Mitochondrial Phylogenomics Suggests Complex Evolutionary Pattern of Pronotal Foliaceous Mimicry in Hierodulinae (Mantodea: Mantidae), with Description of a New Species of Rhombodera Burmeister, 1838 from China †
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. DNA Extraction and Mitogenome Sequencing
2.3. Phylogenetic Analysis
3. Results
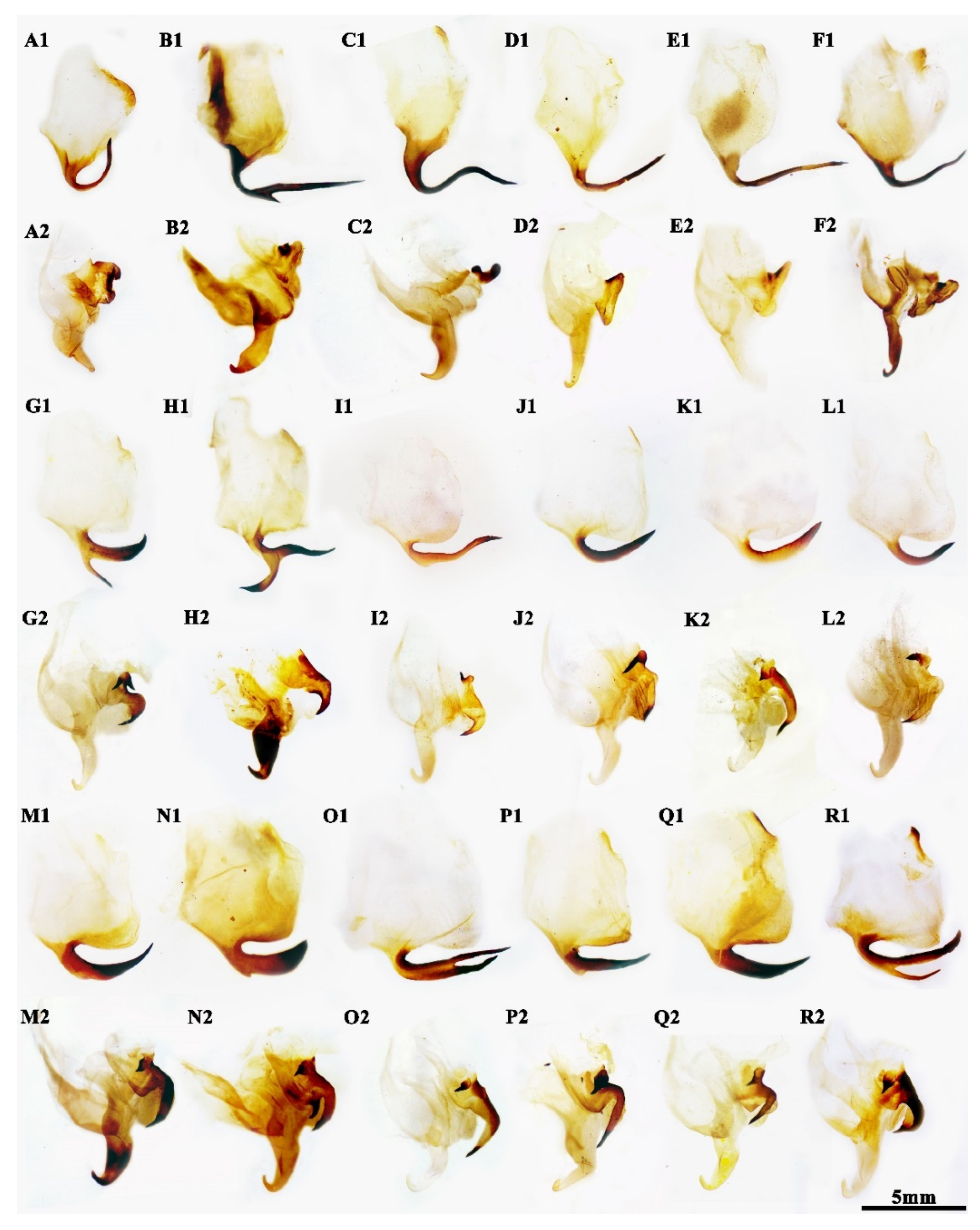
3.1. Taxonomy
3.2. Phylogenetic Analysis
3.3. General Features of Newly Sequenced Mitogenomes
4. Discussion
4.1. Taxonomic Implication for the Future Revision
4.2. Evolutionary Significance of the New Species
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Otte, D.; Spearman, L.; Stiewe, M.B.D. Mantodea Species File Online. Version 5.0/5.0. Available online: http://Mantodea.SpeciesFile.org (accessed on 25 June 2022).
- Burmeister, H.C. Handbuch der Entomologie. Fangschrecken, Mantodea. Handb. Ent. Burm. 1838, 2, 397–756. [Google Scholar] [CrossRef]
- Brunner de Wattenwyl, K. Révision du système des Orthoptères et description des espèces rapportées par M. Leonardo Fea de Birmanie. Annali Mus. Civ. Stor. Nat. Genova 1893, 13, 1–230. [Google Scholar] [CrossRef]
- Giglio-Tos, E. Mantidi esotici. V. Mantes, Tenoderae, Hierodulae et Rhomboderae. Bull. Soc. Ent. Ital. 1912, 43, 3–167. [Google Scholar]
- Giglio-Tos, E. Mantidi esotici. Generi e specie nuove. Bull. Soc. Ent. Ital. 1917, 48, 43–108. [Google Scholar]
- Giglio-Tos, E. Orthoptera Mantidae. In Das Tierreich, Eine Zusammenstellung und Kennzeichnung der Rezenten Tierformen; Schulze, F.E., Kükenthal, W., Eds.; Walter de Gruyter & Co.: Berlin, Germany, 1927; pp. 1–707. [Google Scholar] [CrossRef]
- Werner, F. Zweiter Beitrag zur Kenntnis der Mantodeen von Niederländisch-Indien. Treubia 1923, 3, 387–404. [Google Scholar]
- Beier, M. Mantodea, Fam. Mantidae, Subfam. Mantinae. In Genera Insectorum; Wytsman, P., Ed.; Verteneuil & Desmet: Brussels, Belgium, 1935; Volume 203, pp. 1–146. [Google Scholar]
- Beier, M. Neue und seltene Mantodeen aus deutschen Museen. Ann. Naturhist. Mus. Wien 1942, 52, 126–154. [Google Scholar]
- Schwarz, C.J.; Roy, R. The systematics of Mantodea revisited: An updated classification incorporating multiple data sources (Insecta: Dictyoptera). Ann. Soc. Entomol. Fr. 2019, 55, 101–196. [Google Scholar] [CrossRef]
- Schwarz, C.J.; Konopik, O. An annotated checklist of the praying mantises (Mantodea) of Borneo, including the results of the 2008 scientific expedition to Lanjak Entimau Wildlife Sanctuary, Sarawak. Zootaxa 2014, 3797, 130–168. [Google Scholar] [CrossRef]
- Liu, Q.P.; Liu, Z.J.; Wang, G.L.; Yin, Z.X. Taxonomic revision of the praying mantis subfamily Hierodulinae of China (Mantodea: Mantidae). Zootaxa 2021, 4951, 401–433. [Google Scholar] [CrossRef]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Gao, Y.R.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Novel tRNA gene arrangements in the mitochondrial genomes of praying mantises (Mantodea: Mantidae): Translocation, duplication and pseudogenization. Int. J. Biol. Macromol. 2021, 185, 403–411. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L.; Barker, S.C.; Whiting, M.F. Mitochondrial genomics and the new insect order Mantophasmatodea. Mol. Phylogenet. Evol. 2006, 38, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Ye, F. Comparative mitogenomic analyses of praying mantises (Dictyoptera, Mantodea): Origin and evolution of unusual intergenic gaps. Int. J. Mol. Sci. 2017, 13, 367–382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, L.P.; Yu, D.N.; Storey, K.B.; Cheng, H.Y.; Zhang, J.Y. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny with Mantodea. Int. J. Biol. Macromol. 2018, 111, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.P.; Cai, Y.Y.; Yu, D.N.; Storey, K.B.; Zhang, J.Y. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera Hauseri (Mantodea: Toxoderidae). PeerJ 2018, 6, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, Q.P.; Luo, L.; Yuan, Z.L. Characterization of the complete mitochondrial genome sequence of Asiadodis yunnanensis (Mantidae: Choeradodinae) and phylogenetic analysis. Mitochondr. DNA Part B 2019, 4, 2826–2827. [Google Scholar] [CrossRef]
- Shi, Y.; Li, L.Y.; Liu, Q.P.; Ali, M.Y.; Yuan, Z.L.; Smagghe, G.; Liu, T.X. Complete mitochondrial genomes of four species of praying mantises (Dictyoptera, Mantidae) with ribosomal second structure, evolutionary and phylogenetic analyses. PLoS ONE 2021, 16, e0254914. [Google Scholar] [CrossRef]
- Ehrmann, R.; Borer, M. Mantodea (Insecta) of Nepal: An Annotated Checklist. In Biodiversität & Naturausstattung im Himalaya; Hartmann, M., Weipert, J., Eds.; Verein der Freunde und Förderer des Naturkundemuseums Erfurt e.V: Erfurt, Germany, 2015; Volume 5, pp. 227–274. [Google Scholar]
- Brannoch, S.K.; Wieland, F.; Rivera, J.; Klass, K.D.; Béthoux, O.; Svenson, G.J. Manual of praying mantis morphology, nomenclature, and practices (Insecta, Mantodea). ZooKeys 2017, 696, 1–100. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Meng, G.L.; Li, Y.Y.; Yang, C.T.; Liu, S.L. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Drummond, Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast-online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Shi, Y.; Yuan, Z.L. Characterization of the complete mitochondrial genome of the praying mantis Mekongomantis quinquespinosa (Mantodea, Mantidae). Mitochondr. DNA Part B 2019, 4, 3280–3281. [Google Scholar] [CrossRef]
- Wang, W.J.; Wang, H.; Huang, H.M.; Zhao, Y.Z.; Zhou, Z.J. Mitochondrial genomes of 10 Mantidae species and their phylogenomic implications. Arch. Insect Biochem. Physiol. 2022, e21874. [Google Scholar] [CrossRef]
- Vermeersch, X.H.C.; Unnahachote, T. Hierodula confusa sp. nov., a new species of Hierodula Burmeister, 1838 (Mantodea: Mantidae: Hierodulinae: Hierodulini). Belg. J. Entomol. 2020, 103, 1–13. [Google Scholar]
- Liu, Q.P.; Liu, Z.J.; Chen, Z.T.; Yuan, Z.L.; Shi, Y. A new species and two new species records of Hierodulinae from China, with a revision of Hierodula chinensis (Mantodea: Mantidae). Orient. Insects 2020, 55, 1–20. [Google Scholar] [CrossRef]
- Natural History Museum. Specimens (from Collection Specimens) [Data Set Resource]. Natural History Museum. Available online: https://data.nhm.ac.uk/dataset/collection-specimens/resource/05ff2255-c38a-40c9-b657-4ccb55ab2feb (accessed on 25 July 2022).
- Zhang, G.Z. A new species of the genus Hierodula (Mantodea: Mantidae) from China. Entomotaxonomia 1990, 12, 113–114. [Google Scholar]
- Wang, T.Q.; Dong, D.Z. Three new species of mantids from Yunnan province, China (Mantodea). Zool. Res. 1993, 12, 203–207. [Google Scholar]





| Measurements | Holotype ♂ | Paratype1 ♀ | Paratype2 ♀ | Paratype3 ♂ | Paratype4 ♂ |
|---|---|---|---|---|---|
| Total length | 80.16 | 82.88 | — | r81.00 | 84.42 |
| Body length | 61.86 | 73.60 | 74.72 | 61.76 | 71.36 |
| Head width | 9.72 | 11.23 | 11.53 | 9,52 | 9.68 |
| Head height | 7.36 | 9.90 | 9.80 | 7.76 | 7.56 |
| Pronotum length | 19.94 | 24.58 | 25.50 | 20.50 | 21.36 |
| Pronotum width | 14.00 | 18.10 | 19.37 | 13.66 | 15.28 |
| Pronotum narrow width | 4.42 | 4.66 | 4.98 | 4.80 | 4.66 |
| Prozone length | 6.22 | 7.19 | 7.70 | 6.30 | 6.30 |
| Metazone length | 13.72 | 17.54 | 17.80 | 14.20 | 15.06 |
| Forewing length | 60.34 | 55.50 | 53.88 | 59.38 | 62.38 |
| Hindwing length | 54.58 | 49.14 | 51.28 | 54.50 | 56.54 |
| Forecoxa length | 13.92 | 18.34 | 17.39 | 13.66 | 14.80 |
| Forefemur length | 15.10 | 19.12 | 19.99 | 15.32 | 16.38 |
| Foretibia length | 10.58 | 11.78 | 12.18 | 10.76 | 11.52 |
| Foretarsus length | 10.34 | 12.00 | 11.90 | 10.60 | 12.48 |
| Mesofemur length | 14.22 | 16.54 | 15.64 | 14.46 | 15.18 |
| Mesotibia length | 11.00 | 14.42 | 13.60 | 11.14 | 12.14 |
| Mesotarsus length | 8.18 | 8.78 | 7.30 | 8.50 | 8.68 |
| Metafemur length | 15.32 | 19.56 | 17.98 | 16.08 | 17.76 |
| Metatibia length | 16.12 | 20.72 | 17.74 | 16.36 | 18.82 |
| Metatarsus length | 11.16 | 11.26 | 10.90 | 11.18 | 11.82 |
| Species | A + T (%) | AT-Skew | GC-Skew | Total Length |
|---|---|---|---|---|
| Hierodula chinensis (Sichuan) | 75.80 | 0.06 | −0.21 | 16,145 |
| Hierodula confusa | 75.00 | 0.05 | −0.23 | 16,103 |
| Rhombodera longa | 75.90 | 0.06 | −0.21 | 16,093 |
| Hierodula membranacea | 74.87 | 0.05 | −0.23 | 15,824 |
| Hierodula jianfenglingensis | 75.32 | 0.05 | −0.20 | 16,234 |
| Hierodula latipennis | 75.91 | 0.05 | −0.20 | 15,391 |
| Rhombodera zhangi | 75.58 | 0.06 | −0.22 | 15,396 |
| Rhombodera valida (Java) | 74.24 | 0.05 | −0.19 | 17,298 |
| Rhombodera kirbyi | 76.20 | 0.05 | −0.20 | 16,036 |
| Rhombodera latipronotum (Yunnan) | 75.10 | 0.06 | −0.22 | 16,092 |
| Rhombodera latipronotum (Guangxi) | 75.05 | 0.06 | −0.23 | 16,168 |
| Rhombomantis longipennis | 75.70 | 0.04 | −0.21 | 16,056 |
| Rhombodera megaera | 75.33 | 0.06 | −0.20 | 15,875 |
| Rhombodera hyalina | 75.69 | 0.05 | −0.22 | 16,043 |
| Rhombodera stalii | 75.33 | 0.06 | −0.22 | 16,314 |
| Rhombodera valida (Yunnan) | 75.18 | 0.06 | −0.24 | 16,055 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.-P.; Liang, L.; Zhang, X.-Y.; Li, H.-K.; Zhao, C.-X.; Liu, X.-Y. Mitochondrial Phylogenomics Suggests Complex Evolutionary Pattern of Pronotal Foliaceous Mimicry in Hierodulinae (Mantodea: Mantidae), with Description of a New Species of Rhombodera Burmeister, 1838 from China. Insects 2022, 13, 715. https://doi.org/10.3390/insects13080715
Liu Q-P, Liang L, Zhang X-Y, Li H-K, Zhao C-X, Liu X-Y. Mitochondrial Phylogenomics Suggests Complex Evolutionary Pattern of Pronotal Foliaceous Mimicry in Hierodulinae (Mantodea: Mantidae), with Description of a New Species of Rhombodera Burmeister, 1838 from China. Insects. 2022; 13(8):715. https://doi.org/10.3390/insects13080715
Chicago/Turabian StyleLiu, Qin-Peng, Le Liang, Xin-Yang Zhang, Hao-Kun Li, Chu-Xiang Zhao, and Xing-Yue Liu. 2022. "Mitochondrial Phylogenomics Suggests Complex Evolutionary Pattern of Pronotal Foliaceous Mimicry in Hierodulinae (Mantodea: Mantidae), with Description of a New Species of Rhombodera Burmeister, 1838 from China" Insects 13, no. 8: 715. https://doi.org/10.3390/insects13080715
APA StyleLiu, Q.-P., Liang, L., Zhang, X.-Y., Li, H.-K., Zhao, C.-X., & Liu, X.-Y. (2022). Mitochondrial Phylogenomics Suggests Complex Evolutionary Pattern of Pronotal Foliaceous Mimicry in Hierodulinae (Mantodea: Mantidae), with Description of a New Species of Rhombodera Burmeister, 1838 from China. Insects, 13(8), 715. https://doi.org/10.3390/insects13080715







