The Evolution of Glycoside Hydrolase Family 1 in Insects Related to Their Adaptation to Plant Utilization
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Glycoside Hydrolase Family 1 Identification
2.3. Gene Tree Inference
2.4. Gene Duplication and Loss Inference
2.5. Duplicate Mode Inference and Collinearity Analysis
2.6. Statistical Analysis
3. Results
3.1. GH1s in Insects and Other Arthropods
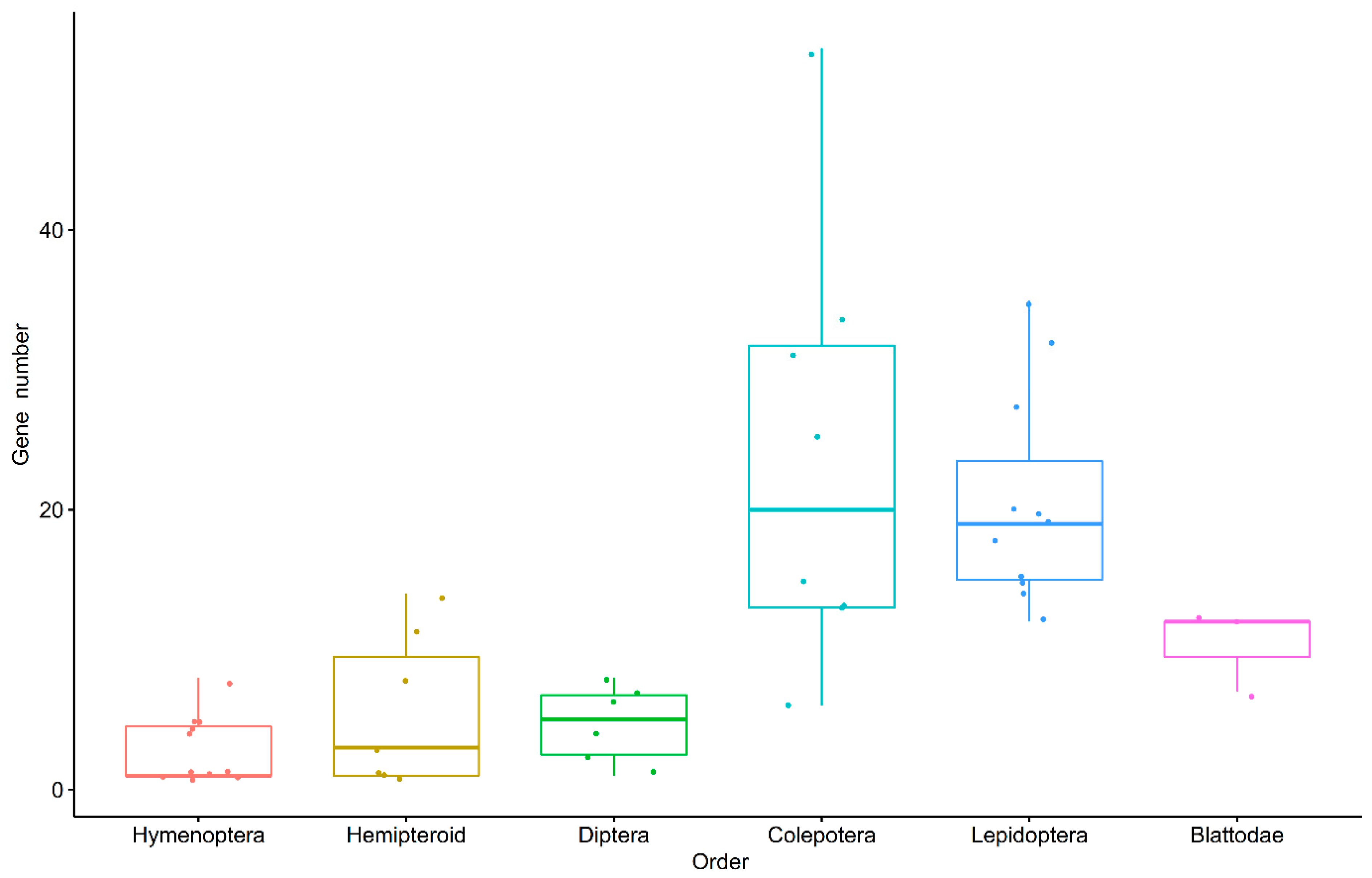
3.2. GH1 Gene Numbers Related to Their Feeding Behaviors
3.3. Phylogenetic Tree of GH1 Genes in Insects
3.4. Reconciliation between the Gene Tree and Species Tree
3.5. Collinearity and Duplication Modes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mello, M.O.; Silva-Filho, M.C. Plant-insect interactions: An evolutionary arms race between two distinct defense mechanisms. BRAZ J. Plant Physiol. 2002, 14, 71–81. [Google Scholar] [CrossRef]
- Tokuda, G. Plant cell wall degradation in insects: Recent progress on endogenous enzymes revealed by multi-omics technologies. Adv. Insect Phys. 2019, 57, 97–136. [Google Scholar]
- Rane, R.V.; Ghodke, A.B.; Hoffmann, A.A.; Edwards, O.R.; Walsh, T.K.; Oakeshott, J.G. Detoxifying enzyme complements and host use phenotypes in 160 insect species. Curr. Opin. Insect Sci. 2019, 31, 131–138. [Google Scholar] [CrossRef]
- Erb, M.; Robert, C.A. Sequestration of plant secondary metabolites by insect herbivores: Molecular mechanisms and ecological consequences. Curr. Opin. Insect Sci. 2016, 14, 8–11. [Google Scholar] [CrossRef]
- Beran, F.; Köllner, T.G.; Gershenzon, J.; Tholl, D. Chemical convergence between plants and insects: Biosynthetic origins and functions of common secondary metabolites. New Phytol. 2019, 223, 52–67. [Google Scholar] [CrossRef]
- Pentzold, S.; Zagrobelny, M.; Rook, F.; Bak, S. How insects overcome two-component plant chemical defence: Plant β-glucosidases as the main target for herbivore adaptation. Biol. Rev. 2014, 89, 531–551. [Google Scholar] [CrossRef]
- Heidel-Fischer, H.M.; Vogel, H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect Sci. 2015, 8, 8–14. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Ferreira, A.H.P.; Marana, S.R.; Terra, W.R.; Ferreira, C. Purification, molecular cloning, and properties of a β-glycosidase isolated from midgut lumen of Tenebrio molitor (Coleoptera) larvae. Insect Biochem. Mol. Biol. 2001, 31, 1065–1076. [Google Scholar] [CrossRef]
- Byeon, G.M.; Lee, K.S.; Gui, Z.Z.; Kim, I.; Kang, P.D.; Lee, S.M.; Sohn, H.D.; Jin, B.R. A digestive β-glucosidase from the silkworm, Bombyx mori: cDNA cloning, expression and enzymatic characterization. Comp. Biochem. Phys. B 2005, 141, 418–427. [Google Scholar] [CrossRef]
- Tokuda, G.; Miyagi, M.; Makiya, H.; Watanabe, H.; Arakawa, G. Digestive β-glucosidases from the wood-feeding higher termite, Nasutitermes takasagoensis: Intestinal distribution, molecular characterization, and alteration in sites of expression. Insect Biochem. Mol. Biol. 2009, 39, 931–937. [Google Scholar] [CrossRef]
- Arakawa, G.; Kamino, K.; Tokuda, G.; Watanabe, H. Purification, characterization, and cDNA cloning of a prominent β-glucosidase from the gut of the xylophagous cockroach Panesthia angustipennis spadica. J. Appl. GlycoSci. 2016, 63, 51–59. [Google Scholar] [CrossRef]
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 2000, 42, 93–114. [Google Scholar] [CrossRef]
- Bridges, M.; Jones, A.M.E.; Bones, A.M.; Hodgson, C.; Cole, R.; Bartlet, E.; Wallsgrove, R.; Karapapa, V.K.; Watts, N.; Rossiter, J.T. Spatial organization of the glucosinolate–myrosinase system in brassica specialist aphids is similar to that of the host plant. P Roy Soc. B-Biol. Sci. 2002, 269, 187–191. [Google Scholar] [CrossRef]
- Francis, F.; Lognay, G.; Wathelet, J.-P.; Haubruge, E. Characterisation of aphid myrosinase and degradation studies of glucosinolates. Arch. Insect Biochem. 2002, 50, 173–182. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, B.W.; Schroeder, F.C.; Jander, G. Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant J. 2008, 54, 1015–1026. [Google Scholar] [CrossRef]
- Kos, M.; Houshyani, B.; Achhami, B.B.; Wietsma, R.; Gols, R.; Weldegergis, B.T.; Kabouw, P.; Bouwmeester, H.J.; Vet, L.E.M.; Dicke, M.; et al. Herbivore-Mediated Effects of Glucosinolates on Different Natural Enemies of a Specialist Aphid. J. Chem. Ecol. 2012, 38, 100–115. [Google Scholar] [CrossRef]
- Beran, F.; Pauchet, Y.; Kunert, G.; Reichelt, M.; Wielsch, N.; Vogel, H.; Reinecke, A.; Svatoš, A.; Mewis, I.; Schmid, D.; et al. Phyllotreta striolata flea beetles use host plant defense compounds to create their own glucosinolate-myrosinase system. Proc. Natl. Acad. Sci. USA 2014, 111, 7349–7354. [Google Scholar] [CrossRef]
- Pentzold, S.; Zagrobelny, M.; Bjarnholt, N.; Kroymann, J.; Vogel, H.; Olsen, C.E.; Møller, B.L.; Bak, S. Metabolism, excretion and avoidance of cyanogenic glucosides in insects with different feeding specialisations. Insect Biochem. Mol. Biol. 2015, 66, 119–128. [Google Scholar] [CrossRef]
- Agnihotri, A.R.; Hulagabali, C.V.; Adhav, A.S.; Joshi, R.S. Mechanistic insight in potential dual role of sinigrin against Helicoverpa armigera. Phytochemistry 2018, 145, 121–127. [Google Scholar] [CrossRef]
- Sporer, T.; Körnig, J.; Beran, F. Ontogenetic differences in the chemical defence of flea beetles influence their predation risk. Funct. Ecol. 2020, 34, 1370–1379. [Google Scholar] [CrossRef] [Green Version]
- Cornette, R.; Farine, J.-P.; Abed-Viellard, D.; Quennedey, B.; Brossut, R. Molecular characterization of a male-specific glycosyl hydrolase, Lma-p72, secreted on to the abdominal surface of the Madeira cockroach Leucophaea maderae (Blaberidae, Oxyhaloinae). Biochem. J. 2003, 372, 535–541. [Google Scholar] [CrossRef]
- Weil, T.; Rehli, M.; Korb, J. Molecular basis for the reproductive division of labour in a lower termite. BMC Genomics 2007, 8, 198. [Google Scholar] [CrossRef]
- Korb, J.; Weil, T.; Hoffmann, K.; Foster, K.R.; Rehli, M. A Gene Necessary for Reproductive Suppression in Termites. Science 2009, 324, 758. [Google Scholar] [CrossRef]
- Matsuura, K.; Yashiro, T.; Shimizu, K.; Tatsumi, S.; Tamura, T. Cuckoo Fungus Mimics Termite Eggs by Producing the Cellulose-Digesting Enzyme β-Glucosidase. Curr. Biol. 2009, 19, 30–36. [Google Scholar] [CrossRef]
- Jones, A.; Bridges, M.; Bones, A.; Cole, R.; Rossiter, J. Purification and characterisation of a non-plant myrosinase from the cabbage aphid Brevicoryne brassicae (L.). Insect Biochem. Mol. Biol. 2001, 31, 1–5. [Google Scholar] [CrossRef]
- Jones, A.; Winge, P.; Bones, A.; Cole, R.; Rossiter, J. Characterization and evolution of a myrosinase from the cabbage aphid Brevicoryne brassicae. Insect Biochem. Mol. Biol. 2002, 32, 275–284. [Google Scholar] [CrossRef]
- Weil, T.; Korb, J.; Rehli, M. Comparison of Queen-Specific Gene Expression in Related Lower Termite Species. Mol. Biol. Evol. 2009, 26, 1841–1850. [Google Scholar] [CrossRef]
- Bujang, N.S.; Harrison, N.A.; Su, N.-Y. Molecular Cloning of Five β-Glucosidases from Four Species of Higher Termites (Blattodea: Termitidae). Ann. EntoMol. Soc. Am. 2014, 107, 251–256. [Google Scholar] [CrossRef]
- Rahfeld, P.; Haeger, W.; Kirsch, R.; Pauls, G.; Becker, T.; Schulze, E.; Wielsch, N.; Wang, D.; Groth, M.; Brandt, W.; et al. Glandular β-glucosidases in juvenile Chrysomelina leaf beetles support the evolution of a host-plant-dependent chemical defense. Insect Biochem. Mol. Biol. 2015, 58, 28–38. [Google Scholar] [CrossRef]
- Pentzold, S.; Jensen, M.K.; Matthes, A.; Olsen, C.E.; Petersen, B.L.; Clausen, H.; Møller, B.L.; Bak, S.; Zagrobelny, M. Spatial separation of the cyanogenic β-glucosidase ZfBGD2 and cyanogenic glucosides in the haemolymph of Zygaena larvae facilitates cyanide release. Roy Soc. Open Sci. 2017, 4, 170262. [Google Scholar] [CrossRef] [Green Version]
- Marques, A.R.; Coutinho, P.M.; Videira, P.; Fialho, A.M.; Sá-Correia, I. Sphingomonas paucimobilis beta-glucosidase Bgl1: A member of a new bacterial subfamily in glycoside hydrolase family 1. Biochem. J. 2003, 370, 793–804. [Google Scholar] [CrossRef]
- Román-Palacios, C.; Scholl, J.P.; Wiens, J.J. Evolution of diet across the animal tree of life. Evol. Lett. 2019, 3, 339–347. [Google Scholar] [CrossRef]
- Husebye, H.; Arzt, S.; Burmeister, W.P.; Härtel, F.V.; Brandt, A.; Rossiter, J.T.; Bones, A.M. Crystal structure at 1.1Å resolution of an insect myrosinase from Brevicoryne brassicae shows its close relationship to β-glucosidases. Insect Biochem. Mol. Biol. 2005, 35, 1311–1320. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, 733–745. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, 95–101. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Thompson, J.D.; Thierry, J.C.; Poch, O. RASCAL: Rapid scanning and correction of multiple sequence alignments. Bioinformatics 2003, 19, 1155–1161. [Google Scholar] [CrossRef]
- Thompson, J.D.; Plewniak, F.; Ripp, R.; Thierry, J.-C.; Poch, O. Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol. 2001, 314, 937–951. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Chen, K.; Durand, D.; Farach-Colton, M. NOTUNG: A progrAm. for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 2000, 7, 429–447. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Gunkel, S.; Meusemann, K.; Kozlov, A.; Podsiadlowski, L.; Petersen, M.; Lanfear, R.; et al. Evolutionary History of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef]
- Johnson, K.P.; Dietrich, C.H.; Friedrich, F.; Beutel, R.G.; Wipfler, B.; Peters, R.S.; Allen, J.M.; Petersen, M.; Donath, A.; Walden, K.K.O.; et al. Phylogenomics and the evolution of hemipteroid insects. Proc. Natl. Acad. Sci. USA 2018, 115, 12775–12780. [Google Scholar] [CrossRef]
- Giribet, G.; Edgecombe, G.D. The Phylogeny and Evolutionary History of Arthropods. Curr. Biol. 2019, 29, 592–602. [Google Scholar] [CrossRef]
- Kawahara, A.Y.; Plotkin, D.; Espeland, M.; Meusemann, K.; Toussaint, E.F.A.; Donath, A.; Gimnich, F.; Frandsen, P.B.; Zwick, A.; Reis, M.d.; et al. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc. Natl. Acad. Sci. USA 2019, 116, 22657–22663. [Google Scholar] [CrossRef]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef]
- Durand, D.; Halldórsson, B.V.; Vernot, B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J. Comput. Biol. 2006, 13, 320–335. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Thomas, G.W.C.; Dohmen, E.; Hughes, D.S.T.; Murali, S.C.; Poelchau, M.; Glastad, K.; Anstead, C.A.; Ayoub, N.A.; Batterham, P.; Bellair, M.; et al. Gene content evolution in the arthropods. Genome Biol. 2020, 21, 15. [Google Scholar] [CrossRef]
- Kao, D.; Lai, A.G.; Stamataki, E.; Rosic, S.; Konstantinides, N.; Jarvis, E.; Di Donfrancesco, A.; Pouchkina-Stancheva, N.; Semon, M.; Grillo, M. The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. eLife 2016, 5, e20062. [Google Scholar] [CrossRef]
- Ramya, S.L.; Venkatesan, T.; Murthy, K.S.; Jalali, S.K.; Varghese, A. Degradation of acephate by Enterobacter asburiae, Bacillus cereus and Pantoea agglomerans isolated from diamondback moth Plutella xylostella (L), a pest of cruciferous crops. J. Environ. Biol. 2016, 37, 611–618. [Google Scholar]
- Xia, X.; Sun, B.; Gurr, G.M.; Vasseur, L.; Xue, M.; You, M. Gut microbiota mediate insecticide resistance in the diamondback moth, Plutella xylostella (L.). Front. MicroBiol. 2018, 9, 25. [Google Scholar] [CrossRef]
- Thakur, A.; Dhammi, P.; Saini, H.S.; Kaur, S. Pathogenicity of bacteria isolated from gut of Spodoptera litura (Lepidoptera: Noctuidae) and fitness costs of insect associated with consumption of bacteria. J. Invertebr. Pathol. 2015, 127, 38–46. [Google Scholar] [CrossRef]
- Vilanova, C.; Baixeras, J.; Latorre, A.; Porcar, M. The generalist inside the specialist: Gut bacterial communities of two insect species feeding on toxic plants are dominated by Enterococcus sp. Front. MicroBiol. 2016, 7, 1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snoeck, S.; Wybouw, N.; Van Leeuwen, T.; Dermauw, W. Transcriptomic plasticity in the arthropod generalist Tetranychus urticae upon long-term acclimation to different host plants. G3-Genes Genom. Genet. 2018, 8, 3865–3879. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, S.; Pavlidi, N.; Pipini, D.; Vontas, J.; Dermauw, W.; Van Leeuwen, T. Substrate specificity and promiscuity of horizontally transferred UDP-glycosyltransferases in the generalist herbivore Tetranychus urticae. Insect Biochem. Mol. Biol. 2019, 109, 116–127. [Google Scholar] [CrossRef]
- Winde, I.; Wittstock, U. Insect herbivore counteradaptations to the plant glucosinolate–myrosinase system. Phytochemistry 2011, 72, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Vyas, D. Myrosinase: Insights on structural, catalytic, regulatory, and environmental interactions. CR Rev. Biotech. 2019, 39, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenberger, A.; Fink, P. Gene expression and activity of digestive enzymes of Daphnia pulex in response to food quality differences. Comp. Biochem. Phys. B 2018, 218, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Fountain, M.T.; Hopkin, S.P. Folsomia candida (Collembola): A “standard” soil arthropod. Annu. Rev. EntoMol. 2005, 50, 201–222. [Google Scholar] [CrossRef]
- Rota-Stabelli, O.; Daley, A.C.; Pisani, D. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr. Biol. 2013, 23, 392–398. [Google Scholar] [CrossRef]
- Moreau, C.S.; Bell, C.D.; Vila, R.; Archibald, S.B.; Pierce, N.E. Phylogeny of the ants: Diversification in the age of angiosperms. Science 2006, 312, 101–104. [Google Scholar] [CrossRef]
- Sann, M.; Niehuis, O.; Peters, R.S.; Mayer, C.; Kozlov, A.; Podsiadlowski, L.; Bank, S.; Meusemann, K.; Misof, B.; Bleidorn, C. Phylogenomic analysis of Apoidea sheds new light on the sister group of bees. BMC Evol. Biol. 2018, 18, 71. [Google Scholar] [CrossRef]
- Eyun, S.-i.; Wang, H.; Pauchet, Y.; Ffrench-Constant, R.H.; Benson, A.K.; Valencia-Jiménez, A.; Moriyama, E.N.; Siegfried, B.D. Molecular Evolution of Glycoside Hydrolase Genes in the Western Corn Rootworm (Diabrotica virgifera virgifera). PLoS ONE 2014, 9, e94052. [Google Scholar] [CrossRef] [PubMed]
- Kunieda, T.; Fujiyuki, T.; Kucharski, R.; Foret, S.; Ament, S.; Toth, A.; Ohashi, K.; Takeuchi, H.; Kamikouchi, A.; Kage, E. Carbohydrate metabolism genes and pathways in insects: Insights from the honey bee genome. Insect Mol. Biol. 2006, 15, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Mathé-Hubert, H.; Colinet, D.; Deleury, E.; Belghazi, M.; Ravallec, M.; Poulain, J.; Dossat, C.; Poirié, M.; Gatti, J.-L. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: A hypothetical role for a GH1 β-glucosidase. Sci. Rep. 2016, 6, 35873. [Google Scholar] [CrossRef]
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.-W.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef]
- Scharf, M.E.; Kovaleva, E.S.; Jadhao, S.; Campbell, J.H.; Buchman, G.W.; Boucias, D.G. Functional and translational analyses of a beta-glucosidase gene (glycosyl hydrolase family 1) isolated from the gut of the lower termite Reticulitermes flavipes. Insect Biochem. Mol. Biol. 2010, 40, 611–620. [Google Scholar] [CrossRef]
- Wang, Q.; Qian, C.; Zhang, X.-Z.; Liu, N.; Yan, X.; Zhou, Z. Characterization of a novel thermostable β-glucosidase from a metagenomic library of termite gut. Enzyme Microb. Tech. 2012, 51, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Maekawa, K. Gene expression and molecular phylogenetic analyses of beta-glucosidase in the termite Reticulitermes speratus (Isoptera: Rhinotermitidae). J. Insect Physiol. 2014, 65, 63–69. [Google Scholar] [CrossRef]
- Singh, G.; Verma, A.K.; Kumar, V. Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech 2016, 6, 3. [Google Scholar] [CrossRef]
- Loehlin, D.W.; Carroll, S.B. Expression of tandem gene duplicates is often greater than twofold. Proc. Natl. Acad. Sci. USA 2016, 113, 5988–5992. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Jiang, B.; Chakraborty, A.; Yu, G. The Evolution of Glycoside Hydrolase Family 1 in Insects Related to Their Adaptation to Plant Utilization. Insects 2022, 13, 786. https://doi.org/10.3390/insects13090786
He S, Jiang B, Chakraborty A, Yu G. The Evolution of Glycoside Hydrolase Family 1 in Insects Related to Their Adaptation to Plant Utilization. Insects. 2022; 13(9):786. https://doi.org/10.3390/insects13090786
Chicago/Turabian StyleHe, Shulin, Bin Jiang, Amrita Chakraborty, and Guozhi Yu. 2022. "The Evolution of Glycoside Hydrolase Family 1 in Insects Related to Their Adaptation to Plant Utilization" Insects 13, no. 9: 786. https://doi.org/10.3390/insects13090786
APA StyleHe, S., Jiang, B., Chakraborty, A., & Yu, G. (2022). The Evolution of Glycoside Hydrolase Family 1 in Insects Related to Their Adaptation to Plant Utilization. Insects, 13(9), 786. https://doi.org/10.3390/insects13090786





