Simple Summary
The honey bee (Apis mellifera) is kept all over the world and plays a dominant role in the pollination of crops. Honey bees may be exposed to different levels of cadmium through the collection of contaminated nectar during their foraging activities. In this study, honey bees were chronically exposed to cadmium to investigate the effects of sublethal cadmium doses on the olfactory learning and brain gene expression profiles in honey bees. Honey bees exhibited significantly impaired olfactory learning performances after being chronically exposed to cadmium and had a significantly lower head weight in comparison with control bees. Furthermore, genes involved in oxidative stress response and odor sensing were dysregulated in the brain of cadmium-treated bees. These results suggest that cadmium exposure exerted oxidative stress and decreased gene expression levels of chemoreceptors in honey bees, which probably resulted in the impaired olfactory learning of honey bees.
Abstract
The honey bee (Apis mellifera) plays vital ecological roles in the pollination of crops and the maintenance of ecological balance, and adult honey bees may be exposed to exogenous chemicals including heavy metals during their foraging activities. Cadmium (Cd) is regarded as a nonessential toxic metal and is readily accumulated in plants; honey bees can therefore acquire Cd through the collection of contaminated nectar. In the present study, honey bees were chronically exposed to Cd to investigate the effects of sublethal cadmium doses on the olfactory learning and brain gene expression profiles of honey bees. The results showed that Cd-treated bees exhibited significantly impaired olfactory learning performances in comparison with control bees. Moreover, the head weight was significantly lower in Cd-treated bees than in control bees after chronic exposure to Cd. Gene expression profiles between the Cd treatment and the control revealed that 79 genes were significantly differentially expressed. Genes encoding chemoreceptors and olfactory proteins were downregulated, whereas genes involved in response to oxidative stress were upregulated in Cd-treated bees. The results suggest that Cd exposure exerts oxidative stress in the brain of honey bees, and the dysregulated expression of genes encoding chemoreceptors, olfactory proteins, and cytochrome P450 enzymes is probably associated with impaired olfactory learning in honey bees.
1. Introduction
Environmental pollution by heavy metals is becoming an increasingly common problem around the world [1,2], and great attention has been paid to the adverse health risks of heavy metal exposure on humans and other organisms [3]. Cadmium (Cd) ranked seventh in toxicity among heavy metals according to the ATSDR [3]. The transfer of Cd from soil to agricultural plants occurs easily [4,5]; therefore, Cd is readily accumulated in plants and can enter the food chain straightforwardly, posing a risk to human and animal health [4,6].
Cd is regarded as a nonessential toxic metal, and it exists in soil, sedimentary rocks, and the lithosphere in the concentration range of 0.2–0.53 mg/kg [7]. Cd concentrations in agricultural soils rarely exceed 10 μg/L; however, anthropic activities, including mining, vehicle exhausts, the excessive use of phosphate fertilizer, and waste disposal have led to increased quantities of Cd released into the soil environment [7,8]. Elevated levels of Cd in soils and its transfer to the human body via the food chain cause health injury including skeletal damage and neurotoxic effects [9,10]. In addition, experimental evidence suggests that mice exhibited impaired cognitive abilities after chronic exposure to Cd at a concentration of 3 mg/L [11]. A higher exposure concentration of Cd contributed to growth limitation and fruit abortion in plants [12].
The honey bee (Apis mellifera) is kept all over the world and plays vital ecological roles in the pollination of crops and the maintenance of ecological balance [13]. Honey bees are a good model for assessing the ecological and health risks of toxic heavy metals on biota [14]. Honey bees are social insects, and worker bees of different ages perform different tasks. Young worker bees perform in-hive tasks, and old worker bees perform out-hive tasks, such as foraging [15]. Honey bee foragers may encounter various environmental factors including pesticides and heavy metals during their foraging activities [14]. Mounting evidence has suggested that forager bees exposed to exogenous chemicals exhibited behavioral and cognitive impairments that may lead to homing failure in forager bees and colony losses [16,17]. The olfactory proboscis extension response (PER) is a behavioral paradigm for evaluating the cognitive ability of honey bees under different conditions [18]. The olfactory learning behavior is closely related to the age of bees and has been shown to be impaired by biotic and abiotic factors [19,20,21].
Previous studies reported that the concentration of Cd in fruits from three different orchards exceeded the tolerance limit for Cd in fruits in Guangzhou, South China [5]. Honey bees can therefore acquire heavy metals through the collection of contaminated nectar, and heavy metal lead accumulated in the nectar of sunflowers decreased bee visit durations [22]. The intake of sucrose solution was decreased in foragers following joint exposure to Cd and copper (Cu), and joint Cd and Cu exposure delayed the development of honey bee larvae [23]. The olfactory learning of honey bees is an essential cognitive function and is required for effective foraging that plays vital roles in plant–honey bee interactions [24]. So far, most studies have found that honey bees are prone to environmental stressors including pesticides and heavy metals that affect the physiology and development of honey bees [23,25,26]. Thus, the honey bee has been regarded as a bioindicator of a polluted environment [27]. However, little is known about the effects of Cd exposure on olfactory learning in honey bees. We, therefore, investigated whether honey bees exposed to chronic Cd exhibited altered olfactory learning, and we further compared the patterns of brain gene expression between cadmium-exposed bees and control bees using RNA-seq analysis. Our findings provide the first evidence that the olfactory learning performances of honey bees can be impaired by chronic cadmium exposure. Furthermore, the dysregulated expression of genes encoding chemoreceptors, olfactory proteins, and cytochrome P450 enzymes was probably associated with Cd-impaired olfactory learning.
2. Materials and Methods
2.1. Honey Bees
Mature brood combs were removed from three different hives and placed in an incubator (34.5 °C, 70% RH). The newly emerged bees were collected and marked on the thorax using water-based paints within 24 h, and the paint-marked bees were then returned to the colony. The paint-marked bees were recovered from the colony after 5 days. A total of 30 same-aged bees were reared in each cage, which we have used previously [28], and eight cages of bees were prepared. The caged bees were placed in an incubator (30 °C, 70% RH) and subjected to Cd exposure.
2.2. Cadmium Exposure
A cadmium chloride (CdCl2) stock solution (25 μg/mL) was prepared by dissolving CdCl2 in sterilized water. The stock solution was diluted 100-fold with 30% sucrose solution (w/v). The cages of bees were randomly divided into two groups with each group consisting of four cages of bees. The bees in the treatment group were fed with 30% sucrose solution containing 0.25 μg/mL CdCl2, and the bees in the control group were fed with 30% sucrose solution. For each cage of honey bees, a 5 mL plastic syringe with the tip cut off was used as the feeder, and the amount of total sugar consumption was recorded directly from the syringe measure. All of the caged bees were fed ad libitum. Sugar consumption and the mortality of three cages of bees of each group were recorded during 18 days of Cd exposure. The sugar consumption per bee per day was calculated by dividing the amount of total sugar consumption by the number of remaining live bees each day. The survival rates were analyzed according to the number of remaining bees from each of the three control cages and the three treatment cages on the last day of the observation.
2.3. Olfactory Learning Experiments
Honey bees from each group were individually collected from the cage after 18 days of Cd exposure, and each bee was placed in a glass vial on ice to anesthetize the bees. After cold anesthesia, each immobilized bee was individually restrained in a customized tube with duct tape, leaving the head fixed and the proboscis moving freely. The harnessed bees were kept in an incubator (30 °C, 70% RH) for 2 h to recover from cold anesthesia before the training test of olfactory learning. The antennae of each bee were touched with a 50% sucrose solution (w/v), and the honey bees not showing proboscis extension were excluded from further behavior analysis. The training test of olfactory learning was adapted from Bitterman et al. [28], and odors A and B (A: 1-nonanol, B: hexanol) were presented in the experiment. Odor A (conditioned stimulus, CS +) was paired with a sugar-water reward (unconditioned stimulus, US), and odor B was paired with no reward (CS −). Each bee was exposed to the two odors according to the pseudorandom order (ABBAAB). One odor was used in each training test, and the test interval was 10 min.
Firstly, each harnessed bee was positioned in front of an olfactometer (kindly provided by M. Giurfa, University of Toulouse, France) and exposed to clean air for 15 s [29]. Odor A was then delivered for 4 s, and the bee was allowed to consume 50% sucrose solution (w/v) for 2 s after 2 s of odor A delivery. Finally, the bee was exposed to clean air for 20 s. An intact olfactory training test lasted 39 s. Odor B was delivered to the bee using the same training protocol, but without pairing with sucrose solution. The number of bees showing olfactory learning to odor A was recorded. The head weight of the honey bees from each group was measured after the training test.
2.4. RNA Isolation and RNA-Seq Library Preparation
A total of thirty honey bees were used for RNA isolation in each group, and the brain of each honey bee was dissected. Brains from ten honey bees were pooled into a single sample, and three pooled brain samples were used for RNA extraction in each group. The total RNA of each pooled brain sample was extracted with Trizol reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. The RNA quality was evaluated with both Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and agarose gel assay. Oligo(dT) beads were used to enrich mRNA from total RNA. The mRNA was fragmented by fragmentation buffer, followed by cDNA synthesis with random primers. DNA polymerase I, RNase H, dNTP, and buffer were used to synthesize the second strand of cDNA. After purification, the cDNA ends were repaired. The poly (A) was added to the cDNA before adapter ligation. The adapter-ligated cDNAs were size selected with agarose gel assay prior to PCR amplification. The cDNAs were sequenced using an Illumina HiSeq 2500 platform by Gene Denovo Biotechnology Co. (Guangzhou, China).
2.5. RNA Sequencing and Analysis
The raw reads were filtered by fastp (version 0.18.0) to remove reads containing adapters and low-quality reads [30]. The raw reads were further processed by Bowtie2 version 2.2.8 to remove rRNA-mapped reads [31]. The remaining paired-end clean reads were mapped to the Apis mellifera L. reference genome (Amel_HAv3.1) using HISAT2. 2.4 [32]. The mapped clean reads were assembled by StringTie v1.3.1, and the expression abundance of each transcript was calculated using the FPKM (fragments per kilobase of transcript per million fragments mapped) method. Analysis of the differentially expressed genes (DEGs) between the two groups of bees was performed by DESeq2 [33]. The Gene Ontology (GO) database was used to determine gene functional classification, and all DEGs were mapped to GO terms in the GO database [34]. Pathway enrichment analyses of DEGs were carried out based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [35].
2.6. Quantitative Real-Time PCR
Nine differentially expressed genes were selected at random to validate the reliability of the RNA-seq results using quantitative real-time PCR. The brains of the honey bees of each group were dissected, and the brains of three honey bees were pooled into a single sample. Seven pooled brain samples were used for RNA extraction in each group. The total RNA was extracted from the brain samples of two groups of bees using the same method mentioned above. The first cDNA strand was synthesized from 1 mg of total RNA using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The qPCR reaction mixture (10 µL) contained 5 µL 2× SYBR Premix Ex Taq II (Tli RNaseH Plus), 0.2 µL of each PCR primer (10 µM) (Table S1) [36], 1 µL of diluted cDNA (1:3 dilution), and 3.6 µL of RNase free water. All reactions were run in triplicate in a Bio-Rad CFX 384 Real-time system. The reaction conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 15 s at 95 °C and 30 s at 55 °C, followed by melting curve analysis. The relative expression levels of each target gene were analyzed using the 2−∆∆CT method [37].
2.7. Statistics
The unpaired Student’s t-test was used to determine the statistical significance in survival rates, sugar consumption, head weight, and the relative gene expression level of nine target genes between control and cadmium-exposed bees. Statistical significance in olfactory learning performances was determined by Fisher’s exact test. For RNA-Seq data analysis, the transcripts with p-value < 0.05 and absolute fold change > 2 were considered differentially expressed transcripts between the two groups of bees. Significantly enriched GO terms and KEGG pathways related to DEGs were determined by a hypergeometric test.
3. Results
3.1. Syrup Consumption and Survival Analysis
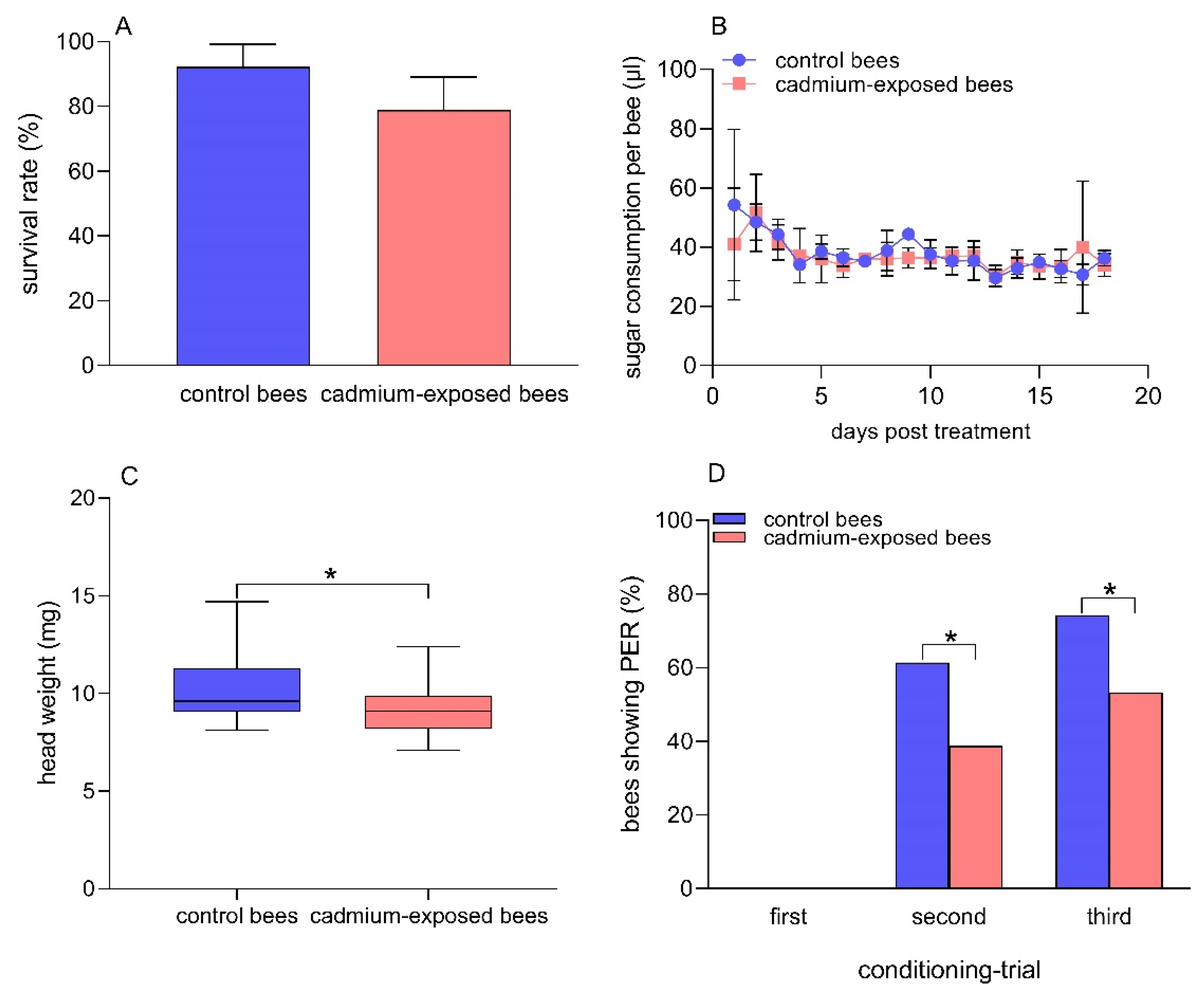
After 18 days of exposure to Cd, the mean survival rates of the control bees and the cadmium-exposed bees were 92.2% and 78.9%, respectively (Figure 1A). There was no significant difference in the mean survival rate between the two groups of bees (t = 1.9, df = 4, p = 0.13). The average sugar consumption per bee per day was 37.8 ± 6.3 µL and 37.0 ± 4.6 µL in the control bees and the cadmium-exposed bees for the 18 days of exposure to Cd, respectively (Figure 1B). There was no significant difference in the sugar consumption per bee between the two groups of bees (t= −0.42, df = 34, p = 0.18). The data suggested that 30% sucrose solution containing 0.25 μg/mL CdCl2 showed no significant repellent effects on honey bees, and the dose of CdCl2 used in our studies showed no acute toxic effects on the survival of honey bees.
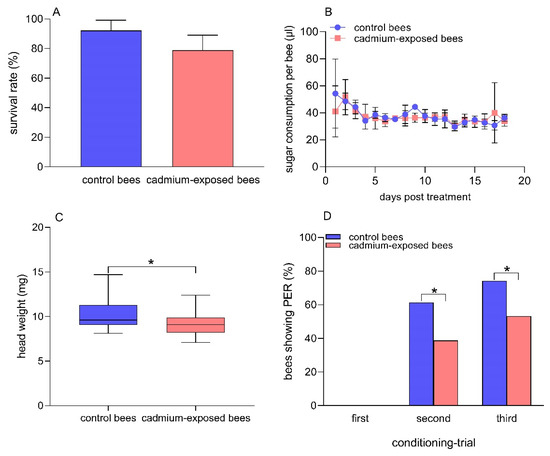
Figure 1.
Comparison between cadmium-exposed and control bees in survival rate (A) sugar consummation per bee; (B) head weight; (C) Student’s t-test, n = 21 in both groups of bees, and the percentage of bees showing PER when presented with the CS +, 1-nonanol; (D) Fisher’s exact test, n = 70 for control bees, n = 62 for cadmium-exposed bees after chronic Cd exposure. (*) p < 0.05.
3.2. Head Weight and Olfactory Learning Performances
The average head weight of the control bees and the cadmium-exposed bees was 10.4 mg and 9.4 mg, respectively (Figure 1C). The head weight was significantly lower in the cadmium-exposed bees than in the control bees after chronic exposure to Cd (t = 2.2, df = 40, p = 0.037). We observed that there was no PER triggered by the odor 1-nonanol (odor A, CS +) in either group of bees in the first learning test. The percentage of honey bees showing PER to 1-nonanol was 61.4% and 38.9% in the control bees and the cadmium-exposed bees in the second learning test, respectively. In addition, the percentage of honey bees showing PER to 1-nonanol was 74.3% and 53.2% in the control bees and the cadmium-exposed bees in the third learning test, respectively. The control bees showed significantly better olfactory learning abilities than chronic Cd-exposed bees in the second (Fisher’s exact test, p = 0.014 ) and third (Fisher’s exact test, p = 0.018) learning tests (Figure 1D). Moreover, we found that the control bees did not show any response to the odor hexanol (odor B, CS −) during the three conditioning trials. The percentage of cadmium-exposed bees showing PER to hexanol was 3.2% and 1.6% in the first and second conditioning trial, respectively, and the cadmium-exposed bees showed no response to hexanol in the third conditioning trial (Figure S1). No significant differences were found between the two groups regarding the response to hexanol (Fisher’s exact test, first conditioning trial: p = 0.22; second conditioning trial: p = 0.47), suggesting the stability and specificity of the behavioral response in the two groups of bees.
3.3. Analysis of Brain Gene Expression Profiles
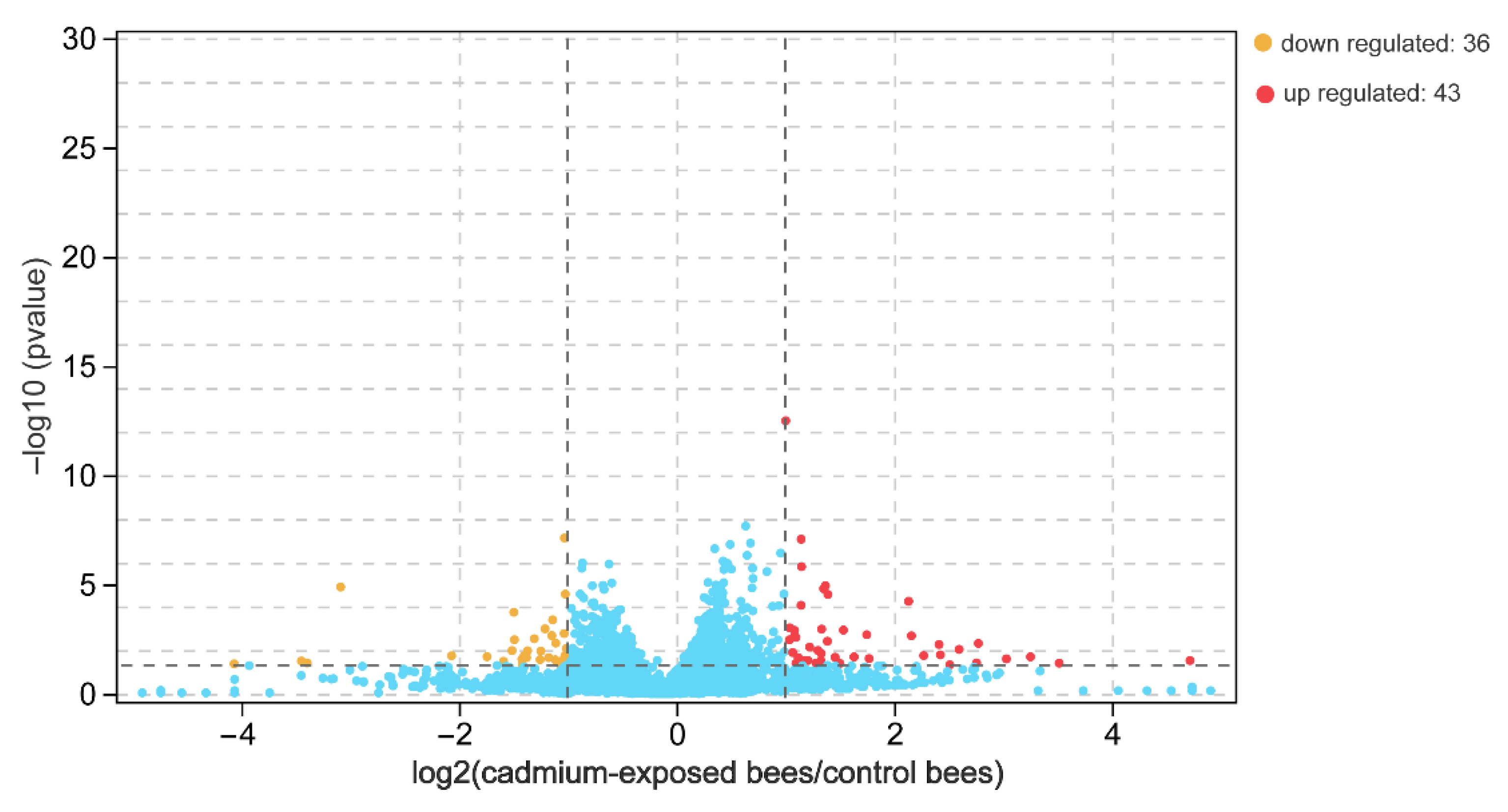
The number of paired-end clean reads mapped to the Apis mellifera L. reference genome was about 14.8 and 18.8 million for the sequencing libraries of the cadmium-exposed and control bees, respectively. The whole sequence reads have been deposited in the Genome Sequence Archive (GSA accession number CRA006897). We obtained a total of 10,488 genes, 596 of which were novel genes after mapping to the reference genome. A total of 79 significantly differentially expressed genes were obtained in the brain between the cadmium-exposed and the control bees based on the p-value and fold-change criteria (Table S2). Of these, 43 and 36 were upregulated and downregulated in the brains of the cadmium-exposed bees, respectively (Figure 2). Cd exposure downregulated genes encoding chemoreceptors and olfactory proteins that include odorant binding protein 14 (LOC677673) and odorant receptor Or1 (LOC100576462). The expression levels of the genes encoding the two proteins exhibited about an 8–10 fold decrease in the brain of the cadmium-exposed bees compared with that of the control bees. In addition, Cd exposure upregulated genes engaged in the response to oxidative stress, and these upregulated genes include cytochrome b5 (LOC726850), cytochrome P450 6a14 (LOC550965), tyrosine aminotransferase (LOC725204), and trehalase (LOC410484). The expression levels of the genes encoding these proteins exhibited about a 3–26 fold increase in the brains of the cadmium-exposed bees compared with that of the control bees.
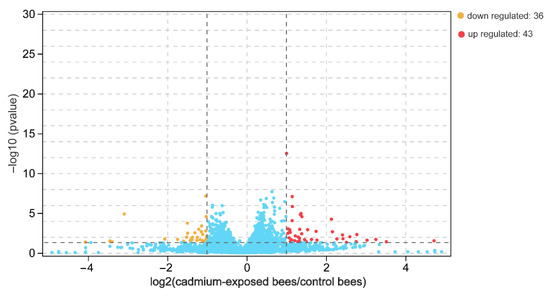
Figure 2.
Differentially expressed genes in the brain between cadmium-exposed bees and control bees based on a p-value < 0.05 and an absolute fold-change > 2. Red and yellow indicate the up- and downregulated genes in cadmium-exposed bees, respectively; blue indicates no differentially expressed genes.
3.4. GO and KEGG Enrichment Analysis
We listed the top ten significantly enriched GO terms associated with 13 unique differentially expressed genes (Table 1), and 2 and 11 genes were downregulated and upregulated in the brains of the cadmium-exposed bees, respectively. These significantly enriched GO terms include monooxygenase activity (GO:0004497), heme binding (GO:0020037), iron ion binding (GO:0005506), etc. All four genes (LOC100577883, LOC413908, LOC550965, and LOC551179), encoding different forms of cytochrome P450 enzymes involved in the monooxygenase activity, were significantly upregulated in the Cd-exposed bees compared to the control bees. Nine significant KEGG pathways associated with the 16 unique differentially expressed genes were identified in the cadmium-exposed bees, and 7 and 9 genes were downregulated and upregulated in the brains of the cadmium-exposed bees, respectively (Table 2). The pathways mainly include starch and sucrose metabolism, insect hormone biosynthesis, lysine biosynthesis, etc.

Table 1.
Enrichment analysis of top ten GO terms related to upregulated and downregulated genes in the brains of cadmium-exposed bees.

Table 2.
Significantly enriched KEGG pathways related to upregulated and downregulated genes in the brains of cadmium-exposed bees.
3.5. Validation of DEGs by qPCR
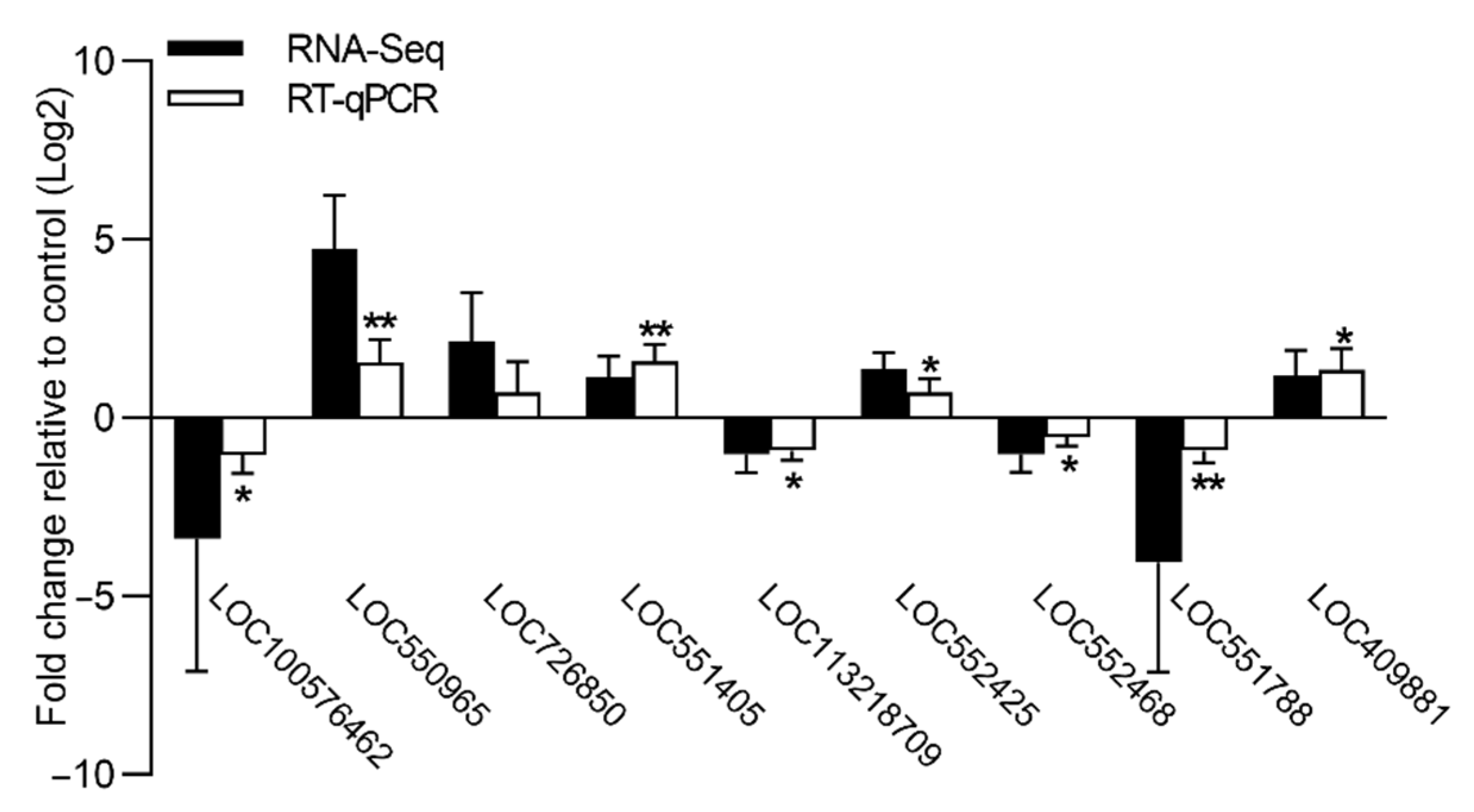
The relative expression levels of nine DEGs were further validated by qPCR (Figure 3). Consistent with the FPKM expression value generated by RNA-seq, the qPCR results revealed that all nine genes exhibited similar gene expression profiles in the brains of the cadmium-exposed bees relative to the controls. The genes encoding odorant receptor 1 (LOC100576462), inactive peptidyl-prolyl cis-trans isomerase shutdown (LOC113218709), MATH and LRR domain-containing protein PFE0570w-like (LOC552468), and katanin p60 ATPase-containing subunit A-like 2 (LOC551788) showed a significantly lower level of expression in the brains of the cadmium-exposed bees relative to the controls. The genes encoding cytochrome P450 6a14 (LOC550965), 4-nitrophenylphosphatase (LOC551405), glucose dehydrogenase (LOC552425), and myosin regulatory light chain 2 (LOC409881) showed a significantly higher level of expression in the brains of the cadmium-exposed bees relative to the controls. Therefore, the validation results of the nine DEGs confirmed the reliability of the transcriptome data.
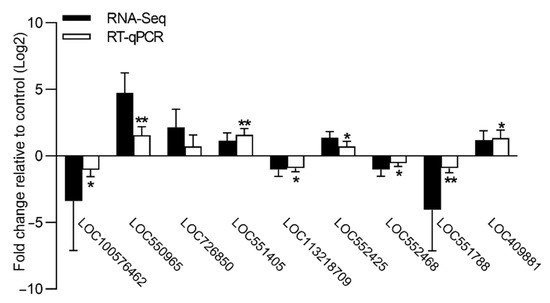
Figure 3.
Validation of FPKM expression value (RNA-seq) for nine genes by RT-qPCR. The abscissa indicates the nine DEGs, and the ordinate indicates the relative expression levels of nine DEGs in the brains of the cadmium-exposed bees relative to the controls. The error bar indicates SD; the asterisk denotes significant differences (Student’s t-test, * p < 0.05; ** p < 0.01).
4. Discussion
Burden et al. [38] measured the sucrose sensitivity of honey bees to five different doses of CdCl2 ranging from 0.001 μg/mL to 10 μg/mL and found that honey bees did not show any significant rejection response to sugar-water contaminated with CdCl2. However, the honey bees exhibited a reduction in consuming the sucrose containing not less than 1 ug/mL of CdCl2; although non-significant, there exists the possibility that sucrose containing more than 1 ug/mL of Cd may affect honey bee sucrose responsiveness that is essential for olfactory learning in honey bees [38,39]. Besides, honey bees fed diets containing 0.24 ug/mL of CdCl2 for up to 7 days had an alteration in the composition of the microbiome [40]. Therefore, a cadmium exposure dose of 0.25 ug/mL was used in our studies. Our data showed that there were no significant differences in both syrup consumption and survival rate between the cadmium-exposed and control bees, thereby further confirming that the Cd dose used in our studies is sublethal and exerts chronic toxic effects on honey bees. Di et al. [41] studied the acute toxic effects of CdCl2 on honey bees and found that honey bees fed Cd exhibited a significant reduction in syrup consumption compared to controls. The lowest Cd dose (26 mg/L) used in their studies was about 100-fold higher than the dose (0.25 μg/mL) used in our studies. A higher dose of Cd may cause malaise in honey bees, thus affecting the syrup consumption of honey bees [38]. In addition, it has been known that Cd exposure induces developmental defects including head deformities and causes a significant decrease in the total protein content in a broad range of vertebrate species [42,43,44,45]. Our studies showed that the head weight was significantly lower in cadmium-exposed bees when compared to controls, suggesting chronic Cd exposure may have adverse effects on the total protein content of the mandibular and hypopharyngeal glands in the heads of adult honey bees, thus resulting in a decrease in head weight in Cd-exposed bees. Previous studies reported that head size was significantly reduced in chronic lead-exposed honey bees that showed impaired olfactory learning performances [46]. Heavy metals including lead and Cd, therefore, have similar adverse effects on the cognitive abilities of honey bees. Odorant binding proteins (OBPs) play important roles in odor sensing in insects [47], and the interaction between OBPs and imidacloprid decreased the binding affinity of OBPs to a floral odor [48]. In addition, significantly downregulated expression levels of genes encoding OBPs were found in imidacloprid-treated bees exhibiting impaired olfactory learning [21]. A significant 8.5-fold decrease in the expression level of the gene encoding the odorant-binding protein 14 (Obp 14) was observed in the brains of cadmium-exposed bees, suggesting that the ability of odor recognition in cadmium-exposed bees may be weakened. It remains to be determined whether cadmium may exhibit a similar binding mode to the OBPs as neonicotinoid imidacloprid in the brains of honey bees. In addition, the expression level of odorant receptor 1 (Or1) was downregulated more than 10-fold in the brains of the cadmium-exposed bees in comparison to the control bees. The odorant receptor was regarded as the primary receptor involved in the sensory detection of external chemical stimuli in insects [49]. Fibers originating from the Or1-expressing neurons in the pore plate sensilla of honey bees may enter the brain and innervate the glomeruli of the antennal lobe of the honey bee brain [50,51]. The dysregulation of Or1 expression in the nerve fibers may have neurologically adverse effects on the transmission of odor information in the brains of honey bees; therefore, the decreased expression levels of both Obp 14 and Or1 probably resulted in the impaired olfactory learning of honey bees. However, further studies are required to elucidate the exact roles of Obp 14 and Or1 in the process of olfactory learning in honey bees. In addition, trehalase plays vital biological roles in recovery after abiotic stress through hydrolyzing trehalose in insects [52]. Cadmium exposure significantly increased the expression level of the gene encoding trehalase in the brain of honey bees, indicating that honey bees may need extra energy to combat cadmium stress. Diacylglycerol kinases (DGKs) play important roles in neuron physiology and behavior regulation, and the gene Dgks are highly expressed in the brain of mammals [53,54]. The knockout of certain Dgk isoforms resulted in substantially impaired cognitive functions in mice [54,55]. The expression level of the gene encoding the η isoform of DGK was significantly decreased in the brains of the cadmium-exposed bees. Like in mammals, the downregulation of Dgk may possibly affect the olfactory learning of honey bees.
Previous studies have demonstrated that cadmium exposure exerts oxidative stress in several tissues and causes neurotoxicity in organisms [10,56], and the brain is highly prone to oxidative damage [57]. Tyrosine aminotransferase (TATN) plays critical roles in oxidative stress response in organisms [58]. The gene encoding TATN was significantly upregulated in the brain of cadmium-exposed bees, suggesting that the upregulation of this gene may provide a protective function against oxidative stress induced by cadmium exposure. Furthermore, it is noteworthy that oxidative stress-related genes were all significantly upregulated in the brains of the cadmium-exposed bees, and these genes include cytochrome b5 (LOC726850), cytochrome P450 4aa1-like (LOC100577883), cytochrome P450 6A1 (LOC413908), cytochrome P450 6a14 (LOC550965) and cytochrome P450 15A1 (LOC551179). Cytochrome b5 (Cyt-b5) has been identified as a neuroprotective factor targeting oxidative damage in Drosophila melanogaster [59]. The expression level of Cyt-b5 was significantly upregulated in the brains of the cadmium-exposed bees in comparison to the control bees, and the upregulation of Cyt-b5 may play neuroprotective roles against oxidative stress induced by cadmium. Significantly increased levels of genes encoding cytochrome P450 (CYP) enzymes were observed in the brains of the cadmium-exposed bees, suggesting that the brain CYPs may play key roles in metabolizing the reactive oxygen species induced by Cd and influencing behavior and cognition in honey bees [60]. Further research is needed to investigate the interplay between the levels of brain CYPs and olfactory learning performances in honey bees. In addition, the starch and sucrose metabolism was shown to be involved in alleviating the toxic effects of Cd on poplars [61]. Two genes associated with the pathway were significantly upregulated in the cadmium-exposed bees, suggesting that this pathway may play a similar protective role for honey bees following Cd exposure. Additionally, previous studies demonstrated that hormone-induced antioxidant responses played important roles in the defense of abiotic and biotic stressors in insects [62]. Two genes associated with the insect hormone biosynthesis pathway were significantly upregulated in cadmium-exposed bees, indicating that the hormonal regulation of antioxidant response may also contribute to defense against Cd-induced oxidative stress in the brain of honey bees.
Taken together, our results indicated that Cd exposure exerted oxidative stress and decreased the gene expression levels of chemoreceptors in the brain of honey bees, which probably resulted in impaired olfactory learning in honey bees. Furthermore, increased expression levels of genes engaged in coping with oxidative stress may serve as a protector to combat oxidative stress and alleviate toxic effects caused by Cd exposure.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13110988/s1, Figure S1: The percentage of bees showing PER to CS -; Table S1: The primer sequences used in qRT-PCR; Table S2. A list of differentially expressed genes in the brains of cadmium-exposed and control bees.
Author Contributions
Conceptualization, Z.L. and S.S.; methodology, Z.L., Y.Q. and S.S.; validation, Y.Q., J.L. and K.W.; formal analysis, Z.L., S.S., Y.Q. and H.N.; investigation, Z.L., Y.Q., J.L. and K.W.; resources, S.S.; writing—original draft preparation, Z.L. and Y.Q.; writing—review and editing, S.S. and H.N.; supervision, Z.L. and S.S.; funding acquisition, Z.L. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Fujian Province (No. 2021J01078), the National Natural Science Foundation of China (No. 31702192), and the earmarked fund for the Modern Agroindustry Technology Research System (No. CARS-44-KXJ4).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Shubao Chen, Zhiyuan Feng, Runlang Su, and Yuxiao Zhang for their help in beekeeping.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, e04691. [Google Scholar] [CrossRef] [PubMed]
- Zwolak, A.; Sarzyńska, M.; Szpyrka, E.; Stawarczyk, K. Sources of soil pollution by heavy metals and their accumulation in vegetables: A review. Water Air Soil Pollut. 2019, 230, 164. [Google Scholar] [CrossRef]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, H.; Kopittke, P.M.; Zhao, F.-J. Cadmium contamination in agricultural soils of China and the impact on food safety. Environ. Pollut. 2019, 249, 1038–1048. [Google Scholar] [CrossRef]
- Li, J.; Qiu, J.; Wang, X.; Zhong, Y.; Lan, C.; Shu, W. Cadmium contamination in orchard soils and fruit trees and its potential health risk in Guangzhou, China. Environ. Pollut. 2006, 143, 159–165. [Google Scholar] [CrossRef]
- Jackson, A.P.; Alloway, B.J. The Transfer of Cadmium from Agricultural Soils to the Human Food Chain. In Biogeochemistry of Trace Metals; Adriano, D.C., Ed.; Lewis Publishers: Boca Raton, FL, USA, 1992; pp. 109–152. [Google Scholar]
- Vig, K.; Megharaj, M.; Sethunathan, N.; Naidu, R. Bioavailability and toxicity of cadmium to microorganisms and their activities in soil: A review. Adv. Environ. Res. 2003, 8, 121–135. [Google Scholar] [CrossRef]
- Manta, D.S.; Angelone, M.; Bellanca, A.; Neri, R.; Sprovieri, M. Heavy metals in urban soils: A case study from the city of Palermo (Sicily), Italy. Sci. Total Environ. 2002, 300, 229–243. [Google Scholar] [CrossRef]
- Åkesson, A.; Bjellerup, P.; Lundh, T.; Lidfeldt, J.; Nerbrand, C.; Samsioe, G.; Skerfving, S.; Vahter, M. Cadmium-induced effects on bone in a population-based study of women. Environ. Health Perspect. 2006, 114, 830–834. [Google Scholar] [CrossRef]
- Méndez-Armenta, M.; Rios, C. Cadmium neurotoxicity. Environ. Toxicol. Pharmacol. 2007, 23, 350–358. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Abel, G.M.; Storm, D.R.; Xia, Z. Cadmium exposure impairs cognition and olfactory memory in male C57BL/6 mice. Toxicol. Sci. 2018, 161, 87–102. [Google Scholar] [CrossRef]
- Hediji, H.; Djebali, W.; Cabasson, C.; Maucourt, M.; Baldet, P.; Bertrand, A.; Zoghlami, L.B.; Deborde, C.; Moing, A.; Brouquisse, R. Effects of long-term cadmium exposure on growth and metabolomic profile of tomato plants. Ecotoxicol. Environ. Saf. 2010, 73, 1965–1974. [Google Scholar] [CrossRef] [PubMed]
- Winfree, R.; Gross, B.J.; Kremen, C. Valuing pollination services to agriculture. Ecol. Econ. 2011, 71, 80–88. [Google Scholar] [CrossRef]
- Goretti, E.; Pallottini, M.; Rossi, R.; La Porta, G.; Gardi, T.; Goga, B.C.; Elia, A.; Galletti, M.; Moroni, B.; Petroselli, C. Heavy metal bioaccumulation in honey bee matrix, an indicator to assess the contamination level in terrestrial environments. Environ. Pollut. 2020, 256, 113388. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-Y.; Robinson, G.E. Regulation of honey bee division of labor by colony age demography. Behav. Ecol. Sociobiol. 1996, 39, 147–158. [Google Scholar] [CrossRef]
- Henry, M.; Beguin, M.; Requier, F.; Rollin, O.; Odoux, J.-F.; Aupinel, P.; Aptel, J.; Tchamitchian, S.; Decourtye, A. A common pesticide decreases foraging success and survival in honey bees. Science 2012, 336, 348–350. [Google Scholar] [CrossRef]
- Tosi, S.; Nieh, J.C.; Brandt, A.; Colli, M.; Fourrier, J.; Giffard, H.; Hernández-López, J.; Malagnini, V.; Williams, G.R.; Simon-Delso, N. Long-term field-realistic exposure to a next-generation pesticide, flupyradifurone, impairs honey bee behaviour and survival. Commun. Biol. 2021, 4, 805. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Menzel, R.; Sandoz, J.-C.; Giurfa, M. Revisiting olfactory classical conditioning of the proboscis extension response in honey bees: A step toward standardized procedures. J. Neurosci. Methods 2012, 211, 159–167. [Google Scholar] [CrossRef]
- Iqbal, J.; Mueller, U. Virus infection causes specific learning deficits in honeybee foragers. Proc. R. Soc. B Biol. Sci. 2007, 274, 1517–1521. [Google Scholar] [CrossRef]
- Ray, S.; Ferneyhough, B. The effects of age on olfactory learning and memory in the honey bee Apis mellifera. Neuroreport 1997, 8, 789–793. [Google Scholar] [CrossRef]
- Li, Z.; Yu, T.; Chen, Y.; Heerman, M.; He, J.; Huang, J.; Nie, H.; Su, S. Brain transcriptome of honey bees (Apis mellifera) exhibiting impaired olfactory learning induced by a sublethal dose of imidacloprid. Pestic. Biochem. Physiol. 2019, 156, 36–43. [Google Scholar] [CrossRef]
- Sivakoff, F.S.; Gardiner, M.M. Soil lead contamination decreases bee visit duration at sunflowers. Urban Ecosyst. 2017, 20, 1221–1228. [Google Scholar] [CrossRef]
- Di, N.; Zhang, K.; Hladun, K.R.; Rust, M.; Chen, Y.-F.; Zhu, Z.-Y.; Liu, T.-X.; Trumble, J.T. Joint effects of cadmium and copper on Apis mellifera forgers and larvae. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 237, 108839. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Cabirol, A.; Devaud, J.-M.; Barron, A.B.; Lihoreau, M. Why bees are so vulnerable to environmental stressors. Trends Ecol. Evol. 2017, 32, 268–278. [Google Scholar] [CrossRef]
- Gizaw, G.; Kim, Y.; Moon, K.; Choi, J.B.; Kim, Y.H.; Park, J.K. Effect of environmental heavy metals on the expression of detoxification-related genes in honey bee Apis mellifera. Apidologie 2020, 51, 664–674. [Google Scholar] [CrossRef]
- Polykretis, P.; Delfino, G.; Petrocelli, I.; Cervo, R.; Tanteri, G.; Montori, G.; Perito, B.; Branca, J.J.V.; Morucci, G.; Gulisano, M. Evidence of immunocompetence reduction induced by cadmium exposure in honey bees (Apis mellifera). Environ. Pollut. 2016, 218, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Celli, G.; Maccagnani, B. Honey bees as bioindicators of environmental pollution. Bull. Insectology 2003, 56, 137–139. [Google Scholar]
- Li, Z.; He, J.; Yu, T.; Chen, Y.; Huang, W.-F.; Huang, J.; Zhao, Y.; Nie, H.; Su, S. Transcriptional and physiological responses of hypopharyngeal glands in honeybees (Apis mellifera L.) infected by Nosema ceranae. Apidologie 2019, 50, 51–62. [Google Scholar] [CrossRef]
- Villar, M.E.; Marchal, P.; Viola, H.; Giurfa, M. Redefining single-trial memories in the honeybee. Cell Rep. 2020, 30, 2603–2613. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Marco Antonio, D.S.; Guidugli-Lazzarini, K.R.; Do Nascimento, A.M.; Simões, Z.L.P.; Hartfelder, K. RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften 2008, 95, 953–961. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Burden, C.M.; Morgan, M.O.; Hladun, K.R.; Amdam, G.V.; Trumble, J.J.; Smith, B.H. Acute sublethal exposure to toxic heavy metals alters honey bee (Apis mellifera) feeding behavior. Sci. Rep. 2019, 9, 4253. [Google Scholar] [CrossRef]
- Scheiner, R.; Page Jr, R.E.; Erber, J. The effects of genotype, foraging role, and sucrose responsiveness on the tactile learning performance of honey bees (Apis mellifera L.). Neurobiol. Learn. Mem. 2001, 76, 138–150. [Google Scholar] [CrossRef]
- Rothman, J.A.; Leger, L.; Kirkwood, J.S.; McFrederick, Q.S. Cadmium and selenate exposure affects the honey bee microbiome and metabolome, and bee-associated bacteria show potential for bioaccumulation. Appl. Environ. Microbiol. 2019, 85, e01411–e01419. [Google Scholar] [CrossRef]
- Di, N.; Hladun, K.R.; Zhang, K.; Liu, T.-X.; Trumble, J.T. Laboratory bioassays on the impact of cadmium, copper and lead on the development and survival of honeybee (Apis mellifera L.) larvae and foragers. Chemosphere 2016, 152, 530–538. [Google Scholar] [CrossRef]
- Cheng, S.H.; Wai, A.W.K.; So, C.H.; Wu, R.S.S. Cellular and molecular basis of cadmium-induced deformities in zebrafish embryos. Environ. Toxicol. Chem. Int. J. 2000, 19, 3024–3031. [Google Scholar] [CrossRef]
- Kippler, M.; Tofail, F.; Gardner, R.; Rahman, A.; Hamadani, J.D.; Bottai, M.; Vahter, M. Maternal cadmium exposure during pregnancy and size at birth: A prospective cohort study. Environ. Health Perspect. 2012, 120, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Varoni, M.V.; Pasciu, V.; Gadau, S.D.; Baralla, E.; Serra, E.; Palomba, D.; Demontis, M.P. Possible antioxidant effect of Lycium barbarum polysaccharides on hepatic cadmium-induced oxidative stress in rats. Environ. Sci. Pollut. Res. 2017, 24, 2946–2955. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Novelli, E.; Silva, M.D.P.; Júnior, R.A. Environmental cadmium exposure and metabolic responses of the Nile tilapia, Oreochromis niloticus. Environ. Pollut. 2001, 114, 169–175. [Google Scholar] [CrossRef]
- Monchanin, C.; Blanc-Brude, A.; Drujont, E.; Negahi, M.M.; Pasquaretta, C.; Silvestre, J.; Baqué, D.; Elger, A.; Barron, A.B.; Devaud, J.-M. Chronic exposure to trace lead impairs honey bee learning. Ecotoxicol. Environ. Saf. 2021, 212, 112008. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-binding proteins as sensing elements for odour monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef]
- Li, H.; Wu, F.; Zhao, L.; Tan, J.; Jiang, H.; Hu, F. Neonicotinoid insecticide interact with honeybee odorant-binding protein: Implication for olfactory dysfunction. Int. J. Biol. Macromol. 2015, 81, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, J.; Pregitzer, P.; Breer, H.; Krieger, J. Access to the odor world: Olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 2018, 75, 485–508. [Google Scholar] [CrossRef]
- Kelber, C.; Rössler, W.; Kleineidam, C.J. Multiple olfactory receptor neurons and their axonal projections in the antennal lobe of the honeybee Apis Mellifera. J. Comp. Neurol. 2006, 496, 395–405. [Google Scholar] [CrossRef]
- Vosshall, L.B.; Wong, A.M.; Axel, R. An olfactory sensory map in the fly brain. Cell 2000, 102, 147–159. [Google Scholar] [CrossRef]
- Shukla, E.; Thorat, L.J.; Nath, B.B.; Gaikwad, S.M. Insect trehalase: Physiological significance and potential applications. Glycobiology 2015, 25, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, V.B.; Lord, J.S.; Diering, G.H.; Zylka, M.J. Mania-and anxiety-like behavior and impaired maternal care in female diacylglycerol kinase eta and iota double knockout mice. Genes Brain Behav. 2020, 19, e12570. [Google Scholar] [CrossRef] [PubMed]
- Mérida, I.; Arranz-Nicolás, J.; Rodríguez-Rodríguez, C.; Ávila-Flores, A. Diacylglycerol kinase control of protein kinase C. Biochem. J. 2019, 476, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Ishisaka, M.; Hara, H. The roles of diacylglycerol kinases in the central nervous system: Review of genetic studies in mice. J. Pharmacol. Sci. 2014, 124, 336–343. [Google Scholar] [CrossRef]
- Branca, J.J.; Fiorillo, C.; Carrino, D.; Paternostro, F.; Taddei, N.; Gulisano, M.; Pacini, A.; Becatti, M. Cadmium-induced oxidative stress: Focus on the central nervous system. Antioxidants 2020, 9, 492. [Google Scholar] [CrossRef]
- Farooqui, T. Iron-induced oxidative stress modulates olfactory learning and memory in honeybees. Behav. Neurosci. 2008, 122, 433. [Google Scholar] [CrossRef]
- Ipson, B.R.; Green, R.A.; Wilson, J.T.; Watson, J.N.; Faull, K.F.; Fisher, A.L. Tyrosine aminotransferase is involved in the oxidative stress response by metabolizing meta-tyrosine in Caenorhabditis elegans. J. Biol. Chem. 2019, 294, 9536–9554. [Google Scholar] [CrossRef]
- Chen, X.; Hall, H.; Simpson, J.P.; Leon-Salas, W.D.; Ready, D.F.; Weake, V.M. Cytochrome b5 protects photoreceptors from light stress-induced lipid peroxidation and retinal degeneration. NPJ Aging Mech. Dis. 2017, 3, 18. [Google Scholar] [CrossRef]
- Ferguson, C.S.; Tyndale, R.F. Cytochrome P450 enzymes in the brain: Emerging evidence of biological significance. Trends Pharmacol. Sci. 2011, 32, 708–714. [Google Scholar] [CrossRef]
- He, F.; Zhao, Q.; Huang, J.-L.; Niu, M.-X.; Feng, H.-C.; Shi, Y.-J.; Zhao, K.-J.; Cui, X.-L.; Wu, X.-L.; Mi, J.-X. External application of nitrogen alleviates toxicity of cadmium on poplars via starch and sucrose metabolism. Tree Physiol. 2021, 41, 2126–2141. [Google Scholar] [CrossRef]
- Kodrík, D.; Bednářová, A.; Zemanová, M.; Krishnan, N. Hormonal regulation of response to oxidative stress in insects—An update. Int. J. Mol. Sci. 2015, 16, 25788–25816. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).