisms are complete [5]. Cryptic species complex, a group of genetically different but morphologically indistinctive species, is an emerging biological problem also observed in whiteflies (Hemiptera: Aleyrodidae). Increasing reports suggest the effects of Wolbachia infection on the mitochondrial diversity and evolution of hosts, supporting the hypothesis that cryptic speciation is related to Wolbachia infections [6,7,8,9,10].
High vigilance must be given to the increasing facts about intercepted whiteflies at the plant quarantine that might also be invasive species, as they would have major environmental and economic consequences. A case in point is the interception of the whitefly Aleurocanthus spiniferus in Japan. This species was first found in 1919 in Kagoshima Prefecture. Due to a lack of natural enemies, it subsequently became a serious pest in citrus orchards on Kyushu Island, Japan [11,12]. Interestingly, some secondary symbionts are supportive agents for whitefly cryptic species complex invasion, such as the sweet potato whitefly Bemisia tabaci [13]. They confer adaptive responses that eventually support the invasion of this pest. For example, Wolbachia promotes fitness and provides some protection against the parasitism of parasitoid wasps [14]. However, it is yet to be determined just how common these phenotypic effects are to be found in other whiteflies.
The Camellia spiny whitefly Aleurocanthus camelliae (Hemiptera: Aleyrodidae) cryptic species complex is a pest to the Theaceae plants that originated from China and is currently considered to be an invasive species, as it has been detected in Japan (2004), the Netherlands (2018), Italy (2020), and Indonesia (2020) [15,16,17,18,19]. The A. camelliae cryptic species complex consists of at least three related species (Aleurocanthus woglumi, Aleurocanthus spiniferus, and A. camelliae) [16] and five associated haplotypes (A. camelliae haplotypes B1–B3 and A. spiniferus haplogroup A1 and A2) [19,20]. A. spiniferus is extremely polyphagous [21]. Conversely, A. camelliae prefers mostly Theaceae plants and is not inhabit Citrus plants (Rutaceae) as their host [22], although they could also be found in Zanthoxylum piperitum (Rutaceae) [23]. Thus, their dispersion was strongly associated with Theaceae mobility through human activities, such as the global trading of Theaceae plants, such as Camellia sinensis, Camellia japonica, Camellia sasanqua, and Eurya japonica. However, the association between A. camelliae cryptic species complex and bacterial symbionts is poorly understood. There are limited studies related to this topic and other close species that have been examined, such as A. woglumi [24] and A. spiniferus [25].
Therefore, this study aimed to examine the infection status and diversity of Wolbachia in the A. camelliae cryptic species complex in Japan, including A. camelliae haplotype B1, A. spiniferus haplogroup A1, and a novel cryptic species complex (Aleurocanthus cf. A. spiniferus). In addition, to detect the possibility of the horizontal transfer mechanism of Wolbachia, the associated population of insects such as Pealius euryae, another Theaceae whitefly that was newly found to inhabit C. sinensis in the fields (Shizuoka and Kyoto Prefectures) and parasitoid wasps. The infection and diversity of Wolbachia in A. camelliae cryptic species complex were determined using single-gene typing and multilocus sequence typing (MLST). Moreover, its phenotypic characteristics were examined via molecular detection of CI-related genes.
2. Materials and Methods
2.1. Sample Collection
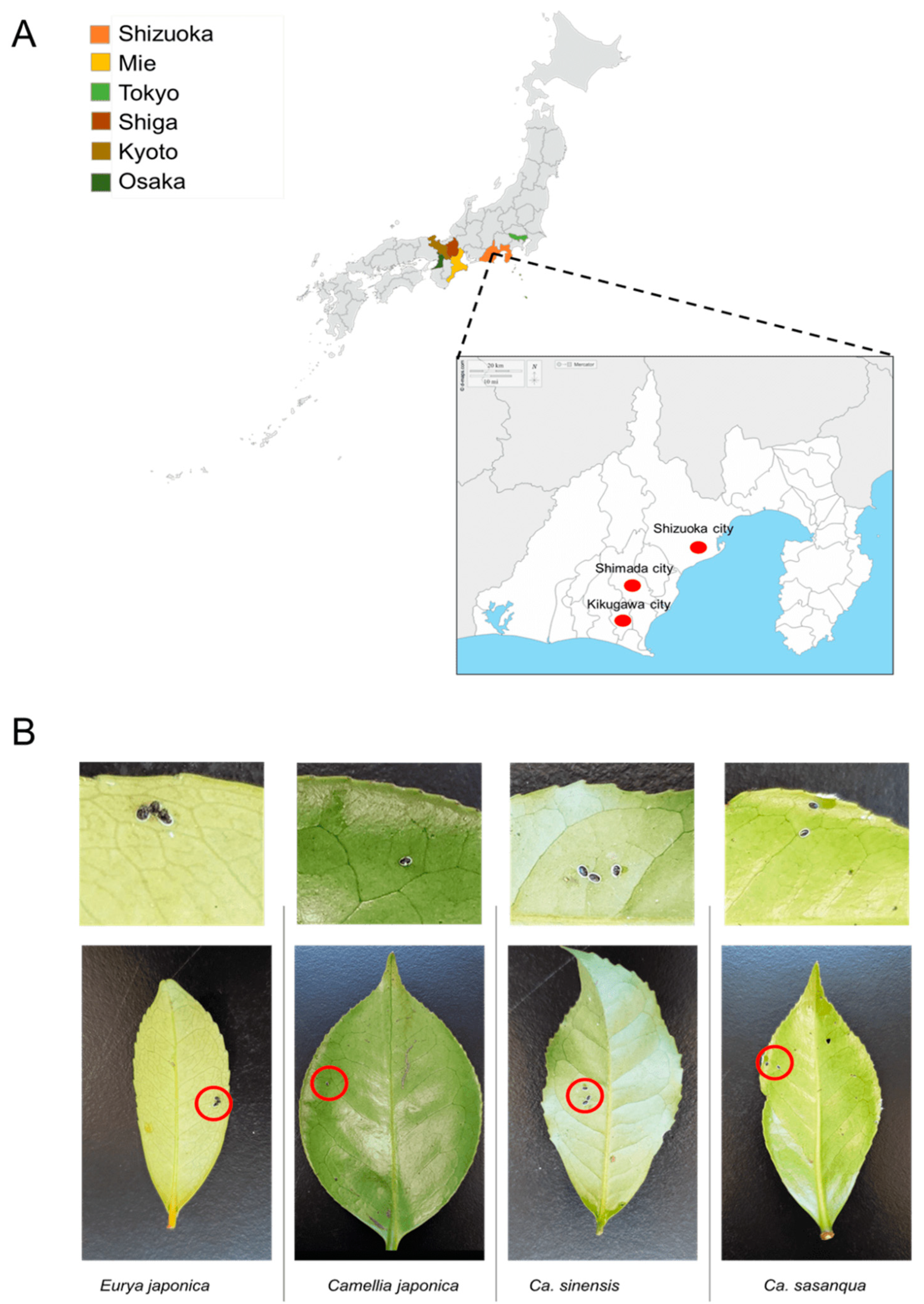
From 2017 to 2022, samples were collected in six Prefectures in Japan from tea (C. sinensis) fields and Theaceae plants, including E. japonica, C. sasanqua, and C. japonica. Samples included in the sample collection stocks were collected between 2009 and 2011 from the Laboratory of Applied Entomology, Shizuoka University [26]. The survey was conducted in Shizuoka Prefecture, Shizuoka City, Shimada City, and Kikugawa City. Other prefectures, such as Osaka, Kyoto, Tokyo, Shiga, and Mie, were also evaluated (Figure 1A). From March 2021 to February 2022, systematic random sampling was employed in a tea field in which many tea varieties (C. sinensis) grow to estimate the dynamics of the positivity rate of Wolbachia infection in the field. This field belongs to the National Agriculture and Food Research Station in Kanaya–Shimada, Shizuoka Prefecture. The leaves infested by a small number of whiteflies were selected as representative samples (Figure 1B; Table 1) and assumed to be a single colony of individuals from different parents. The specimens were stored in a freezer at −20 °C for future deoxyribonucleic acid DNA extraction.
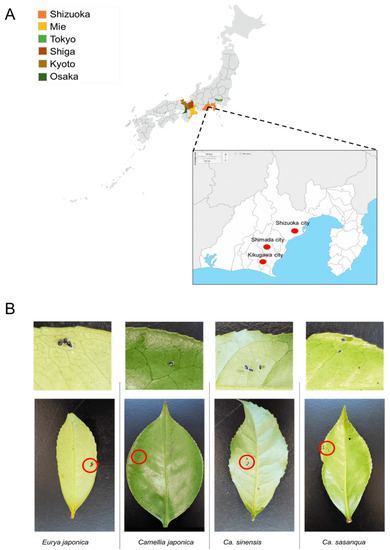
Figure 1.
Sample collection: (A) sampling sites; (B) representative samples of Camellia spiny whitefly and A. camelliae nymphs for molecular assessment.

Table 1.
Whitefly collection.
2.2. DNA Extraction
The DNA of Wolbachia and its hosts was extracted using a slightly modified HotShot method [27] in two steps using Alkaline Buffer (25 mM NaOH and 0.2 EDTA) and a neutralizing solution (40 mM Tris-HCl pH 5.5). Using power masher II for Biomasher II, one individual nymph of whiteflies was crushed in an Eppendorf tube containing 50 μL of Alkaline Buffer. Therefore, aliquots of ~30 μL were transferred into 200 μL tubes and placed in a thermocycler at 95 °C for 15 min. The temperature was reduced to 4 °C, and 30 μL of the neutralizing solution was added and vortexed for 10 s.
2.3. Morphomolecular Identification
Morphological identification was performed using keys on species of the genus Aleurocanthus [28] to determine the species. Morphological comparison between A. spiniferus and A. camelliae described by Kanmiya et al. [15], and simplified keys designated by Jansen and Porcelli [16] were employed to distinguish between Camellia and Citrus spiny whiteflies.
To confirm the morphological identification of mitochondrial DNA markers of cytochrome c oxidase I (COI-1) using the LCO1490/HCO2198 primer set [29], C1-J-2195/L2-N-3014 (COI-2; [30]) and cytochrome b (COB) were used. Species-specific primers designed by Uesugi and Sato [23] were also applied to avoid misamplification due to the parasitism of parasitoid wasps. In addition, haplotype-specific primers were designed to confirm strain A. camelliae without sequencing based on the sequence data accession nos. LCO88497.1, AB786712.1, AB786713.1, and AB786714.1 (AC-55F: AGRAGTGAGTCTGGTAAGTTGG/ACB1-267R: ACCACCTAGAGTTGCCAACC). PCR conditions were set as follows: pre-denaturation at 95 °C for 2 min, continued with 35 cycles of denaturation at 98 °C for 10 s, annealing temperature 50 °C–52 °C for 30 s, and 72 °C for 1 min, with an extension period at 72 °C for 4 min.
2.4. Nested PCR for Determining Wolbachia Infections and MLST Sequencing
Wolbachia surface protein (wsp) typing was performed to detect Wolbachia infections using primer 81F/691R [31]. To confirm the negative results and obtain a fair sequence length of ~500 bp, nested PCR was also performed using primer wspNesF/wspNesR [32] to avoid false-negative results from PCR [32]. The monthly positivity rates of Wolbachia were monitored from March 2021 to February 2022. The monthly average temperature data were retrieved from Japan Meteorological Agency (https://www.data.jma.go.jp/; accessed on 31 March 2022) for Kikukawa–Makinohara (Shimada city, Shizuoka Prefecture). The associations between Wolbachia positivity rates and the average temperatures in the location sample (Shimada city) were estimated using the logistic regression analysis in the R software. Generalized linear models (GLMs; logit link and a binomial distribution) were constructed using the positivity rate as the response variable and the average temperature as an explanatory variable. The p-values for logistic regression were tested using the Wald test, with the level of significance set at p ≤ 0.05).
The single-gene profiling of the 16S rRNA gene of Wolbachia was conducted for comparison using the wspecF/wspecR primer [33]. The diversity of Wolbachia was evaluated by profiling five housekeeping genes using a primer combination designed by [34] and using the ftsZUniF/ftsZUniR primer [33].
PCR was conducted in a total volume of 20 μL GoTaq® Green Master Mix (1 μL DNA template, 1 μL of each primer, 7 μL of double-distilled H2O, and 10 μL of GoTaq). The PCR process used in this study included several steps, starting with pre-denaturation at 98 °C for 2 s, followed by 35 cycles at 98 °C for 10 s. It had an annealing temperature for 50 s, and 72 °C for 1 min, with a final extension period at 72 °C for 4 min. The PCR products were visualized via 1.5% agarose gel electrophoresis. The PCR products were direct-forward-sequenced after purification using ExoSAP-IT (Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania).
2.5. Bacteriophage Detection and Wolbachia Phenotypic Characteristic Determination
The bacteriophage of Wolbachia (phage WO) was detected by targeting the capsid protein gene orf7 of phage WO, WO-F/R [35] and WO-SUF/R [36] comparison phage WO diversity. The genes related to the CI and feminization, such as ankyrin genes pk1 and pk2 [37,38] and non-ankyrin genes cifA and cifB [39,40], were targeted for the detection of a possible mechanism of speciation with the Wolbachia CI strain.
2.6. DNA Sequencing and Phylogenetic Analysis
The amplified fragments of representative samples were directly sequenced by a commercial Sanger sequencing service (Fasmac; Atsugi, Japan), and further analysis was conducted from the obtained sequences. Sequence similarity was analyzed using BLAST [41] on the nucleotide sequences deposited in the NCBI GenBank databases. Sequences were aligned with ClustalW using MEGA X [42]. Phylogenetic analyses were conducted using the maximum likelihood (ML) method [43], and 1000 bootstrap replicates were performed. Evolutionary analysis via the ML method (timetree) was generated using the RelTime method [44], calculated with the ML method, and the Tamura–Nei model [43] using MEGA X.
2.7. Genetic Differentiation, Network Analysis, and Recombination Test of Wolbachia
The net genetic divergence between and within groups (p-distance) of wsp and 16S rRNA of Wolbachia was estimated using MEGA X [42]. The genetic parameters of the population, the number of segregating sites [45], the number of haplotypes (h), haplotype diversity (Hd) [46], and nucleotide diversity (π/bp) [46] were estimated using DNASP version 6 [47]. Using this software, a neutrality test was conducted, which examined population expansion by analyzing deviations from selective neutrality using Tajima’s D [48] and Fu and Li’s D* and F tests [49]. A median-joining 16S rRNA of the Wolbachia haplotype network was constructed using the Network 10 software [50]. The negative Tajima’s D and Fu and Li’s D* and F* values, according to Tseng et al. [51], may indicate a recent population expansion, purifying selection, or genetic hitchhiking, whereas positive values are more likely to indicate a population bottleneck, genetic structure, and/or balancing selection.
Putative recombinant strains in multiple sequence alignments from single-gene typing and MLST were analyzed using RDP5 [52]. Nine methods were employed in the analysis as follows: RDP [53], GENECONV [54], BootsScan [55], MaxChi [56], ChiMaera [57], SiScan [58], Phylpro [59], LARD [60], and 3Seq [61]. The default search parameters of the program were used. The acceptable p-value was <0.05
3. Results
3.1. Morphomolecular Identification
The molecular identification of A. camelliae cryptic species complex using universal primers targeting mitochondrial genes, such as COI and COB, was sensitive to the amplification of genes of parasitoid wasps rather than whiteflies. Parasitoid wasps belonging to the genera Encarsia and Eretmocerus were detected on most representative samples from the fields, such as A1V20, A1W20, B1V20, A1V20, F2X20, A1X21, and A2Z21 (Table 2). Only a few of them were closely related to the sequence data of whiteflies. Using COI-1 typing, A. camelliae haplotype B1 (A1W20-A7) was 99.7% identical to A. spiniferus (no. KJ437166.1), whereas A. spiniferus demonstrated 83.18% reference to Aleurocanthus aracae (no. MZ301225.1). Therefore, Aleurocanthus species-specific (TSW and OSW) primers [19,23] and haplotype-specific (AC55F/ACB1-267R) primers are useful to overcome this obstacle.

Table 2.
Identification of mitochondrial genes using BLAST and the Wolbachia infection status.
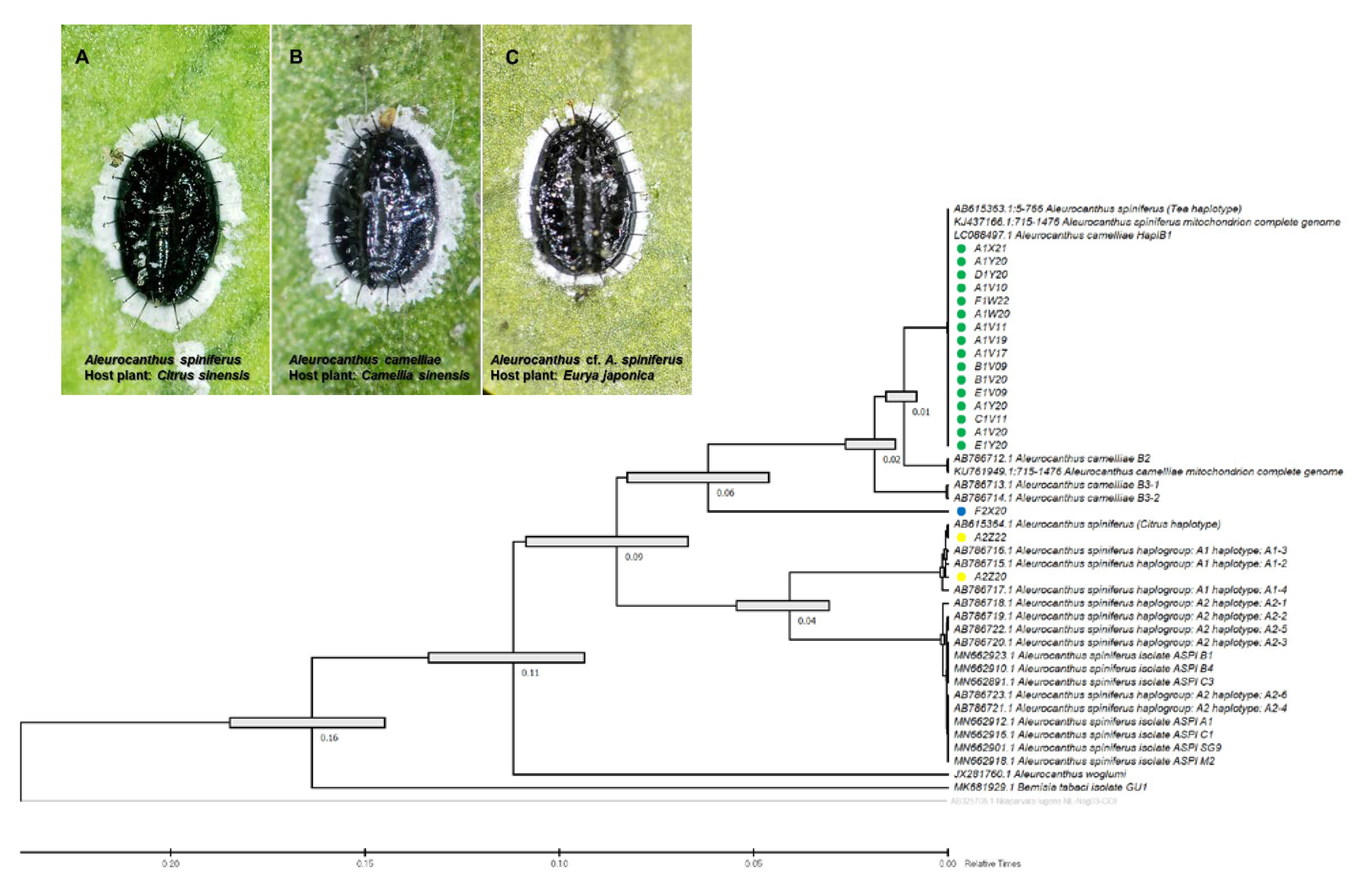
The species-specific (TSW) and haplotype-specific primer (ACF55/ACB1267) primers were unable to confirm one isolate from E. japonica in Tokyo (F2X20) as A. camelliae haplotype B1. Despite the failure to amplify DNA using the TSW primer, the COI gene sequence obtained using the general primer tended to be grouped with A. camelliae (Figure 2). Thus, morphological confirmation was conducted, and it was found that isolate F2X20 was related to A. spiniferus instead of A. camelliae, with features such as a zig-zag arrangement of submedian abdominal spines and having more than 200 marginal teeth. Therefore, this isolate conformed to A. spiniferus (Aleurocanthus cf. A. spiniferus). The F2X20 isolate or Aleurocanthus cf. A. spiniferus sequence was identical to Aleurocanthus sp. (no. KY835557.1 and no. KY836994.1), with >81% similarity. Using COI-2, this isolate was referred to as Tetraleurodes acaciae (no. MT901108.1).
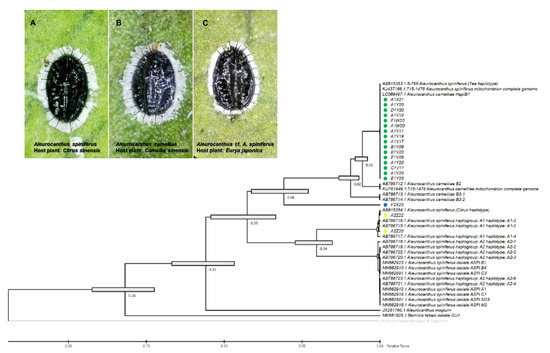
Figure 2.
Evolutionary analysis via maximum likelihood method (timetree) based on the partial sequence mitochondrial COI (COI-2) gene of the A. camelliae cryptic species complex. Yellow circles are isolates of the A. spiniferus haplogroup A1 (A); green circle isolates are the A. camelliae haplotype B1 (B); and the blue circle is an isolate of the Aleurocanthus cf. A. spiniferus (C). Nodes with error bars were indicated in grey bars. Nilaparvata lugens (no. AB325705.1) were assigned as an outgroup. The evolutionary time was predicted by the relative time (Rt) scale bar.
3.2. Positivity and Infection Rates of Wolbachia
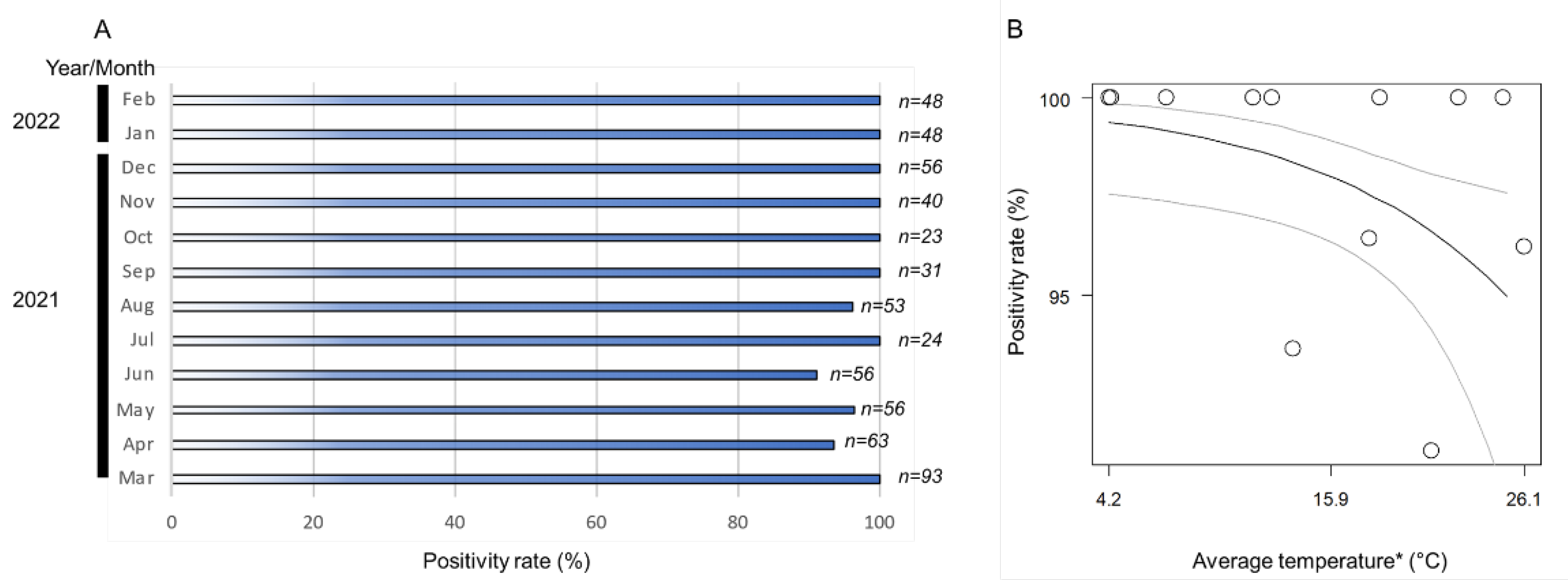
The monthly positivity rates (ratio of positive samples per assessed samples) of Wolbachia in the A. camelliae haplotype B1 ranged from 91% to 100% (Figure 3A). The positive rates remained high across the seasonal temperature, but as the temperature increased (>26 °C), the positive rates tended to decrease (Figure 3B). The high monthly positivity rate confirmed a high infection rate (overall samples assessed) detected in A. camelliae from C. sinensis (96.5%), while a medium rate was detected in C. japonica (40%), and a low rate was detected in C. sasanqua (6.7%) (Table 3). As only a single isolate was examined from E. japonica, it was difficult to estimate their actual infection rate. A. spiniferus is an uninfected population, as individuals were trans-parasitized by Eretmocerus under laboratory conditions, as strongly indicated by their identical strain, Wolbachia, despite some individual nymphs being positively infected (A2Z20-1; see Table 2). A similar case might have also occurred in Aleurocanthus cf. A. spiniferus. Only one individual (F2X20-3; see Table 3) was confirmed to be infected by Wolbachia and simultaneously parasitized by the parasitoid wasp.
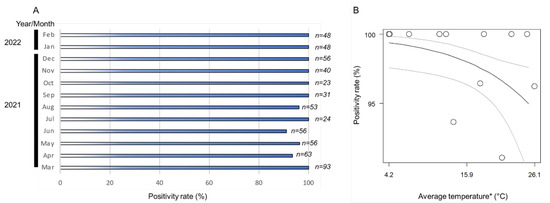
Figure 3.
(A) Nested PCR detection of the wsp gene revealed a positivity rate range of 91–100%; (B) logistic regression analysis on fixed infection across the seasonal temperature. Black line indicates regression line, while grey lines are upper and lower thresholds of 95% confidence interval of predicted line. Regression coefficient was significant (Wald test; p < 0.05). (*) Monthly average temperature data were retrieved from the Japan Meteorological Agency (https://www.data.jma.go.jp/; accessed on 31 March 2022) for Kikukawa–Makinohara (Shimada city, Shizuoka Prefecture).

Table 3.
Infection status of Wolbachia using nested PCR.
3.3. Genetic Diversity of Wolbachia
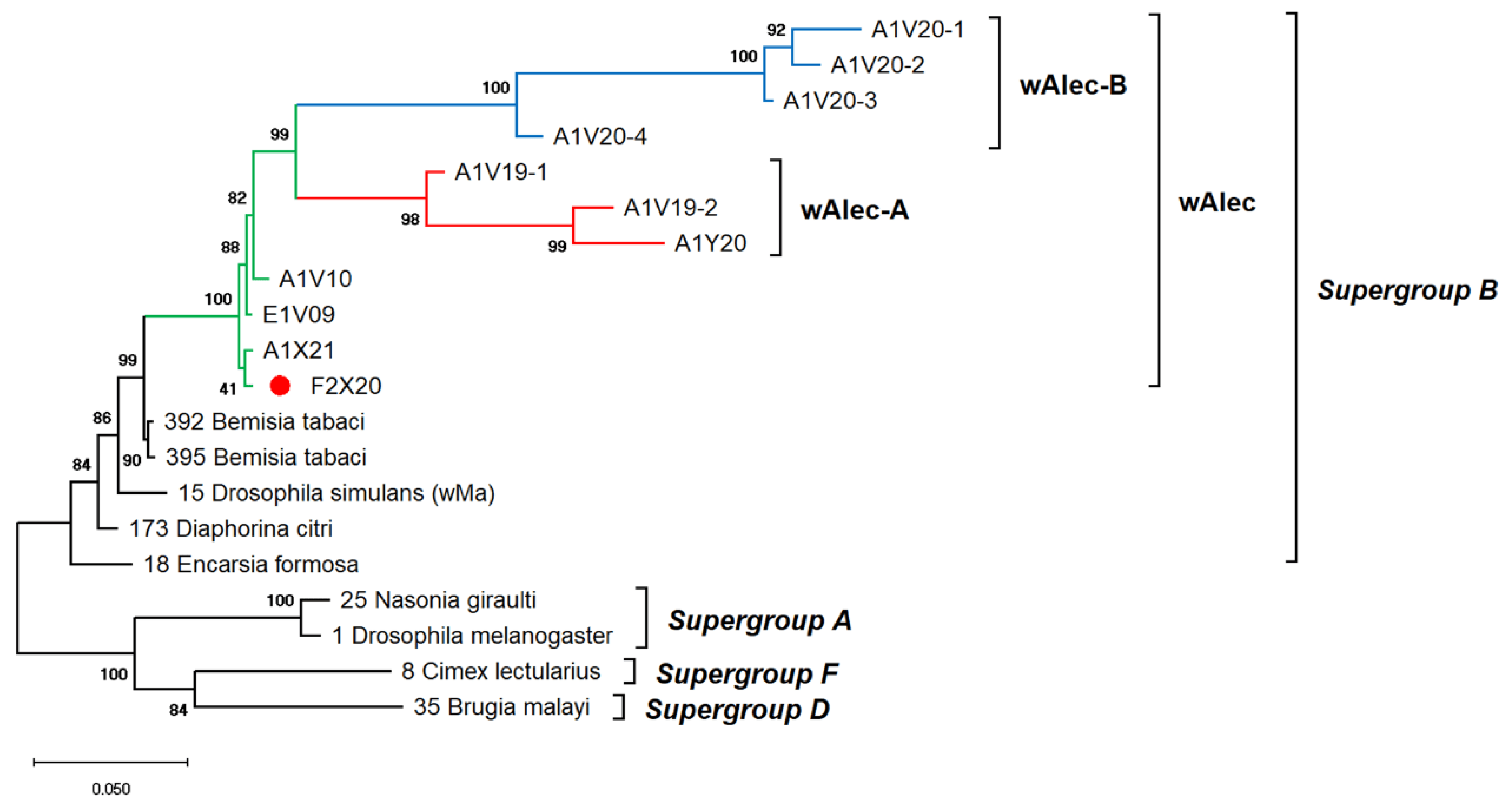
The genetic diversity of Wolbachia infects A. camelliae is difficult to estimate. Single-gene typing using wsp indicated an exceptionally low diversity of Wolbachia, which only consisted of three haplotypes (Hd: 0.1) and nucleotide diversity (π: 0.00099). Other genes, such as the 16S rRNA of Wolbachia, detected among A. camelliae populations, were found to be extremely diverse (Hd: 0.8), with 21 haplotypes and diversity among nucleotides (π: 0.02292) (Table 4). Through MLST, Aleurocanthus spp., notably A. camelliae haplotype B1 and Aleurocanthus cf. A. spiniferus, seemed to harbor a single group of Wolbachia, namely, wAlec, as indicated by the monophyletic clade among these strains. The wAlec strains developed subgroups A and B (Figure 4). These strains were grouped into the Wolbachia supergroup B with other strains such as wBtab, wMa, wDcit, and wEfor.

Table 4.
Haplotype diversity of Wolbachia in A. camelliae haplotype B1 was estimated from a 364 bp wsp and 385 bp 16S rRNA of Wolbachia gene fragments.
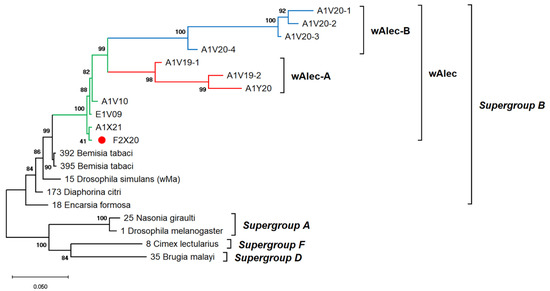
Figure 4.
ML phylogenetic tree of Wolbachia MLST genes. The tree was constructed based on multiple alignments of concatenated DNA sequences encoding gatB, coxA, hcpA, ftsZ, and fbpA in ~2 kbp. Bootstrap values are shown for all nodes. A single lineage of wAlec (green line) evolved into two distinct branches of recombinant strains subgroups A (red) and B (blue). The wAlec also infected Aleurocanthus cf. A. spiniferus (red circle).
3.4. Phage WO Detection and Wolbachia Phenotypic Screening
The low genetic distance or sequence dissimilarity (<1%) of phage WO-infected Wolbachia in A. camelliae from C. sinensis (A1V20) and E. japonica (A1X21), along with Aleurocanthus sp. in E. japonica (F2X20), indicated that they harbored a single strain of phage WO, namely, WOAlec (Table 5). This was also confirmed by the sequences obtained using the new primer set of WOSUF/R. The genes that regulated CI phenotypes in Wolbachia from ankyrin and non-ankyrin genes were not detected in the phage WO strain.

Table 5.
Diversity of phage WO (WOAlec) and phenotypic screening.
3.5. Recombination and Haplotype Diversity of Wolbachia
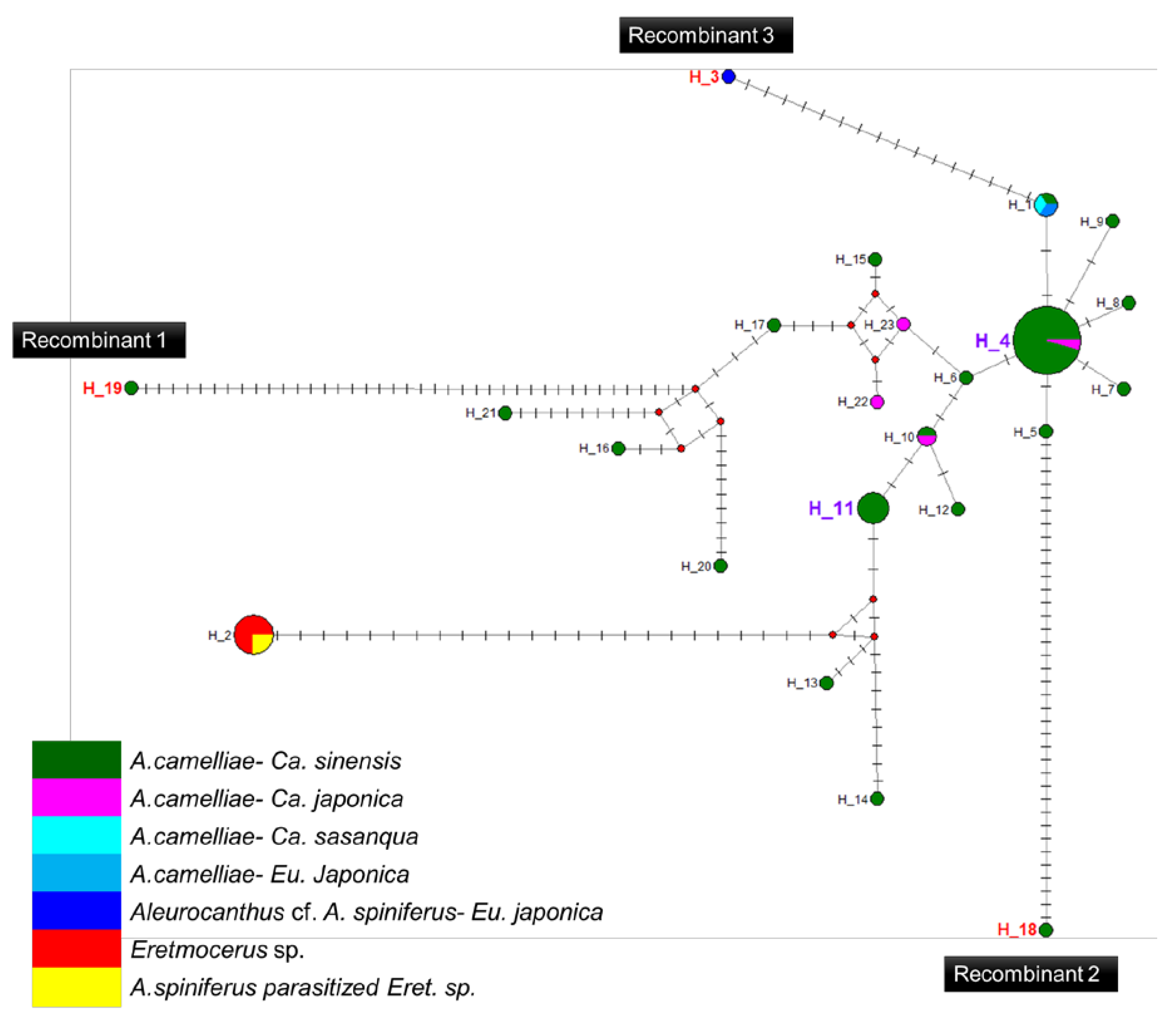
A high prevalence of putative recombinant strains was consistently detected using the GENECONV, ChiMaera, and Phylpro tests. Ten strains were identified in single-gene and MLST-aligned sequences. Other tests, such as RDP, BootScan, ChiMaera, and SiScan, confirmed four to nine recombination events (Table 6). Recombination was observed in the wsp of E3V20 or Pealius euryae in C. sinensis from Kyoto with the main parent, A. camelliae, from the same host and location (E1V20; Table 6). In addition, haplotype 18 (A1V20-27), haplotype 19 (A1V20-10), and haplotype 3 (F2X20) experienced recombination on their 16S rRNA of the Wolbachia gene (Figure 5; Table 6). Based on the MLST sequences, the major parent of recombinant strains of wAlec subgroup B (A1V20-3, A1V20-2, A1V20-4, and A1V20-1) was the strain from wAlec subgroup A (A1V19-2 and A1V19-1) with similarity of 93.1–96.4% (Figure 4; Table 6). In addition, wAlec subgroup A (A1V19-2 and A1V19-1) seemed to have A1Y20 from the same subgroup as their major parent (Figure 5; Table 6).

Table 6.
Intragenic recombination in wAlec by using nine different methods implemented in RDP5 software.
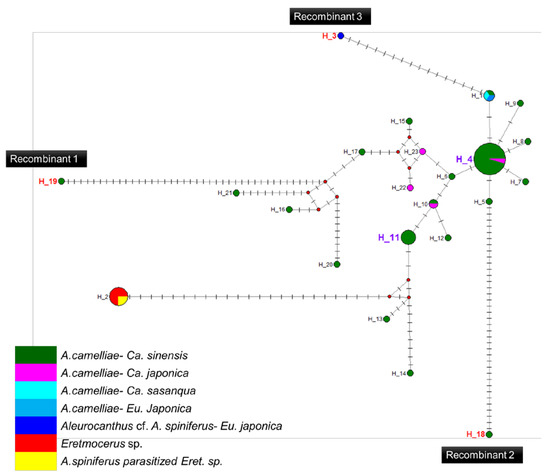
Figure 5.
Haplotype network diagram inferred from the 16S rRNA gene of Wolbachia. Red nodes are median vectors. Striped lines indicate the number of nucleotide mutations.
4. Discussion
Whiteflies are sap-sucking insects belonging to the family Aleyrodidae, which consists of >1550 species, mostly belonging to the subfamilies Aleurodicinae and Aleyrodinae [62]. The morphological identification of whiteflies (Hemiptera: Aleyrodidae), which focused on the characteristics of puparium, has been suggested to be limited and might not even be genus-specific [63] to the Aleurocanthus genus [16]. The current morphological characteristics, number of submarginal spines, number of marginal teeth, arrangement of submedian abdominal spines, and microscopic papillae failed to separate Aleurocanthus cf. A. spiniferus (F2X20) from A. spiniferus, which is genetically different from A. spiniferus and A. camelliae (Figure 2). This confirms the existence of the novel cryptic species complex of A. camelliae in Japan.
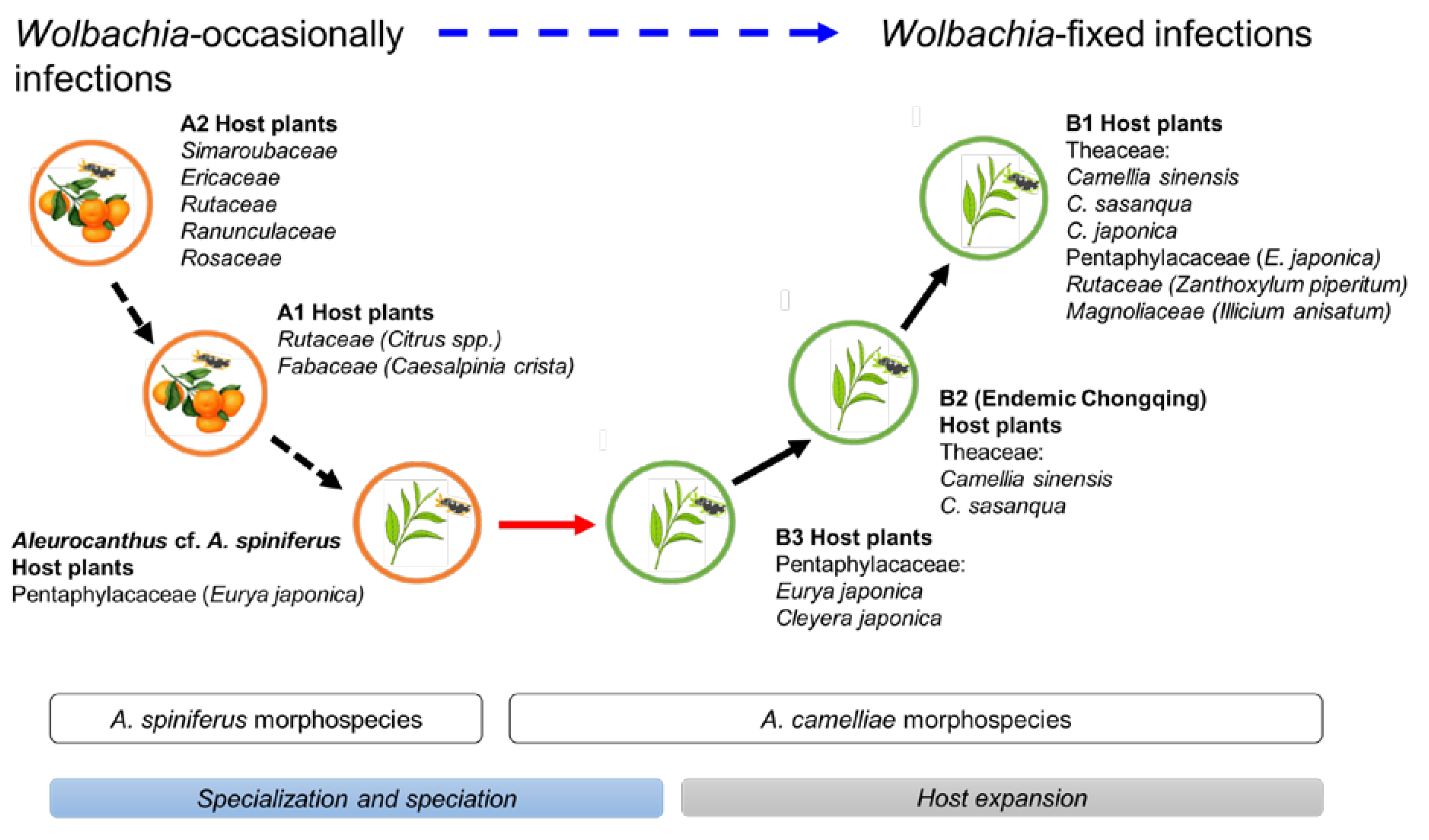
The Aleurocanthus genus consists of at least 78 recorded species, and most species are specific to one or two families of host plants [64]. Among those species, A. woglumi and A. spiniferus are well-known as extremely polyphagous whiteflies that are widely distributed worldwide. A. woglumi inhabits more than 37 host plants, while A. spiniferus inhabits more than 19 families of host plants [64]. The oscillation hypothesis suggests a link between the host plant and geographical range as a contributing factor in increasing diversification rates [65], indicating that the occurrence of diversity in phytophagous insects may be promoted through oscillation in the host plant range. We believe that the current findings also support this hypothesis (Figure 6). The discovery of the novel cryptic species, Aleurocanthus cf. A. spiniferus, linked the history of adaptation among A. spiniferus and A. camelliae, suggesting that the most recent common ancestor of A. camelliae morphospecies is the A. spiniferus morphospecies that inhabits Theaceae sensu lato (Pentaphylacaceae). The cladogenesis of Aleurocanthus cf. A. spiniferus tended to lean toward A. camelliae instead of A. spiniferus (Figure 2), perhaps correlating to the host plants’ group. Theaceae and Pentaphylacaceae are plant families that belong to the same order of Ericales [66]. Therefore, further research on the oscillation hypothesis for the cryptic speciation of A. camelliae may benefit from investigations of how A. spiniferus inhabits another plant of the order Ericales such as Diospyros khaki (Ebenaceae) in Japan [67].
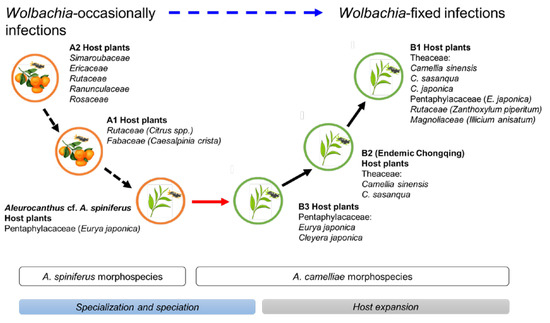
Figure 6.
Hypothetical diagram of the evolutionary history of A. camelliae cryptic species. Predicted speciation time (see Figure 2) among A. spiniferus morphospecies occurred at the relatively same time (Rt 0.06) and was significantly separated from the predicted speciation time of A. camelliae morphospecies (Rt 0.01–0.02). The oscillation in the host plant range represents specialization (black-dashed arrow) and speciation (red arrow; blue bar) to host expansion (black arrow; gray bar). The hierarchical infection status of Wolbachia might be associated with the morphospecies (blue-dashed arrow).
In the cryptic species complex of A. camelliae, a different pattern of Wolbachia infection was found (Table 3). Whiteflies morphologically identical to A. spiniferus (Aleurocanthus sp. in E. japonica and A. spiniferus in Citrus) were grouped into uninfected populations, whereas A. camelliae B1 from C. sinensis was considered the Wolbachia-infected population. Wolbachia infections have been known to significantly affect the structure and mitochondrial diversity of host insects [10,68], leading to cryptic speciation [3]. A similar case has recently been reported in the Wiebesia pumilae cryptic species (Hymenoptera: Agaonidae), which produce hierarchical Wolbachia infection patterns [69]. The spread barrier produced by cryptic species or a different ancestor host population containing Wolbachia CI strains may be the reason for the distinct infection status among cryptic species. However, wAlec is not a Wolbachia CI strain (Table 5), but it does not rule out the possibility that wAlec had a role in speciation since the retention of Wolbachia CI strains for long-term prognosis following secondary contact and spatial reunification of two allopatrically separated populations of a species is normally not favorable. The wAlec CI strains may exist and could have aided the emergence of further reproductive isolation through the process of reinforcement [70] and maintained population differentiation [71].
The intraspecies or intrapopulation infection rates might also vary following the host preferences of A. camelliae itself. Lower infection rates were found in A. camelliae-B1-infesting alternative hosts, such as C. japonica and C. sasanqua. Wolbachia titer is not only maternally inherited, but it can also be horizontally transmitted [71] or eventually lost [72]. Fixed infection in A. camelliae haplotype B1 inhibited C. sinensis (Figure 3A), suggesting that wAlec might have nutritional mutualism such as synthesizing biotin, which might explain the transition from facultative symbiosis to obligate mutualism [73].
This study also provided novel evidence of the recombination event of Wolbachia in the whitefly community in C. sinensis. Wolbachia-strain-infected P. euryae (E3V20) was derived from Wolbachia-infected A. camelliae (E1V20). Both were collected from Kyoto. The recombination was also observed in the population of A. camelliae that were infected by the wAlec group strains. Notably, wAlec subgroup B (A1V20-3, A1V20-2, A1V20-4, and A1V20-1) was derived from wAlec subgroup A (A1V19-2 and A1V19-1) as major parents, and the samples were collected in 2020 and 2019 from the same location, respectively. The recombination is likely to be essential for Wolbachia adaptation to escape Muller’s ratchet, a process leading to the accumulation of mildly deleterious alleles, which is a problem for symbionts that face a population bottleneck in each generation [74,75]. Production of new recombinants results in Wolbachia strains with fewer harmful mutations and greater genetic variety, allowing them to use a wider range of hosts. This phenomenon is also well-known in pathogenic bacteria [76,77,78]. High recombination rates might also indicate a high incidence of horizontal transmission. Bacterial symbionts often maintain intermediate symbiont genome sizes and substantial functional genetic variation through horizontal transmission and recombination [79]. Further analysis is required to determine whether the mechanism of high recombination in wAlec results in the loss of CI strains. The bioassay confirmation of the CI phenotype of wAlec and/or trans-infection of Wolbachia CI strains, e.g., wMel [80], might be useful as a biological control method to contain the A. camelliae cryptic species complex [80].
The detection of positive infection in some parasitized nymphs of the A. spiniferus morphospecies and Eretmocerus sp. (Table 2 and Table 3) revealed the possibility of parasitoids as vectors of Wolbachia [81,82] or the reverse transmission pathway from hosts to parasitoids [83]. Eretmocerus sp. parasitizing A. camelliae is a newly recorded occurrence in Japan. Historically, Encarsia smithi is the only parasitoid wasp of the black spiny whitefly species (A. camelliae and A. spiniferus) in Japan [19,84,85,86,87]. Thus, further studies are needed to identify the Eretmocerus species parasitizing A. camelliae and their origin in order to provide comprehensive information regarding the potential natural enemies of A. camelliae.
Author Contributions
Conceptualization, E.A. and A.K.; methodology, E.A.; software, E.A.; validation, E.A. and A.K.; formal analysis, E.A.; investigation, E.A.; resources, A.K.; data curation, E.A.; writing—original draft preparation, E.A.; writing—review and editing, E.A. and A.K.; visualization, E.A.; supervision, A.K.; project administration, A.K.; funding acquisition, A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study.
Acknowledgments
We thank Tagami Yohsuke, Hitoshi Sawada, and Koji Tsuchida for their valuable comments and suggestions during the study. We also thank Tsutomu Saito and Jessica A. Kapojos for permitting us to examine their collections.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Werren, J.H. Biology of Wolbachia. Annu. Rev. Entomol. 1997, 42, 587–609. [Google Scholar] [CrossRef] [PubMed]
- Saridaki, A.; Bourtzis, K. Wolbachia: More than just a bug in insects genitals. Curr. Opin. Microbiol. 2010, 13, 67–72. [Google Scholar] [CrossRef]
- Rokas, I. Wolbachia as a speciation agent. Trends Ecol. Evol. 2000, 15, 44–45. [Google Scholar] [CrossRef]
- Gill, A.C.; Darby, A.C.; Makepeace, B.L. Iron necessity: The secret of Wolbachia’s success? PLoS Negl. Trop. Dis. 2014, 8, e3224. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, D.D.; Katju, V.; Jaenike, J. Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution 1999, 53, 1157–1164. [Google Scholar] [CrossRef]
- Cariou, M.; Duret, L.; Charlat, S. The global impact of Wolbachia on mitochondrial diversity and evolution. J. Evol. Biol. 2017, 30, 2204–2210. [Google Scholar] [CrossRef]
- Bartoňová, A.S.; Konvička, M.; Marešová, J.; Wiemers, M.; Ignatev, N.; Wahlberg, N.; Schmitt, T.; Faltýnek Fric, Z. Wolbachia affects mitochondrial population structure in two systems of closely related Palaearctic blue butterflies. Sci. Rep. 2021, 11, 3019. [Google Scholar] [CrossRef]
- Avtzis, D.N.; Doudoumis, V.; Bourtzis, K. Wolbachia infections and mitochondrial diversity of two Chestnut feeding Cydia species. PLoS ONE 2014, 9, e112795. [Google Scholar] [CrossRef]
- Xiao, J.H.; Wang, N.X.; Murphy, R.W.; Cook, J.; Jia, L.Y.; Huang, D.W. Wolbachia infection and dramatic intraspecific mitochondrial DNA divergence in a Fig wasp. Evolution 2012, 66, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Schuler, H.; Köppler, K.; Daxböck-Horvath, S.; Rasool, B.; Krumböck, S.; Schwarz, D.; Hoffmeister, T.S.; Schlick-Steiner, B.C.; Steiner, F.M.; Telschow, A.; et al. The hitchhiker’s guide to Europe: The infection dynamics of an ongoing Wolbachia invasion and mitochondrial selective sweep in Rhagoletis cerasi. Mol. Ecol. 2016, 25, 1595–1609. [Google Scholar] [CrossRef]
- Kodama, Y. Studies on Aleurocanthus spiniferus Quaint; Kagoshima Prefectural Office of Internal Affairs: Kagoshima, Japan, 1931. (In Japanese) [Google Scholar]
- Clausen, C.P. Introduced Parasites and Predators of Arthropod Pests and Weeds: A World Review; US Department of Agriculture Handbook No. 480; Agricultural Research Service, US Department of Agriculture: Washington, DC, USA, 1978; ISBN 001-000-03739-1.
- Lu, M.; Hulcr, J.; Sun, J. The role of symbiotic microbes in insect invasions. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 487–505. [Google Scholar] [CrossRef]
- Xue, X.; Li, S.J.; Ahmed, M.Z.; de Barro, P.J.; Ren, S.X.; Qiu, B.L. Inactivation of Wolbachia reveals its biological roles in whitefly host. PLoS ONE 2012, 7, e48148. [Google Scholar] [CrossRef] [PubMed]
- Kanmiya, K.; Ueda, S.; Kasai, A.; Yamashita, K.; Sato, Y.; Yoshiyasu, Y. Proposal of new specific status for tea-infesting populations of the nominal Citrus spiny whitefly Aleurocanthus spiniferus (Homoptera: Aleyrodidae). Zootaxa 2011, 2797, 25–44. [Google Scholar] [CrossRef]
- Jansen, M.; Porcelli, F. Aleurocanthus camelliae (Hemiptera: Aleyrodidae), a species possibly new for the European fauna of a genus in great need of revision. Tijdschr. Entomol. 2018, 161, 63–78. [Google Scholar] [CrossRef]
- Rizzo, D.; Suma, P.; Rossi, E.; Farina, P.; da Lio, D.; Bartolini, L.; Salemi, C.; Farina, A.; Rapisarda, C. First record of Aleurocanthus camelliae Kanmiya & Kasai, 2011 (Hemiptera, Aleyrodidae) from Italy, on Ornamental Camellia spp. plants. EPPO Bull. 2021, 51, 333–339. [Google Scholar] [CrossRef]
- Adi, M.; Susanti, D. Short communication: First record of Aleurocanthus camelliae (Homoptera: Aleyrodidae) in Indonesia, an invasive pest on various medicinal plants. TOI 2020, 13, 94–100. [Google Scholar] [CrossRef]
- Uesugi, R.; Sato, Y.; Han, B.Y.; Huang, Z.D.; Yara, K.; Furuhashi, K. Molecular evidence for multiple phylogenetic groups within two species of invasive Spiny whiteflies and their parasitoid wasp. Bull. Entomol. Res. 2016, 106, 328–340. [Google Scholar] [CrossRef]
- Nugnes, F.; Laudonia, S.; Jesu, G.; Jansen, M.G.M.; Bernardo, U.; Porcelli, F. Aleurocanthus spiniferus (Hemiptera: Aleyrodidae) in some European countries: Diffusion, hosts, molecular characterization, and natural enemies. Insects 2020, 11, 42. [Google Scholar] [CrossRef]
- Cioffi, M.; Cornara, D.; Corrado, I.; Gerardus, M.; Jansen, M.; Porcelli, F. The status of Aleurocanthus spiniferus from its unwanted introduction in Italy to date. Bull. Insectol. 2013, 66, 273–281. [Google Scholar]
- Kasai, A.; Yamashita, K.; Yoshiyasu, Y. Tea-infesting population of the Citrus spiny whitefly, Aleurocanthus spiniferus (Homoptera: Aleyrodidae), does not accept Citrus leaves as host plants. Jpn. J. Appl. Entomol. Zool. 2010, 54, 140–143. [Google Scholar] [CrossRef]
- Uesugi, R.; Sato, Y. Differentiation of the Tea-infesting population of Citrus spiny whitefly Aleurocanthus spiniferus (Homoptera: Aleyrodidae) from the Citrus-infesting population in Japan on the basis of differences in the mitochondrial Cytochrome c oxidase subunit I gene. Jpn. J. Appl. Entomol. Zool. 2011, 55, 155–161. [Google Scholar] [CrossRef][Green Version]
- Pandey, N.; Singh, A.; Rana, V.S.; Rajagopal, R. Molecular characterization and analysis of bacterial diversity in Aleurocanthus woglumi (Hemiptera: Aleyrodidae). Environ. Entomol. 2013, 42, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Bubici, G.; Prigigallo, M.I.; Garganese, F.; Nugnes, F.; Jansen, M.; Porcelli, F. First report of Aleurocanthus spiniferus on Ailanthus altissima: Profiling of the insect microbiome and MicroRNAs. Insects 2020, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Takatsuka, J.; Shimazu, M. Characterization of Paecilomyces cinnamomeus from the Camellia whitefly, Aleurocanthus camelliae (Hemiptera: Aleyrodidae), infesting Tea in Japan. J. Invertebr. Pathol. 2012, 110, 14–23. [Google Scholar] [CrossRef][Green Version]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality Mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Gillespie, P.S. A review of the whitefly Genus Aleurocanthus Quaintance & Baker (Hemiptera: Aleyrodidae) in Australia. Zootaxa 2012, 3252, 1–42. [Google Scholar]
- Folmer, O.F.; Black, M.B.; Hoeh, W.R.; v Lutz, R.; Vrijenhoek, R.C. DNA primers for amplification of mitochondrial Cytochrome c oxidase subunit I from diverse Metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Mol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Zhou, W.; Rousset, F.; O’Neil, S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 1998, 265, 509–515. [Google Scholar] [CrossRef]
- Ji, H.L.; Qi, L.D.; Hong, X.Y.; Xie, H.F.; Li, Y.X. Effects of host sex, plant species, and putative host species on the prevalence of Wolbachia in natural populations of Bemisia tabaci (Hemiptera: Aleyrodidae): A modified nested PCR study. J. Econ. Entomol. 2015, 108, 210–218. [Google Scholar] [CrossRef]
- Werren, J.H.; Windsor, D.M. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc. Biol. Sci. 2000, 267, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Baldo, L.; Hotopp, J.C.D.; Jolley, K.A.; Bordenstein, S.R.; Biber, S.A.; Choudhury, R.R.; Hayashi, C.Y.; Maiden, M.C.J.; Tettelin, H.; Werren, J.H. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006, 72, 7098–7110. [Google Scholar] [CrossRef] [PubMed]
- Masui, S.; Kuroiwa, H.; Sasaki, T.; Inui, M.; Kuroiwa, T.; Ishikawa, H. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of Arthropods. Biochem. Biophys. Res. Commun. 2001, 283, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Su, C.Y.; Zhu, D.H.; Yang, X.H. Design and testing of effective primers for amplification of the orf7 gene of phage WO associated with Andricus hakonensis. Insects 2021, 12, 713. [Google Scholar] [CrossRef]
- LePage, D.P.; Metcalf, J.A.; Bordenstein, S.R.; On, J.; Perlmutter, J.I.; Shropshire, J.D.; Layton, E.M.; Funkhouser-Jones, L.J.; Beckmann, J.F.; Bordenstein, S.R. Prophage WO genes recapitulate and enhance Wolbachia-induced Cytoplasmic incompatibility. Nature 2017, 543, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Pichon, S.; Bouchon, D.; Liu, C.; Chen, L.; Garrett, R.A.; Grève, P. The expression of one ankyrin Pk2 allele of the WO Prophage is correlated with the Wolbachia feminizing effect in Isopods. BMC Microbiol. 2012, 12, 55. [Google Scholar] [CrossRef]
- Walker, T.; Klasson, L.; Sebaihia, M.; Sanders, M.J.; Thomson, N.R.; Parkhill, J.; Sinkins, S.P. Ankyrin repeat do-main-encoding genes in the WPip Strain of Wolbachia from the Culex pipiens Group. BMC Biol. 2007, 5, 39. [Google Scholar] [CrossRef]
- Shropshire, J.D.; On, J.; Layton, E.M.; Zhou, H.; Bordenstein, S.R. One prophage WO gene rescues Cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2018, 115, 4987–4991. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [PubMed]
- Watterson, G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975, 7, 256–276. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; ISBN 9780231886710. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Tseng, S.P.; Wetterer, J.K.; v Suarez, A.V.; Lee, C.Y.; Yoshimura, T.; Shoemaker, D.W.; Yang, C.S. Genetic diversity and Wolbachia infection patterns in a globally distributed invasive ant. Front. Genet. 2019, 10, 838. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new Gemini viruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Posada, D.; Crandall, K.A.; Williamson, C. A modified Bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 2005, 21, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Weiller, G.F. Phylogenetic Profiles: A graphical method for detecting genetic recombinations in homologous sequences. Mol. Biol. Evol. 1998, 15, 326–335. [Google Scholar] [CrossRef]
- Holmes, E.C.; Worobey, M.; Rambaut, A. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 1999, 16, 405–409. [Google Scholar] [CrossRef]
- Lam, H.M.; Ratmann, O.; Boni, M.F. Improved algorithmic complexity for the 3SEQ recombination detection algorithm. Mol. Biol. Evol. 2018, 35, 247–251. [Google Scholar] [CrossRef]
- Martin, J.H.; Mound, L.A. An annotated check list of the World’s whiteflies (Insecta: Hemiptera: Aleyrodidae). Zootaxa 2007, 1492, 1–84. [Google Scholar] [CrossRef]
- Manzari, S.; Quicke, D.L.J. A cladistic analysis of whiteflies, Subfamily Aleyrodinae (Hemiptera: Sternorrhyncha: Aleyrodidae). J. Nat. Hist. 2006, 40, 2423–2554. [Google Scholar] [CrossRef]
- Evans, G. The Whiteflies (Hemiptera: Aleyrodidae) of the World and Their Host Plants and Natural Enemies. 2007. Available online: http://keys.lucidcentral.org/keys/v3/whitefly/PDF_PwP%20ETC/world-whitefly-catalog-Evans.pdf (accessed on 29 June 2022).
- Janz, N.; Nylin, S. The Oscillation Hypothesis of Host-Plant Range and Speciation; Tilmon, K.J., Ed.; University of California Press: London, UK, 2008; pp. 203–215. [Google Scholar]
- Angiosperm Phylogeny Group. An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar] [CrossRef]
- Miyatake, Y. A list of the whiteflies of Japan with their host plant and distribution data (Homoptera: Aleyrodidae). Rostria 1980, 32, 291–330. (In Japanese) [Google Scholar]
- Narita, S.; Nomura, M.; Kato, Y.; Fukatsu, T. Genetic structure of sibling butterfly species affected by Wolbachia infection sweep: Evolutionary and biogeographical implications. Mol. Ecol. 2006, 15, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tong, X.; Li, Y.Y.; Sun, Q.; Gao, Y.; Zhang, S.H.; Wang, R.; Chen, X.Y. Presence of cryptic species in host insects forms a hierarchical Wolbachia infection pattern. Entomol. Genet. 2022, 42, 571–578. [Google Scholar] [CrossRef]
- Jaenike, J.; Dyer, K.A.; Cornish, C.; Minhas, M.S. Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol. 2006, 4, 325. [Google Scholar] [CrossRef] [PubMed]
- Bruzzese, D.J.; Schuler, H.; Wolfe, T.M.; Glover, M.M.; v Mastroni, J.V.; Doellman, M.M.; Tait, C.; Yee, W.L.; Rull, J.; Aluja, M.; et al. Testing the potential contribution of Wolbachia to speciation when Cytoplasmic incompatibility becomes associated with host-related reproductive isolation. Mol. Ecol. 2022, 31, 2935–2950. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Martins-Simões, P.; Szöllosi, G.J.; Mialdea, G.; Sagot, M.F.; Charlat, S. How long does Wolbachia remain on board? Mol. Biol. Evol. 2017, 34, 1183–1193. [Google Scholar] [CrossRef]
- Nikoh, N.; Hosokawa, T.; Moriyama, M.; Oshima, K.; Hattori, M.; Fukatsu, T. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 2014, 111, 10257–10262. [Google Scholar] [CrossRef]
- Moran, N.A. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 2873–2878. [Google Scholar] [CrossRef]
- Jiggins, F.M.; von der Schulenburg, J.H.; Hurst, G.D.; Majerus, M.E. Recombination confounds interpretations of Wolbachia evolution. Proc. Biol. Sci. 2001, 268, 1423–1427. [Google Scholar] [CrossRef]
- Awadalla, P. The evolutionary genomics of pathogen recombination. Nat. Rev. Genet. 2003, 4, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, H.; Zhou, R. Genome-wide evidence for positive selection and recombination in Actinobacillus pleuropneumoniae. BMC Evol. Biol. 2011, 11, 203. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Zhu, D.H.; Liu, Z.; Zhao, L.; Su, C.Y. High levels of multiple infections, recombination, and horizontal transmission of Wolbachia in the Andricus mukaigawae (Hymenoptera; Cynipidae) communities. PLoS ONE 2013, 8, e78970. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Pepper-Tunick, E.; Svedberg, J.; Byrne, A.; Ruelas Castillo, J.; Vollmers, C.; Beinart, R.A.; Corbett-Detig, R. Horizontal transmission and recombination maintain forever young bacterial symbiont genomes. PLoS Genet. 2020, 16, e1008935. [Google Scholar] [CrossRef]
- Zhou, X.F.; Li, Z.X. Establishment of the Cytoplasmic incompatibility-inducing Wolbachia strain wMel in an important agricultural pest insect. Sci. Rep. 2016, 6, 39200. [Google Scholar] [CrossRef] [PubMed]
- Vavre, F.; Fleury, F.; Lepetit, D.; Fouillet, P.; Boulétreau, M. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 1999, 16, 1711–1723. [Google Scholar] [CrossRef]
- Ahmed, M.Z.; Li, S.J.; Xue, X.; Yin, X.J.; Ren, S.X.; Jiggins, F.M.; Greeff, J.M.; Qiu, B.L. The intracellular bacterium Wolbachia uses parasitoid wasps as phoretic vectors for efficient horizontal transmission. PLoS Pathog. 2015, 10, e1004672. [Google Scholar] [CrossRef]
- Johannesen, J. Tracing the history and ecological context of Wolbachia double infection in a specialist host (Urophora cardui)—Parasitoid (Eurytoma serratulae) system. Ecol. Evol. 2017, 7, 986–996. [Google Scholar] [CrossRef]
- DeBach, P. Biological Control of Insect Pests and Weeds; Reihold: New York, NY, USA, 1964; p. 676. [Google Scholar]
- Ozawa, A.; Uchiyama, T. Parasitism of the tea spiny whitefly, Aleurocanthus camelliae Kanmiya & Kasai, by Encarsia smithi (Silvestri) in tea fields in Shizuoka Prefecture in 2012, two years after the first identification of the pest. Ann. Rept. Kansai Pl. Prot. 2013, 55, 89–91. [Google Scholar]
- Uesugi, R.; Yara, K.; Sato, Y. Changes in population density of Aleurocanthus camelliae (Hemiptera: Aleyrodidae) and parasitism rate of Encarsia smithi (Hymenoptera: Aphelinidae) during the early invasion stages. Appl. Entomol. Zool. 2016, 51, 581–588. [Google Scholar] [CrossRef]
- Kuwana, I. Notes on a newly imported parasite of the Spiny whitefly attacking Citrus in Japan. In Proceedings of the fifth Pacific Science Congress Organized by the Pacific Science Association and the National Research Council of Canada, Victoria and Vancouver, BC, Canada, 1–14 June 1933; University of Toronto: Toronto, ON, Canada, 1934; pp. 3521–3525. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).