Genetic Diversity and Population Structure of Spirobolus bungii as Revealed by Mitochondrial DNA Sequences
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
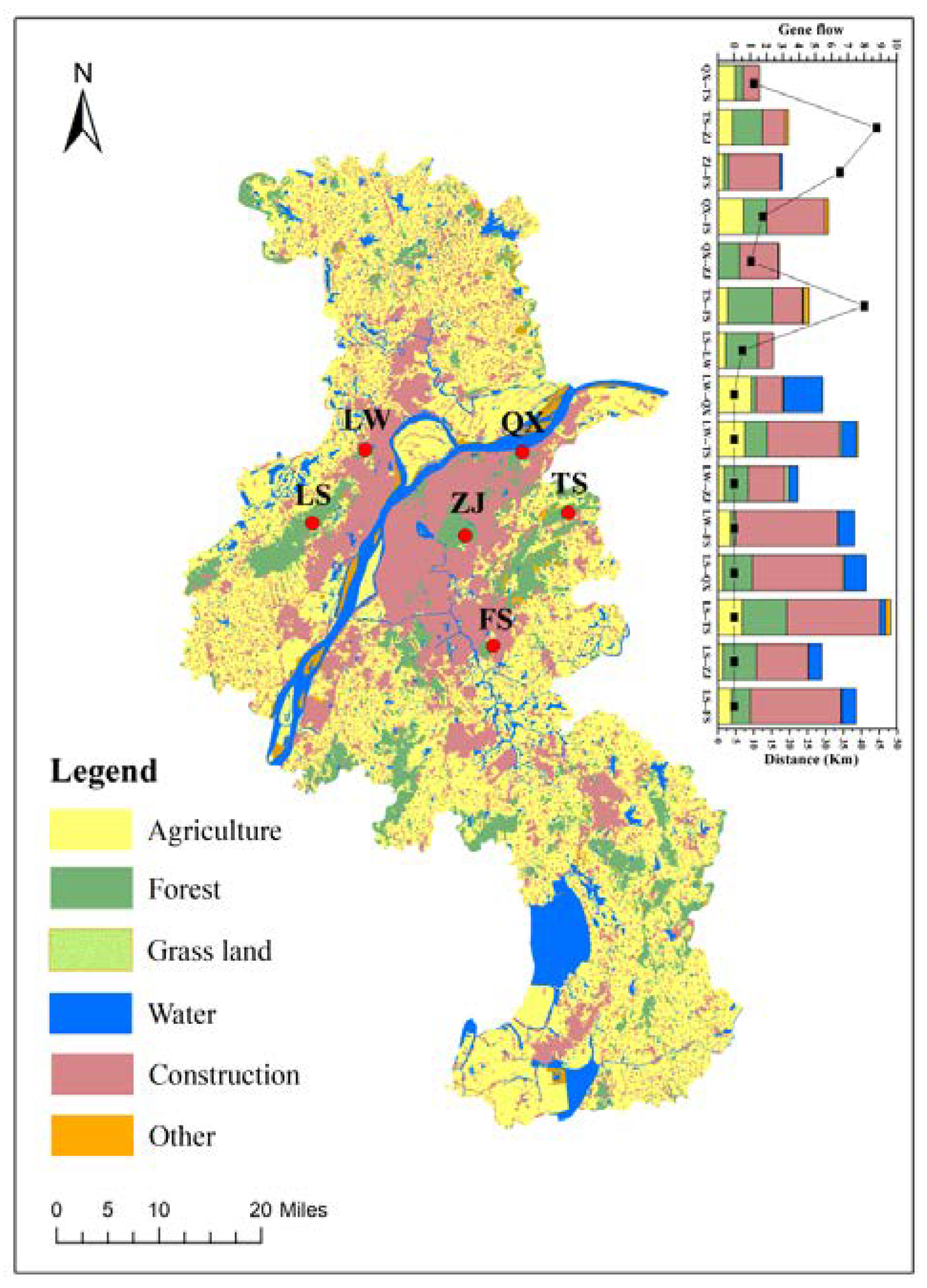
2.1. Sample Collection
2.2. DNA Extraction and MtDNA Amplification and Sequencing
2.3. Soil Detection
2.4. Sequence Analysis
3. Results
3.1. Genetic Diversity of S. bungii
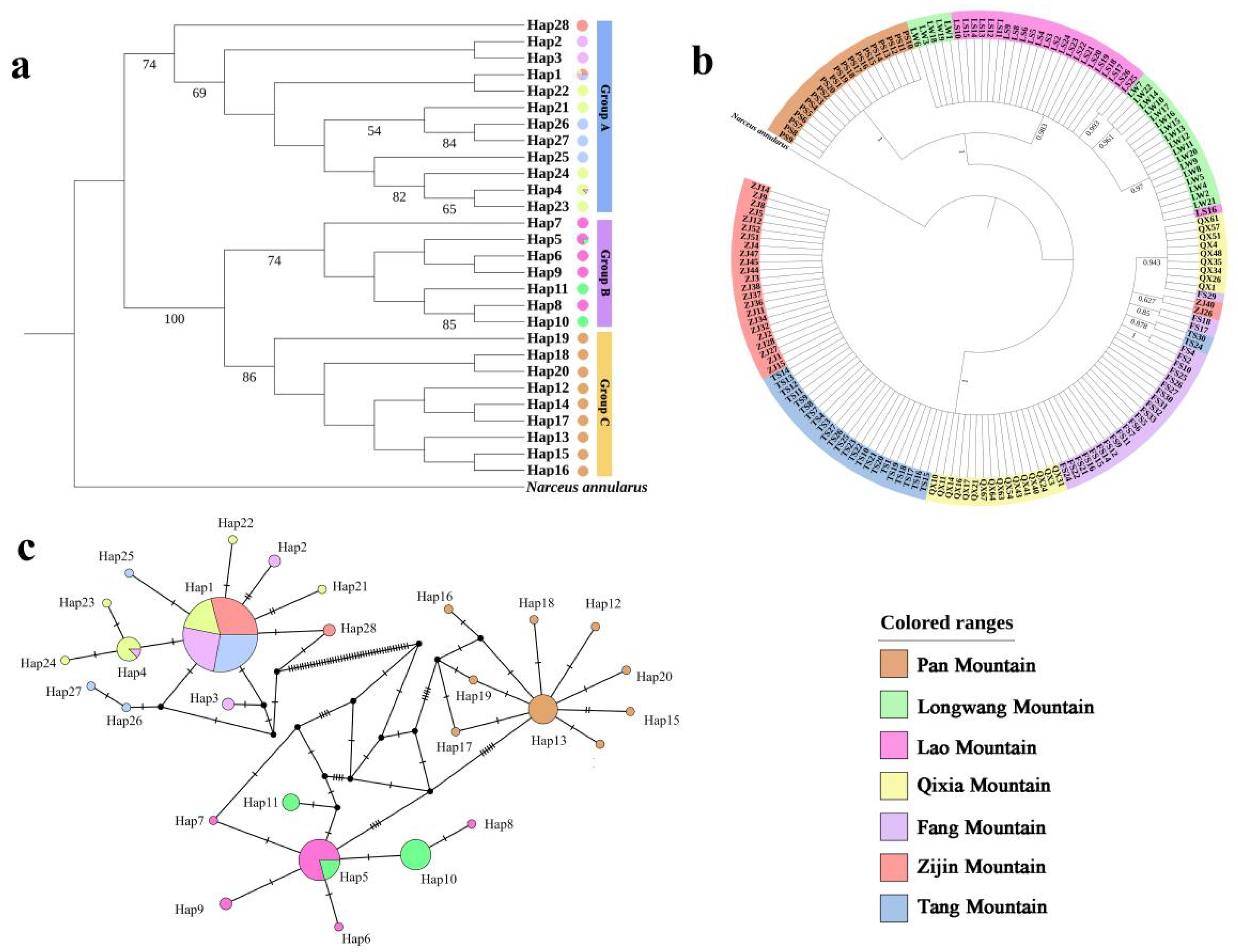
3.2. Population Genetic Structure of S. bungii
3.3. Phylogenetic Analyses and the TCS Haplotype Network
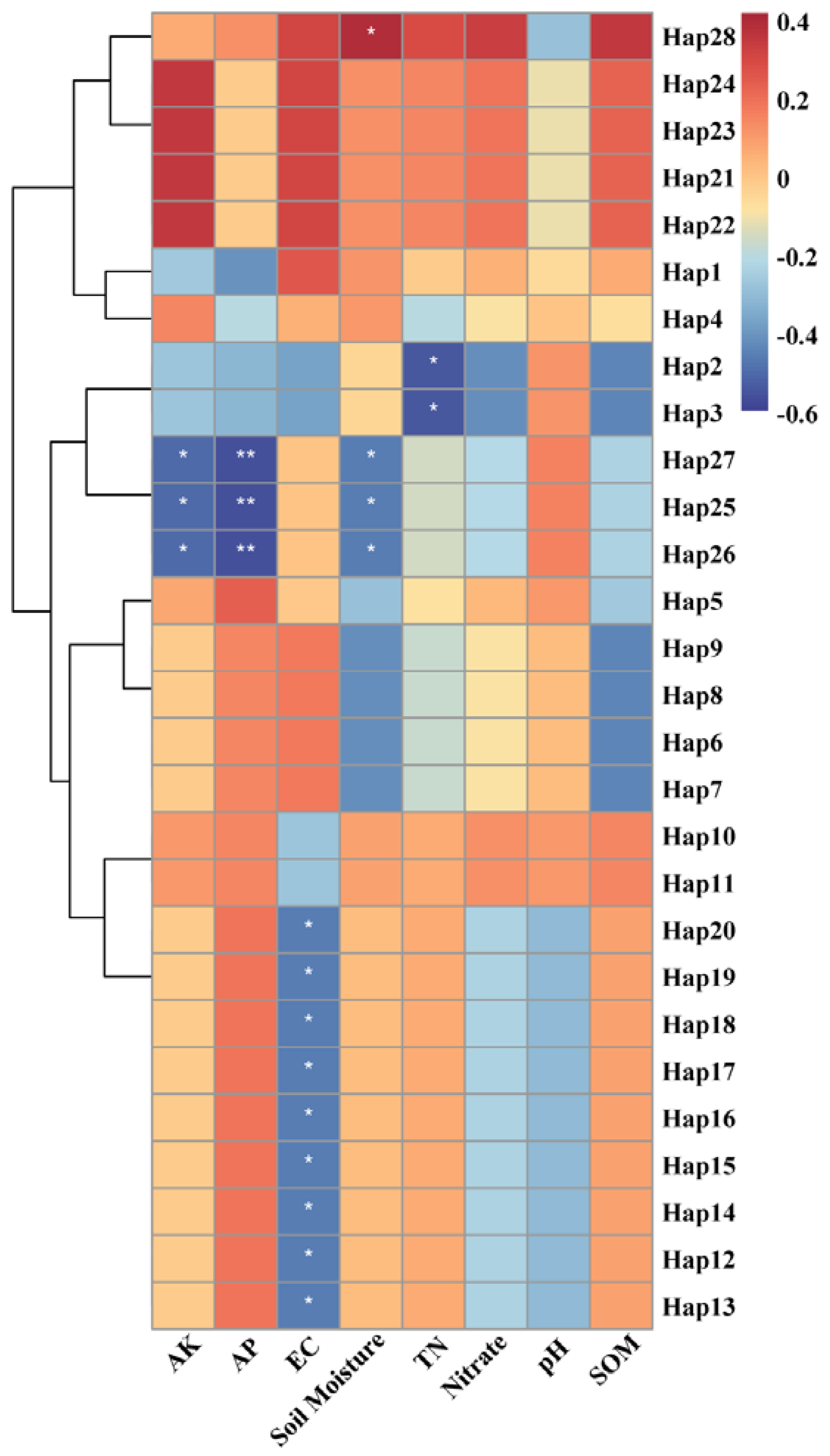
3.4. Relationship between Environmental Factors and Haplotype Distribution
4. Discussion
4.1. Differences in Genetic Structure between Soil Macrofauna
4.2. Gene Flow between S. bungii Populations Blocked by Geographical Barriers
4.3. Environmental Factors on the Genetic Diversity of S. bungii
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fang, Y.; Chen, J.; Ruan, H.H.; Xu, N.; Que, Z.T.; Liu, H.Y. Genetic Diversity and Population Structure of Metaphire vulgaris Based on the Mitochondrial COI Gene and Microsatellites. Front. Genet. 2021, 12, 979. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Z.; Pan, K.W.; Tariq, A.; Zhang, L.; Sun, X.M.; Li, Z.L.; Sun, F.; Xiong, Q.L.; Song, D.G.; Olatunji, O.A. Combined effects of cropping types and simulated extreme precipitation on the community composition and diversity of soil macrofauna in the eastern Qinghai-Tibet Plateau. J. Soils Sediments 2018, 18, 3215–3227. [Google Scholar] [CrossRef]
- Gholami, S.; Sayad, E.; Gebbers, R.; Schirrmann, M.; Joschko, M.; Timmer, J. Spatial analysis of riparian forest soil macrofauna and its relation to abiotic soil properties. Pedobiologia 2016, 59, 27–36. [Google Scholar] [CrossRef]
- Wu, P.F.; Wang, C.T. Differences in spatiotemporal dynamics between soil macrofauna and mesofauna communities in forest ecosystems: The significance for soil fauna diversity monitoring. Geoderma 2019, 337, 266–272. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Zhang, D.H.; Xu, Y.L.; Zhang, G.S.; Sun, Z.J. Effects of fragmentation on genetic variation in populations of the terrestrial earthworm Drawida japonica Michaelsen, 1892 (Oligochaeta, Moniligastridae) in Shandong and Liaodong peninsulas, China. J. Nat. Hist. 2012, 46, 1387–1405. [Google Scholar] [CrossRef]
- Keller, E.L.; Connolly, S.T.; Gorres, J.H.; Schall, J.J. Genetic diversity of an invasive earthworm, Lumbricus terrestris, at a long-term trading crossroad, the Champlain Valley of Vermont, USA. Biol. Invasions 2020, 22, 1723–1735. [Google Scholar] [CrossRef]
- Pitz, K.M.; Sierwald, P. Phylogeny of the millipede order Spirobolida (Arthropoda: Diplopoda: Helminthomorpha). Cladistics 2010, 26, 497–525. [Google Scholar] [CrossRef]
- Pimvichai, P.; Enghoff, H.; Panha, S.; Backeljau, T. Integrative taxonomy of the new millipede genus Coxobolellus, gen. nov. (Diplopoda: Spirobolida: Pseudospirobolellidae), with descriptions of ten new species. Invertebr. Syst. 2020, 34, 591–617. [Google Scholar] [CrossRef]
- Means, J.C.; Hennen, D.A.; Tanabe, T.; Marek, P.K. Phylogenetic systematics of the millipede family Xystodesmidae. Insect Syst. Divers. 2021, 5, 1. [Google Scholar] [CrossRef]
- Pimvichai, P.; Panha, S.; Backeljau, T. Combining mitochondrial DNA and morphological data to delineate four new millipede species and provisional assignment to the genus Apeuthes Hoffman & Keeton (Diplopoda: Spirobolida: Pachybolidae: Trigoniulinae). Invertebr. Syst. 2022, 36, 91–112. [Google Scholar] [CrossRef]
- Brookfield, M.E.; Catlos, E.J.; Suarez, S.E. Myriapod divergence times differ between molecular clock and fossil evidence: U/Pb zircon ages of the earliest fossil millipede-bearing sediments and their significance. Hist. Biol. 2021, 33, 2014–2018. [Google Scholar] [CrossRef]
- Lavelle, P.; Decaens, T.; Aubert, M.; Barot, S.; Blouin, M.; Bureau, F.; Margerie, P.; Mora, P.; Rossi, J.P. Soil invertebrates and ecosystem services. Eur. J. Soil Biol. 2006, 42, S3–S15. [Google Scholar] [CrossRef]
- Snyder, B.A.; Hendrix, P.F. Current and Potential Roles of Soil Macroinvertebrates (Earthworms, Millipedes, and Isopods) in Ecological Restoration. Restor. Ecol. 2008, 16, 629–636. [Google Scholar] [CrossRef]
- Kula, E.; Lazorik, M. Centipedes, millipedes, terrestrial isopods and their relationships to physical and chemical properties of forest soils. Entomol. Fenn. 2016, 27, 33–51. [Google Scholar] [CrossRef]
- Uys, C.; Hamer, M.; Slotow, R. Turnover in flightless invertebrate species composition over different spatial scales in Afrotemperate forest in the Drakensberg, South Africa. Afr. J. Ecol. 2009, 47, 341–351. [Google Scholar] [CrossRef]
- Olson, C.I.; Beaubien, G.B.; Sims, J.L.; Otter, R.R. Mercury Accumulation in Millipedes (Narceus spp.) Living Adjacent to a Southern Appalachian Mountain Stream (USA). Bull. Environ. Contam. Toxicol. 2019, 103, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Naude, A.L.; Snyman, R.G.; Odendaal, J.P. Aluminium and Iron Contamination of Soil, Leaf Litter and Bioindicators in Selected South African Forest Pockets. Water Air Soil Pollut. 2021, 232, 304. [Google Scholar] [CrossRef]
- Brandt, J.L. Tentaminum quorundam monographicorum Insecta Myriapoda Chilognathi Latreillii spectantium prodromus. Bull. Bull. Socieéteé Impeériale Nat. Moscou 1833, 6, 194–209. [Google Scholar]
- Ma, Y.S.; Zeng, Q.L.; Song, B.; Du, J.J.; Yang, F.Q.; Zhao, Y. SHRIMP U-Pb dating of zircon from Panshan granitoid pluton in Yanshan orogenic belt and its tectonic implications. Acta Petrol. Sin. 2007, 23, 547–556. [Google Scholar]
- Chen, J.L.; Gao, J.L.; Yuan, F.; Wei, Y.D. Spatial Determinants of Urban Land Expansion in Globalizing Nanjing, China. Sustainability 2016, 8, 868. [Google Scholar] [CrossRef]
- Liu, H.B.; Liu, Y.F.; Wang, H.N.; Yang, J.Y.; Zhou, X. Research on the coordinated development of greenization and urbanization based on system dynamics and data envelopment analysis-A case study of Tianjin. J. Clean. Prod. 2019, 214, 195–208. [Google Scholar] [CrossRef]
- Yu, W.Q.; Wang, Y.J.; Hu, H.B.; Wang, Y.Q.; Zhang, H.L.; Wang, B.; Liu, Y. Regulations and patterns of soil moisture dynamics and their controlling factors in hilly regions of lower reaches of Yangtze River basin, China. J. Cent. South Univ. 2015, 22, 4764–4777. [Google Scholar] [CrossRef]
- Sha, J.; Zou, J.; Sun, J.N. Observational study of land-atmosphere turbulent flux exchange over complex underlying surfaces in urban and suburban areas. Sci. China-Earth Sci. 2021, 64, 1050–1064. [Google Scholar] [CrossRef]
- Hassan, M.M.; Hassan, M.M. Molecular and morphological identification of some millipedes (Spirostreptida: Spirostreptidae) collected from Taif, Saudi Arabia. Zool. Middle East 2021, 67, 177–185. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Norman, R.J.; Stucki, J.W. The determination of nitrate and nitrite in soil extracts by ultraviolet spectrophotometry. Soil Sci. Soc. Am. J. 1981, 47, 347–353. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Rosenberg, N.A.; Burke, T.; Elo, K.; Feldmann, M.W.; Freidlin, P.J.; Groenen, M.A.M.; Hillel, J.; Maki-Tanila, A.; Tixier-Boichard, M.; Vignal, A.; et al. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 2001, 159, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Floyd, R.; Abebe, E.; Papert, A.; Blaxter, M. Molecular barcodes for soil nematode identification. Mol. Ecol. 2002, 11, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.Y.K.; Carson, J.; Elliott, N.G. 18S ribosomal DNA-based PCR identification of Neoparamoeba pemaquidensis, the agent of amoebic gill disease in sea-farmed salmonids. Dis. Aquat. Org. 2004, 60, 65–76. [Google Scholar] [CrossRef]
- Countway, P.D.; Gast, R.J.; Savai, P.; Caron, D.A. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J. Eukaryot. Microbiol. 2005, 52, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xiong, J.; Yu, Y.H. Taxonomic Resolutions Based on 18S rRNA Genes: A Case Study of Subclass Copepoda. PLoS ONE 2015, 10, e0131498. [Google Scholar] [CrossRef]
- Bohonak, A.J. Dispersal, gene flow, and population structure. Q. Rev. Biol. 1999, 74, 21–45. [Google Scholar] [CrossRef]
- Clobert, J.; Le Galliard, J.F.; Cote, J.; Meylan, S.; Massot, M. Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol. Lett. 2009, 12, 197–209. [Google Scholar] [CrossRef]
- Chatelain, M.; Mathieu, J. How good are epigeic Earthworms at dispersing? An investigation to compare epigeic to endogeic and anecic groups. Soil Biol. Biochem. 2017, 111, 115–123. [Google Scholar] [CrossRef]
- Jenkins, D.G.; Brescacin, C.R.; Duxbury, C.V.; Elliott, J.A.; Evans, J.A.; Grablow, K.R.; Hillegass, M.; LyonO, B.N.; Metzger, G.A.; Olandese, M.L.; et al. Does size matter for dispersal distance? Glob. Ecol. Biogeogr. 2007, 16, 415–425. [Google Scholar] [CrossRef]
- Lester, S.E.; Ruttenberg, B.I.; Gaines, S.D.; Kinlan, B.P. The relationship between dispersal ability and geographic range size. Ecol. Lett. 2007, 10, 745–758. [Google Scholar] [CrossRef]
- Dupont, L.; Pauwels, M.; Dume, C.; Deschins, V.; Audusseau, H.; Gigon, A.; Dubs, F.; Vandenbulcke, F. Genetic variation of the epigeic earthworm Lumbricus castaneus populations in urban soils of the Paris region (France) revealed using eight newly developed microsatellite markers. Appl. Soil Ecol. 2019, 135, 33–37. [Google Scholar] [CrossRef]
- Bruna, E.M.; Izzo, T.J.; Inouye, B.D.; Uriarte, M.; Vasconcelos, H.L. Asymmetric Dispersal and Colonization Success of Amazonian Plant-Ants Queens. PLoS ONE 2011, 6, e22937. [Google Scholar] [CrossRef]
- Suarez, D.; Arribas, P.; Jimenez-Garcia, E.; Emerson, B.C. Dispersal ability and its consequences for population genetic differentiation and diversification. Proc. R. Soc. B-Biol. Sci. 2022, 289, 489. [Google Scholar] [CrossRef]
- Novo, M.; Almodovar, A.; Diaz-Cosin, D.J. High genetic divergence of hormogastrid Earthworms (Annelida, Oligochaeta) in the central Iberian Peninsula: Evolutionary and demographic implications. Zool. Scr. 2009, 38, 537–552. [Google Scholar] [CrossRef]
- Zheng, H.B.; Clift, P.D.; Wang, P.; Tada, R.J.; Jia, J.T.; He, M.Y.; Jourdan, F. Pre-Miocene birth of the Yangtze River. Proc. Natl. Acad. Sci. USA 2013, 110, 7556–7561. [Google Scholar] [CrossRef]
- Fu, X.W.; Zhu, W.L.; Geng, J.H.; Yang, S.Y.; Zhong, K.; Huang, X.T.; Zhang, L.Y.; Xu, X. The present-day Yangtze River was established in the late Miocene: Evidence from detrital zircon ages. J. Asian Earth Sci. 2021, 205, 104600. [Google Scholar] [CrossRef]
- Guo, R.J.; Sun, X.L.; Li, C.A.; Li, Y.W.; Wei, C.Y.; Zhang, Z.J.; Leng, Y.H.; Klotzli, U.; Li, G.N.; Lv, L.Y.; et al. Cenozoic evolution of the Yangtze River: Constraints from detrital zircon U-Pb ages. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2021, 579, 110586. [Google Scholar] [CrossRef]
- Pan, B.T.; Hu, Z.B.; Wang, J.P.; Vandenberghe, J.; Hu, X.F.; Wen, Y.H.; Li, Q.; Cao, B. The approximate age of the planation surface and the incision of the Yellow River. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2012, 356, 54–61. [Google Scholar] [CrossRef]
- Li, B.F.; Sun, D.H.; Xu, W.H.; Wang, F.; Liang, B.Q.; Ma, Z.W.; Wang, X.; Li, Z.J.; Chen, F.H. Paleomagnetic chronology and paleoenvironmental records from drill cores from the Hetao Basin and their implications for the formation of the Hobq Desert and the Yellow River. Quat. Sci. Rev. 2017, 156, 69–89. [Google Scholar] [CrossRef]
- Wang, J.J.; Li, C.; Chen, J.Q.; Wang, J.J.; Jin, J.N.; Jiang, S.Y.; Yan, L.B.; Lin, H.D.; Zhao, J. Phylogeographic structure of the dwarf snakehead (Channa gachua) around Gulf of Tonkin: Historical biogeography and pronounced effects of sea-level changes. Ecol. Evol. 2021, 11, 12583–12595. [Google Scholar] [CrossRef] [PubMed]
- Loretan, G.; Rueda, E.C.; Cabrera, J.M.; Perez-Losada, M.; Collins, P.A.; Giri, F. Geographical isolation and restricted gene flow drive speciation of Aegla singularis (Decapoda: Anomura: Aeglidae) in southern South America. Biol. J. Linn. Soc. 2020, 129, 177–189. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, Z.T.; Duan, P.X.; Xiao, Y.S.; Zhang, H.K.; Huang, Z.X.; Zhou, R.C.; Wen, H.; Wang, K.X.; Wang, D. Whistle signal variations among three Indo-Pacific humpback dolphin populations in the South China Sea: A combined effect of the Qiongzhou Strait’s geographical barrier function and local ambient noise? Integr. Zool. 2021, 16, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.D.; Yu, H.T.; Bi, X.X.; Lai, Y.C.; Jiang, W.; Huang, L. Phylogeography of Chinese house mice (Mus musculus musculus/castaneus): Distribution, routes of colonization and geographic regions of hybridization. Mol. Ecol. 2014, 23, 4387–4405. [Google Scholar] [CrossRef]
- Pan, T.; Yan, P.; Yang, M.; Wang, H.; Ali, I.; Ayub, M.; Zhang, J.H.; Wang, J.J.; Li, E.; Xue, H.; et al. Genetic differentiation of regional populations of the widespread Asiatic toad (Bufo gargarizans), as revealed by development of novel microsatellite markers. Aust. J. Zool. 2018, 66, 335–342. [Google Scholar] [CrossRef]
- Wall, D.H.; Bradford, M.A.; St John, M.G.; Trofymow, J.A.; Behan-Pelletier, V.; Bignell, D.D.E.; Dangerfield, J.M.; Parton, W.J.; Rusek, J.; Voigt, W.; et al. Global decomposition experiment shows soil animal impacts on decomposition are climate-dependent. Glob. Chang. Biol. 2008, 14, 2661–2677. [Google Scholar] [CrossRef]
- Makkonen, M.; Berg, M.P.; Handa, I.T.; Hattenschwiler, S.; van Ruijven, J.; van Bodegom, P.M.; Aerts, R. Highly consistent effects of plant litter identity and functional traits on decomposition across a latitudinal gradient. Ecol. Lett. 2012, 15, 1033–1041. [Google Scholar] [CrossRef]
- Garcia-Palacios, P.; Maestre, F.T.; Kattge, J.; Wall, D.H. Climate and litter quality differently modulate the effects of soil fauna on litter decomposition across biomes. Ecol. Lett. 2013, 16, 1045–1053. [Google Scholar] [CrossRef]
- Bengtsson, J. Disturbance and resilience in soil animal communities. Eur. J. Soil Biol. 2002, 38, 119–125. [Google Scholar] [CrossRef]
- Caruso, T.; Taormina, M.; Migliorini, M. Relative role of deterministic and stochastic determinants of soil animal community: A spatially explicit analysis of oribatid mites. J. Anim. Ecol. 2012, 81, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ding, J.Y.; Lin, S.P.; Xu, R.F.; Liu, H.Y. Comparative mitogenomes of three species in Moenkhausia: Rare irregular gene rearrangement within Characidae. Int. J. Biol. Macromol. 2021, 183, 1079–1086. [Google Scholar] [CrossRef]



| Locality | N | COX2 Sequence | Cytb Sequence | Concatenated Sequence | 18S rRNA Sequence | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | H | Hd | pi | K | S | H | Hd | pi | K | S | H | Hd | pi | K | S | H | Hd | pi | K | ||
| South bank of the Yangtze river | |||||||||||||||||||||
| ZiJin Mountain (ZJ) | 25 | - | 1 | - | - | - | 1 | 2 | 0.153 | 0.00024 | 0.153 | 1 | 2 | 0.153 | 0.00013 | 0.153 | 1 | 2 | 0.080 | 0.00011 | 0.080 |
| Tang Mountain (TS) | 25 | 1 | 2 | 0.080 | 0.00015 | 0.080 | 2 | 3 | 0.157 | 0.00036 | 0.233 | 3 | 4 | 0.230 | 0.00027 | 0.313 | 1 | 2 | 0.153 | 0.00022 | 0.153 |
| QiXia Mountain (QX) | 25 | 4 | 4 | 0.230 | 0.00060 | 0.320 | 2 | 3 | 0.507 | 0.00086 | 0.560 | 6 | 6 | 0.627 | 0.00074 | 0.880 | - | 1 | - | - | - |
| Fang Mountain (FS) | 25 | 1 | 2 | 0.153 | 0.00029 | 0.153 | 3 | 4 | 0.360 | 0.00060 | 0.387 | 4 | 4 | 0.360 | 0.00046 | 0.540 | 1 | 2 | 0.380 | 0.00054 | 0.380 |
| Total of south | 100 | 6 | 6 | 0.117 | 0.00026 | 0.140 | 7 | 8 | 0.321 | 0.00059 | 0.380 | 13 | 12 | 0.371 | 0.00044 | 0.520 | 1 | 2 | 0.165 | 0.00024 | 0.165 |
| North bank of the Yangtze river | |||||||||||||||||||||
| Lao Mountain (LS) | 24 | 2 | 3 | 0.163 | 0.00031 | 0.167 | 3 | 3 | 0.236 | 0.00050 | 0.326 | 5 | 5 | 0.377 | 0.00042 | 0.493 | - | 1 | - | - | - |
| LongWang Mountain (LW) | 22 | - | 1 | - | - | - | 2 | 3 | 0.593 | 0.00126 | 0.818 | 2 | 3 | 0.593 | 0.00069 | 0.818 | - | 1 | - | - | - |
| Total of north | 46 | 2 | 3 | 0.086 | 0.00016 | 0.087 | 4 | 6 | 0.604 | 0.00112 | 0.724 | 6 | 7 | 0.651 | 0.00069 | 0.811 | - | 1 | - | - | - |
| Population in Tianjin | |||||||||||||||||||||
| Pan Mountain (PS) | 20 | 5 | 5 | 0.368 | 0.00094 | 0.500 | 5 | 6 | 0.447 | 0.00077 | 0.500 | 10 | 9 | 0.653 | 0.00085 | 1.000 | - | 1 | - | - | - |
| Total | 166 | 33 | 14 | 0.603 | 0.01933 | 10.324 | 45 | 19 | 0.717 | 0.02437 | 15.793 | 78 | 28 | 0.742 | 0.02210 | 26.117 | 1 | 2 | 0.498 | 0.00071 | 0.498 |
| Population | ZJ | QX | FS | TS | LS | LW | PS |
|---|---|---|---|---|---|---|---|
| ZJ | - | 1.04729 | 6.49946 | 8.74928 | 0.00159 | 0.00238 | 0.00274 |
| QX | 0.19271 | - | 1.75401 | 1.20943 | 0.00336 | 0.00414 | 0.00445 |
| FS | 0.03704 | 0.12475 | - | 8.00083 | 0.0025 | 0.00332 | 0.00366 |
| TS | 0.02778 | 0.17130 | 0.03030 | - | 0.00198 | 0.00277 | 0.00312 |
| LS | 0.99368 | 0.98673 | 0.98995 | 0.99214 | - | 0.51527 | 0.01865 |
| LW | 0.99057 | 0.98372 | 0.98689 | 0.98905 | 0.32668 | - | 0.02193 |
| PS | 0.98914 | 0.98251 | 0.98558 | 0.98768 | 0.93057 | 0.91935 | - |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Scenario I: the populations in the southern bank of the Yangtze River (Groups A) and the northern bank of the Yangtze River (Group B) of S. bungii. | ||||
| Among Groups | 1 | 1603.880 | 25.42563 Va | 98.74 |
| Among populations Within groups | 4 | 7.072 | 0.06143 Vb | 0.24 |
| Within populations | 141 | 36.898 | 0.26355 Vc | 1.02 |
| Total | 146 | 1647.849 | 25.75062 | |
| Scenario II: the populations in southern bank of the Yangtze River + the northern bank of the Yangtze River (Groups A + B) and Tianjin (Group C) of S. bungii. | ||||
| Among Groups | 1 | 497.325 | 6.40580 Va | 32.14 |
| Among populations Within groups | 5 | 1610.952 | 13.23417 Vb | 66.40 |
| Within populations | 160 | 46.398 | 0.29181 Vc | 1.46 |
| Total | 166 | 2154.675 | 19.93178 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, R.; Chen, J.; Pan, Y.; Wang, J.; Chen, L.; Ruan, H.; Wu, Y.; Xu, H.; Wang, G.; Liu, H. Genetic Diversity and Population Structure of Spirobolus bungii as Revealed by Mitochondrial DNA Sequences. Insects 2022, 13, 729. https://doi.org/10.3390/insects13080729
Xu R, Chen J, Pan Y, Wang J, Chen L, Ruan H, Wu Y, Xu H, Wang G, Liu H. Genetic Diversity and Population Structure of Spirobolus bungii as Revealed by Mitochondrial DNA Sequences. Insects. 2022; 13(8):729. https://doi.org/10.3390/insects13080729
Chicago/Turabian StyleXu, Runfeng, Jie Chen, Yu Pan, Jiachen Wang, Lu Chen, Honghua Ruan, Yongbo Wu, Hanmei Xu, Guobing Wang, and Hongyi Liu. 2022. "Genetic Diversity and Population Structure of Spirobolus bungii as Revealed by Mitochondrial DNA Sequences" Insects 13, no. 8: 729. https://doi.org/10.3390/insects13080729
APA StyleXu, R., Chen, J., Pan, Y., Wang, J., Chen, L., Ruan, H., Wu, Y., Xu, H., Wang, G., & Liu, H. (2022). Genetic Diversity and Population Structure of Spirobolus bungii as Revealed by Mitochondrial DNA Sequences. Insects, 13(8), 729. https://doi.org/10.3390/insects13080729






