Characterizing Billbug (Sphenophorus spp.) Seasonal Biology Using DNA Barcodes and a Simple Morphometric Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Adult DNA Barcoding Reference Database
2.2. Larval Species Identification
2.3. Larval Seasonal Phenology Charts
3. Results
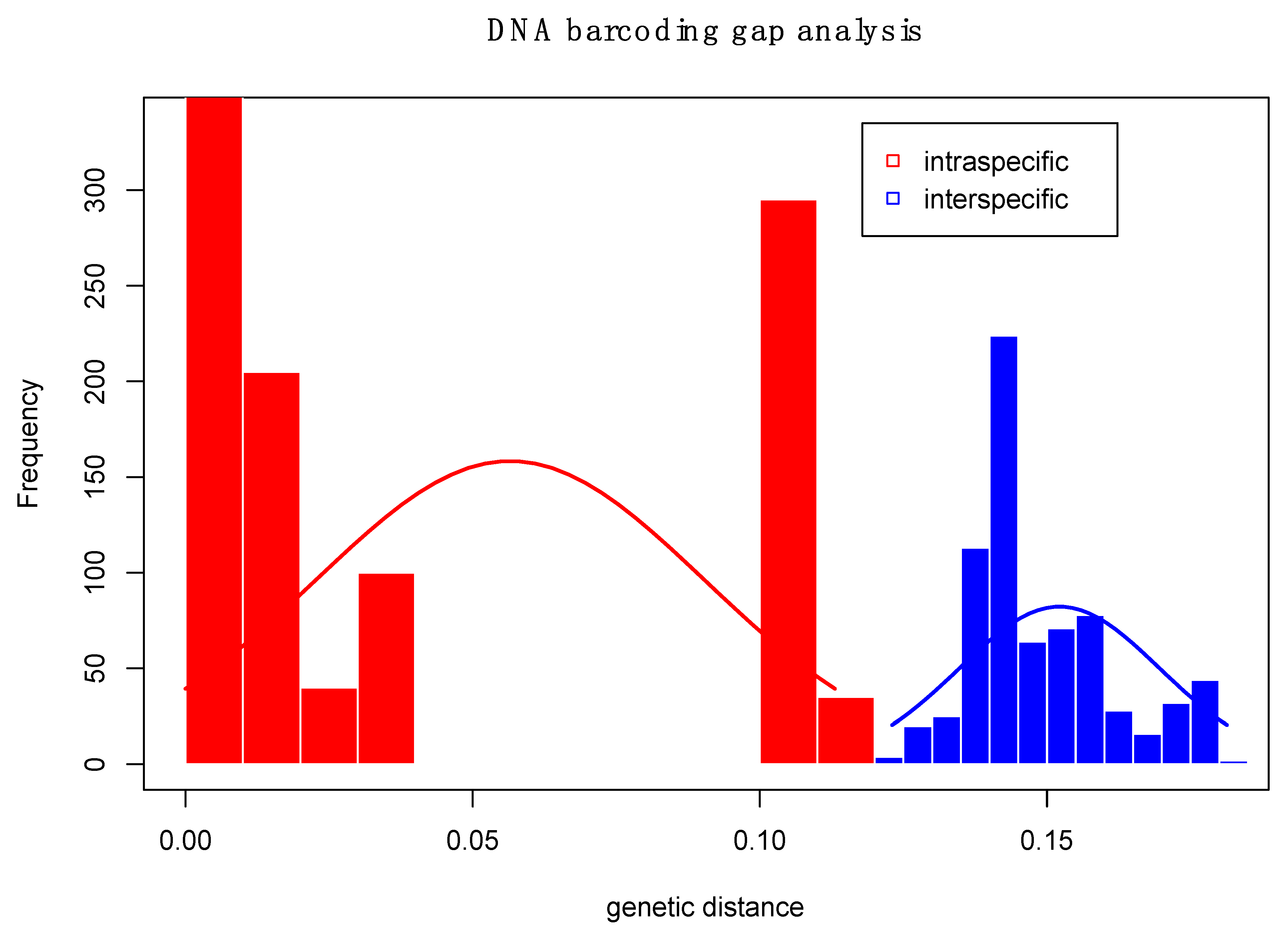
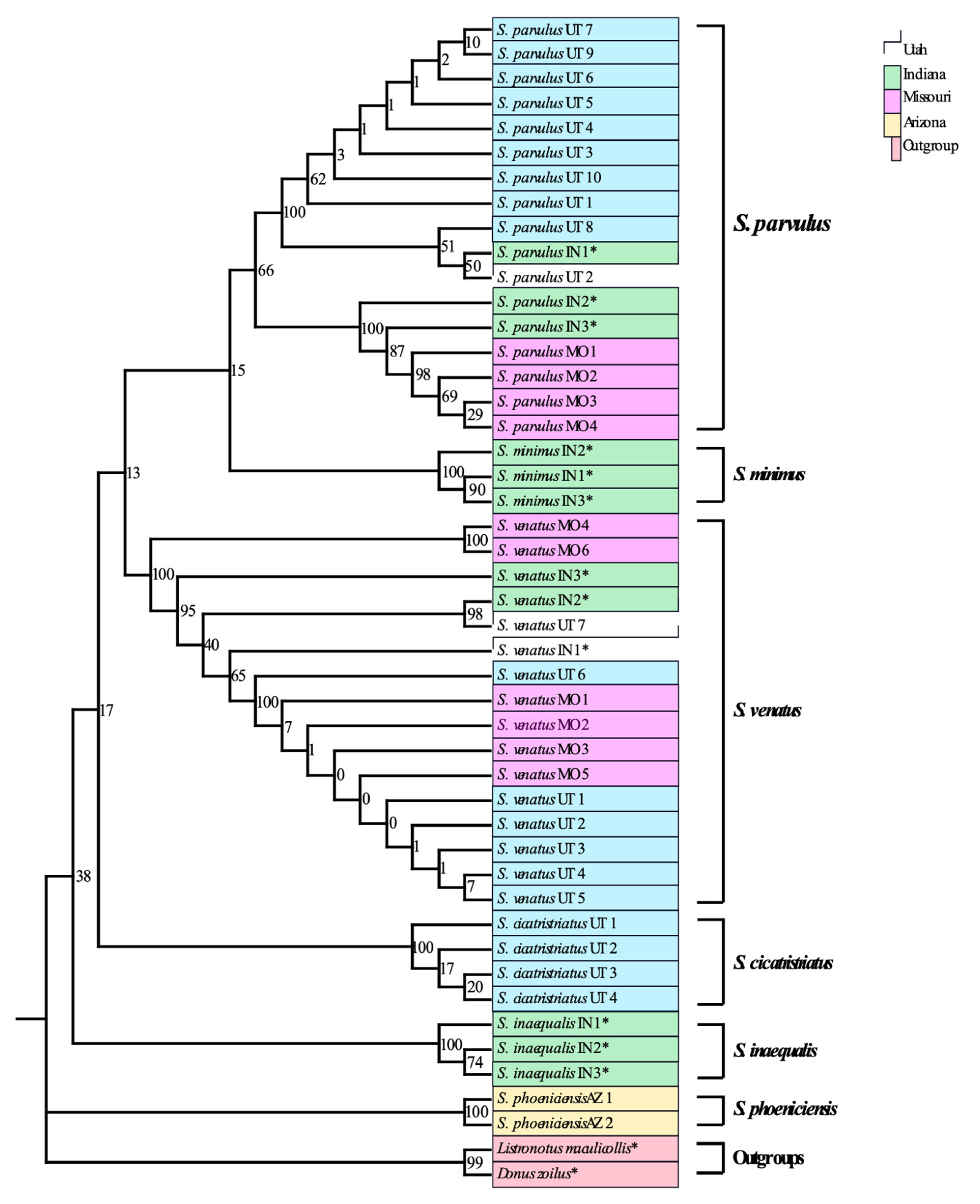
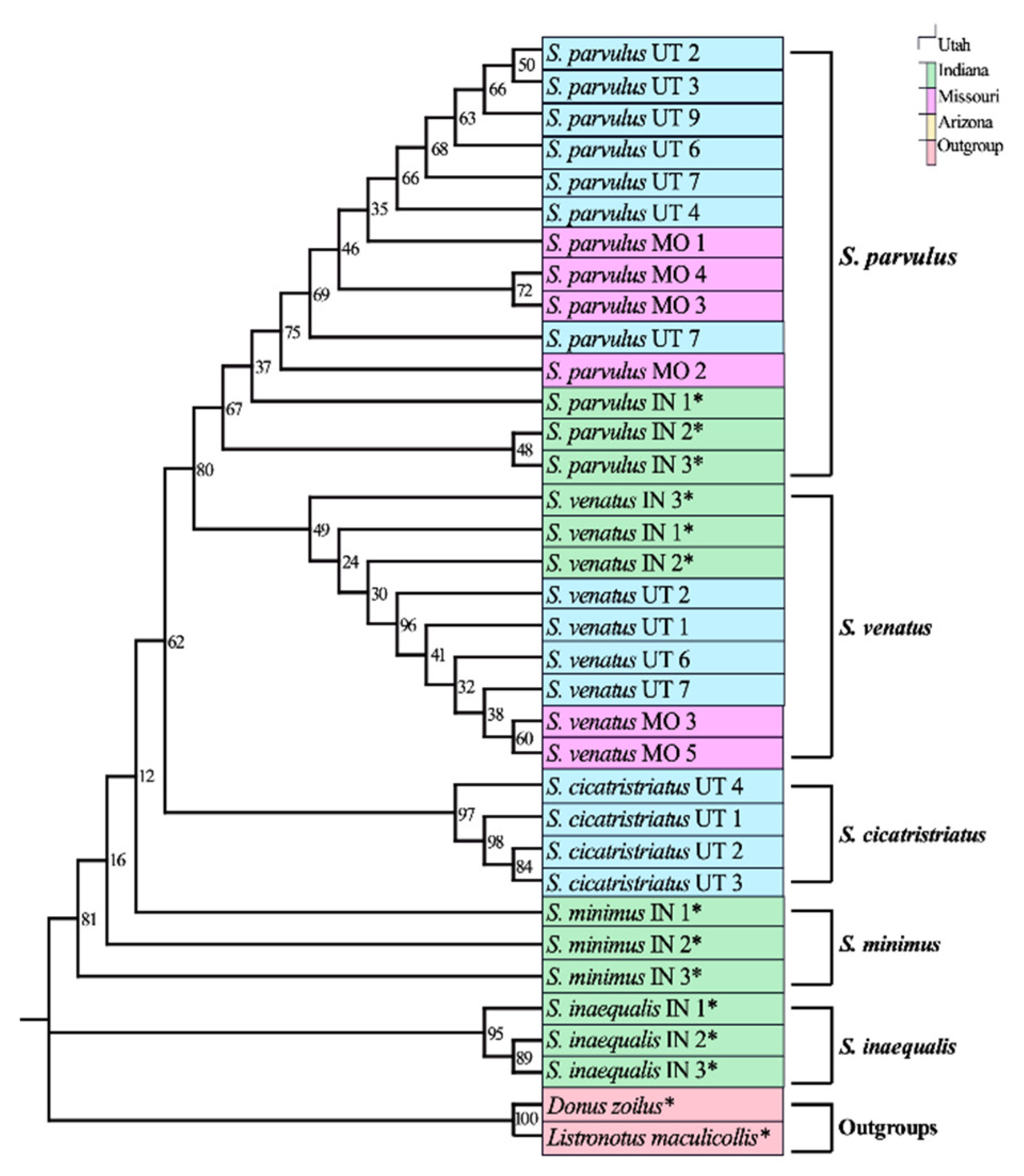
3.1. Adult DNA Barcoding Reference Database
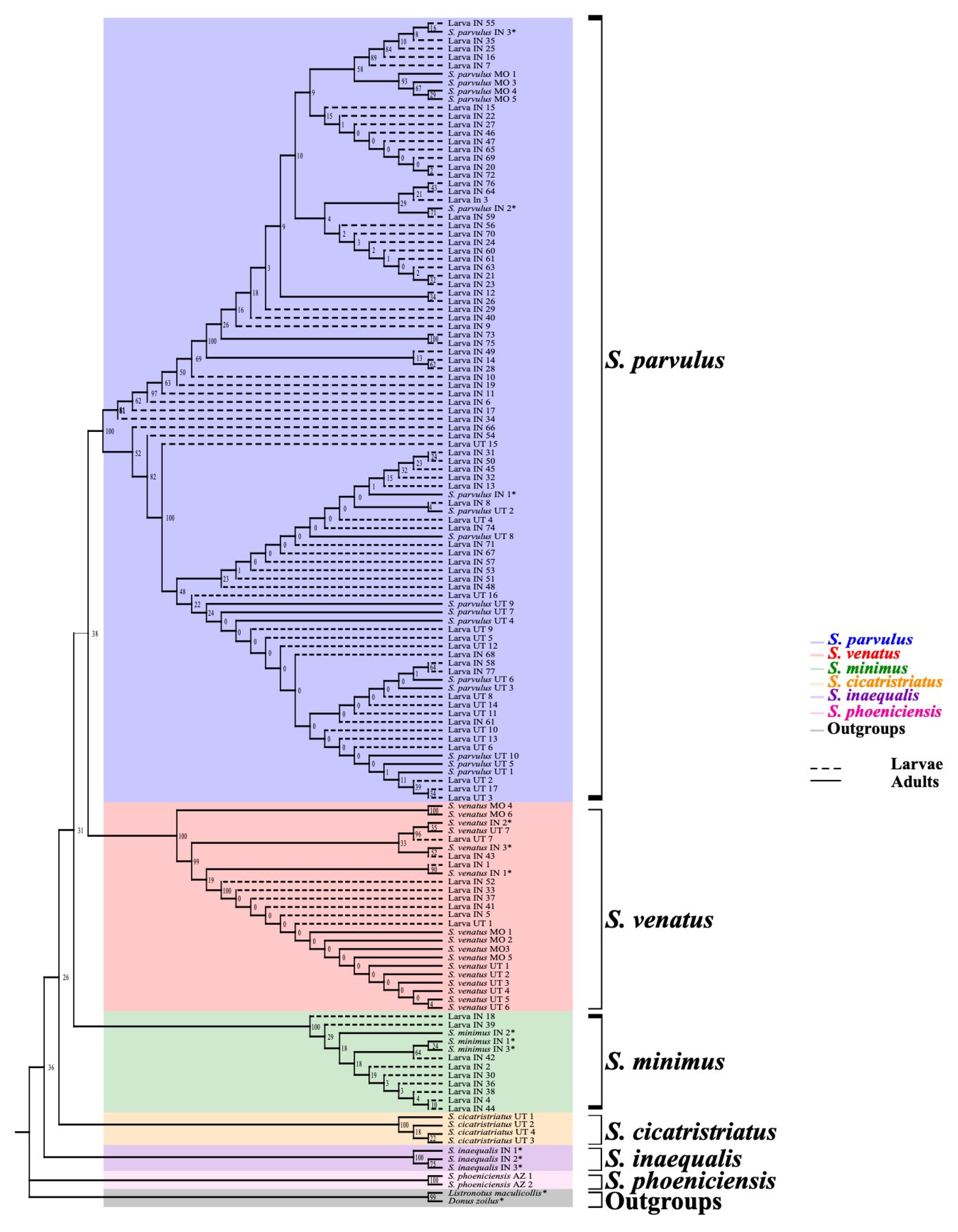
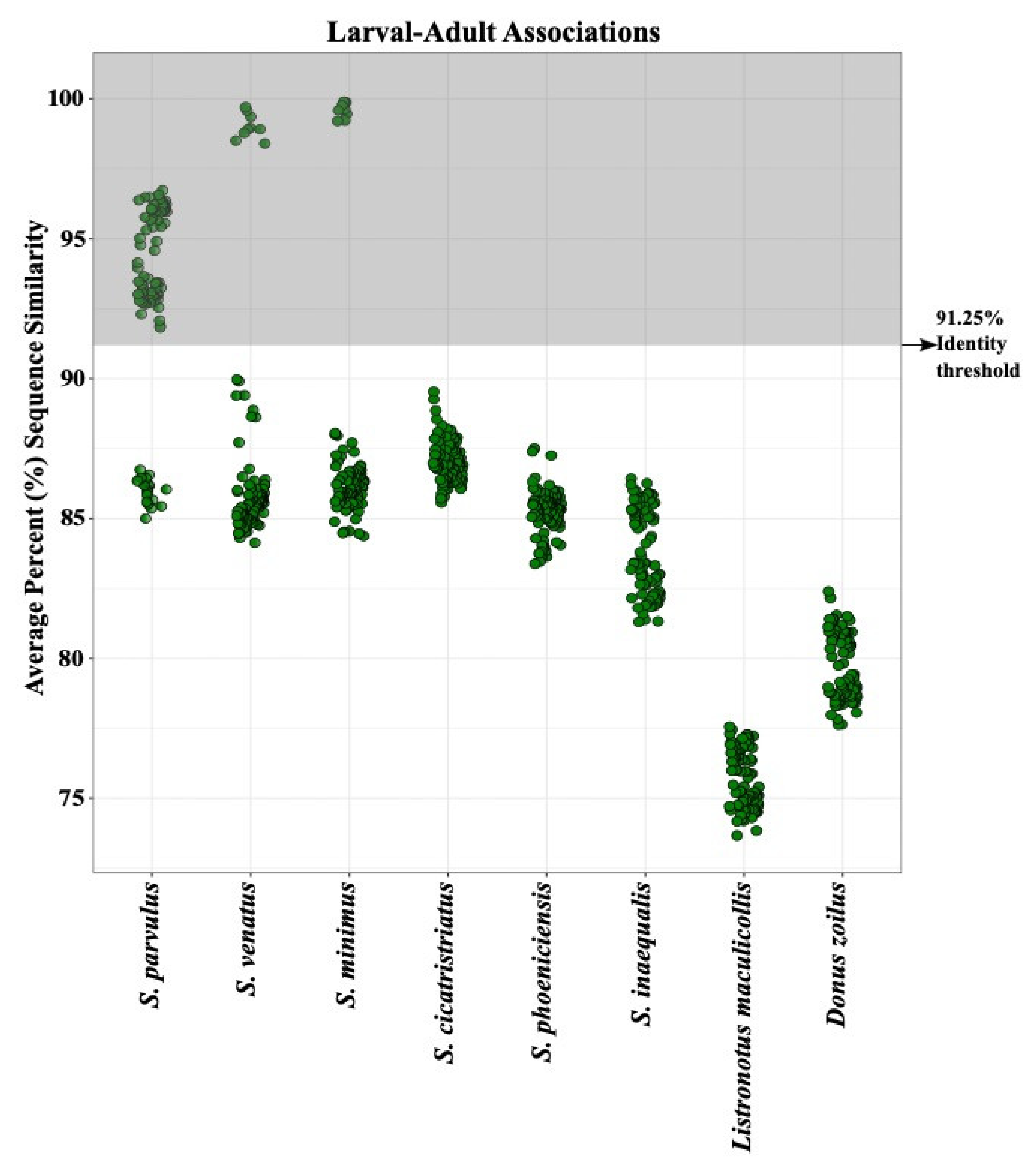
3.2. Larval Species Identification
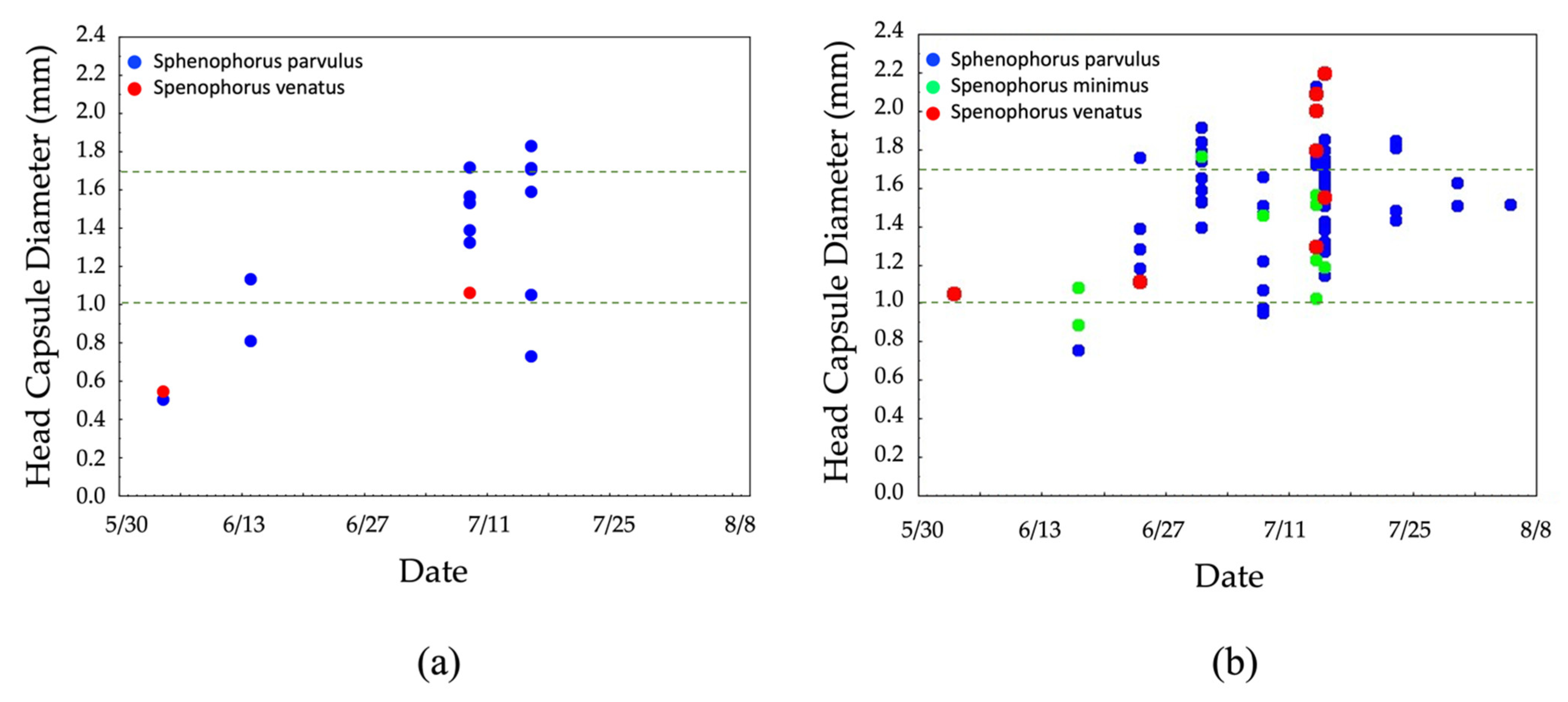
3.3. Larval Seasonal Phenology Charts
4. Discussion
4.1. Development of a Reliable DNA Barcoding Tool for Sphenophorus in the U.S.
4.2. DNA Barcoding Tool Provides Insight into Billbug Biology
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Larval Specimen | Species | State | Collection Location | Grass Type | Collection Date | Head Capsule Width Diameter (mm) |
|---|---|---|---|---|---|---|
| Larva UT 1 | S. parvulus | Utah | GRF | Cool | 4 June 2018 | 0.504 |
| Larva UT 2 | S. venatus | Utah | GRF | Cool | 4 June 2018 | 0.546 |
| Larva UT 3 | S. parvulus | Utah | LCC | Cool | 14 June 2018 | 0.81 |
| Larva UT 4 | S. parvulus | Utah | LCC | Cool | 14 June 2018 | 1.133 |
| Larva UT 6 | S. parvulus | Utah | LCC | Cool | 9 July 2018 | 1.39 |
| Larva UT 9 | S. venatus | Utah | LCC | Cool | 9 July 2018 | 1.062 |
| Larva UT 10 | S. parvulus | Utah | LCC | Cool | 9 July 2018 | 1.567 |
| Larva UT 11 | S. parvulus | Utah | GRF | Cool | 16 July 2018 | 1.715 |
| Larva UT 14 | S. parvulus | Utah | GRF | Cool | 16 July 2018 | 0.73 |
| Larva UT 15 | S. parvulus | Utah | GRF | Cool | 16 July 2018 | 1.591 |
| Larva UT 16 | S. parvulus | Utah | GRF | Cool | 16 July 2018 | 1.051 |
| Larva UT 17 | S. parvulus | Utah | GRF | Cool | 16 July 2018 | 1.711 |
| Larva IN 1 | S. venatus | Indiana | BPC | Warm | 3 June 2020 | 1.054 |
| Larva IN 2 | S. minimus | Indiana | DTRC | Cool | 17 Jun 2020 | 0.891 |
| Larva IN 3 | S. parvulus | Indiana | DTRC | Cool | 17 Jun 2020 | 0.760 |
| Larva IN 4 | S. minimus | Indiana | DTRC | Cool | 17 Jun 2020 | 1.083 |
| Larva IN 5 | S. venatus | Indiana | DTRC | Cool | 24 Jun 2020 | 1.113 |
| Larva IN 6 | S. parvulus | Indiana | DTRC | Cool | 24 Jun 2020 | 1.287 |
| Larva IN 7 | S. parvulus | Indiana | DTRC | Cool | 24 Jun 2020 | 1.762 |
| Larva IN 8 | S. parvulus | Indiana | DTRC | Cool | 24 Jun 2020 | 1.184 |
| Larva IN 9 | S. parvulus | Indiana | DTRC | Cool | 24 Jun 2020 | 1.393 |
| Larva IN 10 | S. parvulus | Indiana | BPC | Warm | 1 July 2020 | 1.919 |
| Larva IN 11 | S. parvulus | Indiana | BPC | Warm | 1 July 2020 | 1.592 |
| Larva IN 12 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.398 |
| Larva IN 13 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.841 |
| Larva IN 14 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.743 |
| Larva IN 15 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.782 |
| Larva IN 16 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.536 |
| Larva IN 17 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.795 |
| Larva IN 18 | S. minimus | Indiana | DTRC | Cool | 1 July 2020 | 1.769 |
| Larva IN 19 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.758 |
| Larva IN 20 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.654 |
| Larva IN 21 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.593 |
| Larva IN 22 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.529 |
| Larva IN 23 | S. parvulus | Indiana | DTRC | Cool | 1 July 2020 | 1.748 |
| Larva IN 24 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 1.512 |
| Larva IN 25 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 1.466 |
| Larva IN 26 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 1.224 |
| Larva IN 27 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 0.953 |
| Larva IN 28 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 1.660 |
| Larva IN 29 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 1.659 |
| Larva IN 30 | S. minimus | Indiana | DTRC | Cool | 8 July 2020 | 1.459 |
| Larva IN 31 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 1.074 |
| Larva IN 32 | S. parvulus | Indiana | DTRC | Cool | 8 July 2020 | 0.975 |
| Larva IN 33 | S. venatus | Indiana | DTRC | Cool | 14 July 2020 | 1.799 |
| Larva IN 34 | S. parvulus | Indiana | DTRC | Cool | 14 July 2020 | 2.130 |
| Larva IN 35 | S. parvulus | Indiana | DTRC | Cool | 14 July 2020 | 1.722 |
| Larva IN 36 | S. minimus | Indiana | DTRC | Cool | 14 July 2020 | 1.568 |
| Larva IN 37 | S. venatus | Indiana | DTRC | Cool | 14 July 2020 | 1.300 |
| Larva IN 38 | S. minimus | Indiana | DTRC | Cool | 14 July 2020 | 1.031 |
| Larva IN 39 | S. minimus | Indiana | DTRC | Cool | 14 July 2020 | 1.517 |
| Larva IN 40 | S. parvulus | Indiana | DTRC | Cool | 14 July 2020 | 1.753 |
| Larva IN 41 | S. venatus | Indiana | DTRC | Cool | 14 July 2020 | 2.091 |
| Larva IN 42 | S. minimus | Indiana | DTRC | Cool | 14 July 2020 | 1.227 |
| Larva IN 43 | S. venatus | Indiana | DTRC | Cool | 14 July 2020 | 2.008 |
| Larva IN 44 | S. minimus | Indiana | DTRC | Warm | 15 July 2020 | 1.192 |
| Larva IN 45 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.643 |
| Larva IN 46 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.323 |
| Larva IN 47 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.752 |
| Larva IN 48 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.797 |
| Larva IN 49 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.574 |
| Larva IN 50 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.668 |
| Larva IN 51 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.655 |
| Larva IN 52 | S. venatus | Indiana | DTRC | Warm | 15 July 2020 | 1.553 |
| Larva IN 53 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.857 |
| Larva IN 54 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.626 |
| Larva IN 55 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.302 |
| Larva IN 56 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.423 |
| Larva IN 57 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.147 |
| Larva IN 58 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.559 |
| Larva IN 59 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.650 |
| Larva IN 60 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.431 |
| Larva IN 61 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.424 |
| Larva IN 62 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.673 |
| Larva IN 63 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.645 |
| Larva IN 64 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.514 |
| Larva IN 65 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.384 |
| Larva IN 66 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.606 |
| Larva IN 67 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.273 |
| Larva IN 68 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.659 |
| Larva IN 69 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.298 |
| Larva IN 70 | S. parvulus | Indiana | DTRC | Warm | 15 July 2020 | 1.723 |
| Larva IN 71 | S. parvulus | Indiana | DTRC | Warm | 23 July 2020 | 1.850 |
| Larva IN 72 | S. parvulus | Indiana | DTRC | Warm | 23 July 2020 | 1.810 |
| Larva IN 73 | S. parvulus | Indiana | DTRC | Warm | 23 July 2020 | 1.438 |
| Larva IN 74 | S. parvulus | Indiana | DTRC | Warm | 23 July 2020 | 1.488 |
| Larva IN 75 | S. parvulus | Indiana | DTRC | Warm | 30 July 2020 | 1.512 |
| Larva IN 76 | S. parvulus | Indiana | DTRC | Warm | 30 July 2020 | 1.631 |
| Larva IN 77 | S. parvulus | Indiana | DTRC | Warm | 5 August 2020 | 1.520 |
References
- Feder, J.L.; Filchak, K.E. It’s about time: The evidence for host plant-mediated selection in the apple maggot fly, Rhagoletis pomonella, and its implications for fitness trade-offs in phytophagous insects. Èntomol. Exp. Appl. 1999, 91, 211–225. [Google Scholar] [CrossRef]
- Berlocher, S.H.; Feder, J.L. Sympatric Speciation in Phytophagous Insects: Moving Beyond Controversy? Ann. Rev. Entomol. 2002, 47, 773–815. [Google Scholar] [CrossRef] [Green Version]
- Farkas, P.; György, Z.; Tóth, A.; Sojnóczki, A.; Fail, J. A simple molecular identification method of the Thrips tabaci (Thysanoptera: Thripidae) cryptic species complex. Bull. Èntomol. Res. 2020, 110, 397–405. [Google Scholar] [CrossRef]
- Ahrens, D.; Monaghan, M.T.; Vogler, A.P. DNA-based taxonomy for associating adults and larvae in multi-species assemblages of chafers (Coleoptera: Scarabaeidae). Mol. Phylogenetics Evol. 2007, 44, 436–449. [Google Scholar] [CrossRef]
- Hackett, B.J.; Gimnig, J.; Guelbeogo, W.M.; Costantini, C.; Koekemoer, L.L.; Coetzee, M.; Collins, F.H.; Besansky, N.J. Ribosomal DNA internal transcribed spacer (ITS2) sequences differentiate Anopheles funestus and An. rivulorum, and uncover a cryptic taxon. Insect Mol. Biol. 2000, 9, 369–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassanzadeh-Avval, M.; Sadeghi-Namaghi, H.; Fekrat, L. Molecular and morphological identification of Anthocoris spp. (Hemiptera: Anthocoridae) predators of three economically important psyllid species in Razavi Khorasan province, Northeastern Iran. Biologia 2020, 75, 1667–1677. [Google Scholar] [CrossRef]
- Soroka, J.; Bennett, A.M.; Kora, C.; Schwarzfeld, M.D. Distribution of alfalfa weevil (Coleoptera: Curculionidae) and its parasitoids on the Canadian Prairies, with a key to described species of Nearctic Bathyplectes (Hymenoptera: Ichneumonidae). Can. Èntomol. 2020, 152, 663–701. [Google Scholar] [CrossRef]
- Tyagi, K.; Kumar, V.; Singha, D.; Chandra, K.; Laskar, B.A.; Kundu, S.; Chakraborty, R.; Chatterjee, S. DNA Barcoding studies on Thrips in India: Cryptic species and Species complexes. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, C.; Hu, B.; Liu, Y.; Walter, G.H.; Hereward, J.P. Gene flow across host-associated populations of the rice stem borer Chilo suppressalis Walker (Lepidoptera: Crambidae): Implications for Bt resistance management in rice. Pest Manag. Sci. 2020, 76, 695–703. [Google Scholar] [CrossRef]
- De Meyer, M.; Delatte, H.; Ekesi, S.; Jordaens, K.; Kalinová, B.; Manrakhan, A.; Mwatawala, M.; Steck, G.; Van Cann, J.; Vaníčková, L.; et al. An integrative approach to unravel the Ceratitis FAR (Diptera, Tephritidae) cryptic species complex: A review. ZooKeys 2015, 540, 405–427. [Google Scholar] [CrossRef] [Green Version]
- Doskocil, J.P.; Walker, N.R.; Bell, G.E.; Marek, S.; Reinert, J.A.; Royer, T. Species Composition and Seasonal Occurrence of Phyllophaga (Coleoptera: Scarabaeidae) Infesting Intensely Managed Bermudagrass in Oklahoma. J. Econ. Èntomol. 2008, 101, 1624–1632. [Google Scholar] [CrossRef]
- Rodriguez-Soto, M.M. Characterizing Billbug (Sphenophorus spp.) Seasonal Biology Using DNA Barcodes and a Simple Morphometric Analysis. Ph.D. Thesis, Purdue University, West Lafayette, IN, USA, 2021. [Google Scholar]
- Johnson-Cicalese, J.M.; Wolfe, G.W.; Funk, C.R. Biology, Distribution, and Taxonomy of Billbug Turf Pests (Coleoptera: Curculionidae). Environ. Èntomol. 1990, 19, 1037–1046. [Google Scholar] [CrossRef]
- Chong, J.-H. Species Richness and Seasonal Abundance of Billbugs, Sphenophorus spp., from South Carolina Golf Courses1. J. Agric. Urban Èntomol. 2015, 31, 20–28. [Google Scholar] [CrossRef]
- Georgis, R.; Koppenhöfer, A.M.; Lacey, L.A.; Bélair, G.; Duncan, L.W.; Grewal, P.S.; Samish, M.; Tan, L.; Torr, P.; van Tol, R.W.H.M. Successes and failures in the use of parasitic nematodes for pest control. Biol. Control. 2006, 38, 103–123. [Google Scholar] [CrossRef]
- Richmond, D. Managing Turfgrass Insects. Available online: https://extension.entm.purdue.edu/publications/E-266/E-266.html (accessed on 12 April 2021).
- Duffy, A.G.; Powell, G.S.; Zaspel, J.M.; Richmond, D.S. Billbug (Coleoptera: Dryophthoridae: Sphenophorus spp.) Seasonal Biology and DNA-Based Life Stage Association in Indiana Turfgrass. J. Econ. Èntomol. 2018, 111, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, M.M.; Ramirez, R.A. Biology and Management of Billbugs (Coleoptera: Curculionidae) in Turfgrass. J. Integr. Pest Manag. 2016, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doskocil, J.P.; Brandenburg, R.L. Hunting billbug (Coleoptera: Curculionidae) life cycle and damaging life stage in North Carolina, with notes on other billbug species abundance. J. Econ. Èntomol. 2012, 105, 2045–2051. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.-I.; Buss, E.A. Billbug (Coleoptera: Curculionidae) species composition, abundance, seasonal activity, and developmental time in Florida. J. Econ. Èntomol. 2009, 102, 309–314. [Google Scholar] [CrossRef]
- Umble, J.; Fisher, G.; Rao, S. Sampling methods and seasonal phenology of Sphenophorus venatus confluens Chittenden (Coleoptera: Curculionidae) in orchardgrass (Dactylis glomerata L.). J. Agric. Urban Entomol. 2005, 22, 79–85. [Google Scholar]
- Etzler, F.E.; Wanner, K.W.; Morales-Rodriguez, A.; Ivie, M.A. DNA Barcoding to Improve the Species-Level Management of Wireworms (Coleoptera: Elateridae). J. Econ. Èntomol. 2014, 107, 1476–1485. [Google Scholar] [CrossRef]
- Pramual, P.; Wongpakam, K. Association of black fly (Diptera: Simuliidae) life stages using DNA barcode. J. Asia-Pacific Èntomol. 2014, 17, 549–554. [Google Scholar] [CrossRef]
- Jinbo, U.; Kato, T.; Ito, M. Current progress in DNA barcoding and future implications for entomology. Èntomol. Sci. 2011, 14, 107–124. [Google Scholar] [CrossRef]
- Brasier, M.J.; Wiklund, H.; Neal, L.; Jeffreys, R.; Linse, K.; Ruhl, H.; Glover, A.G. DNA barcoding uncovers cryptic diversity in 50 % of deep-sea Antarctic polychaetes Subject Category. R. Soc. Open Sci. 2016, 3, 160432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliavia, M.; Nicosia, A.; Salamone, M.; Biondo, G.; Bennici, C.D.; Mazzola, S.; Cuttitta, A. Development of a fast DNA extraction method for sea food and marine species identification. Food Chem. 2016, 203, 375–378. [Google Scholar] [CrossRef]
- Pompanon, F.; Deagle, B.; Symondson, W.O.C.; Brown, D.S.; Jarman, S.; Taberlet, P. Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef] [Green Version]
- Marullo, R.; Mercati, F.; Vono, G. DNA Barcoding: A Reliable Method for the Identification of Thrips Species (Thysanoptera, Thripidae) Collected on Sticky Traps in Onion Fields. Insects 2020, 11, 489. [Google Scholar] [CrossRef]
- Graf, N.; Rebek, E.; Royer, T.; Walker, N. Spatial and Temporal Distribution of spp. Infesting Bermudagrass Stands in Oklahoma. ITSRJ 2017, 13, 489–496. [Google Scholar] [CrossRef]
- Raupach, M.J.; Astrin, J.J.; Hannig, K.; Peters, M.K.; Stoeckle, M.Y.; Wägele, J.-W. Molecular species identification of Central European ground beetles (Coleoptera: Carabidae) using nuclear rDNA expansion segments and DNA barcodes. Front. Zool. 2010, 7, 26. [Google Scholar] [CrossRef] [Green Version]
- Rubinoff, D.; Cameron, S.; Will, K. A Genomic Perspective on the Shortcomings of Mitochondrial DNA for “Barcoding” Identification. J. Hered. 2006, 97, 581–594. [Google Scholar] [CrossRef] [Green Version]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Griekspoor, A.; Groothuis, T. 4Peaks. Available online: https://www.nucleobytes.com/4peaks/index.html (accessed on 12 April 2021).
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A modular system for evolutionary analysis. Evolution 2008, 62, 1103–1118. [Google Scholar]
- Inkscape Project. Inkscape. Available online: https://inkscape.org (accessed on 1 April 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Taylor & Francis: Abingdon, UK, 2016. [Google Scholar]
- Zhang, A.-B.; Hao, M.-D.; Yang, C.-Q.; Shi, Z.-Y. BarcodingR: An integrated r package for species identification using DNA barcodes. Methods Ecol. Evol. 2016, 8, 627–634. [Google Scholar] [CrossRef] [Green Version]
- TIBCO Software Inc. TIBCO Statistica® User’s Guide; TIBCO Software Inc.: Palo Alto, CA, USA, 2017. [Google Scholar]
- Godin, J.; Maltais, P.; Gaudet, S. Head Capsule Width as an Instar Indicator for Larvae of the Cranberry Fruitworm (Lepidoptera: Pyralidae) in Southeastern New Brunswick. J. Econ. Èntomol. 2002, 95, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Held, D.W.; Potter, D.A. Prospects for Managing Turfgrass Pests with Reduced Chemical Inputs. Annu. Rev. Èntomol. 2012, 57, 329–354. [Google Scholar] [CrossRef] [PubMed]
- Vittum, P.J. Turfgrass Insects of the United States and Canada, 3rd ed.; Comstock Publishing Associates: Ithaca, NY, USA, 2020. [Google Scholar]
- Yusseff-Vanegas, S.Z.; Agnarsson, I. DNA-barcoding of forensically important blow flies (Diptera: Calliphoridae) in the Caribbean Region. PeerJ 2017, 5, e3516. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Liu, S.; Zhang, Z. Efficacy of using DNA barcoding to identify parasitoid wasps of the melon-cotton aphid (Aphis gossypii) in watermelon cropping system. BioControl 2018, 63, 677–685. [Google Scholar] [CrossRef]
- Jordal, B.H.; Kambestad, M. DNA barcoding of bark and ambrosia beetles reveals excessive NUMTs and consistent east-west divergence across Palearctic forests. Mol. Ecol. Resour. 2014, 14, 7–17. [Google Scholar] [CrossRef]
- Sethusa, M.; Millar, I.; Yessoufou, K.; Jacobs, A.; Van Der Bank, M.; Van Der Bank, H. DNA Barcode Efficacy for the Identification of Economically Important Scale Insects (Hemiptera: Coccoidea) in South Africa. Afr. Èntomol. 2014, 22, 257–266. [Google Scholar] [CrossRef]
- Lv, J.; Wu, S.; Zhang, Y.; Zhang, T.; Feng, C.; Jia, G.; Lin, X. Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial DNA 2013, 25, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Fagan-Jeffries, E.P.; Cooper, S.J.; Bradford, T.; Austin, A.D. Intragenomic internal transcribed spacer 2 variation in a genus of parasitoid wasps (Hymenoptera: Braconidae): Implications for accurate species delimitation and phylogenetic analysis. Insect Mol. Biol. 2019, 28, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; Armstrong, K.F.; Cruickshank, R.H. Molecular phylogenetics of a South Pacific sap beetle species complex (Carpophilus spp., Coleoptera: Nitidulidae). Mol. Phylogenetics Evol. 2012, 64, 428–440. [Google Scholar] [CrossRef] [PubMed]


| U. S. State | Site (Coordinates) | Abbreviated Name | Turfgrass Cover | Turfgrass Cultivar | Soil Type |
|---|---|---|---|---|---|
| Utah | Logan Country Club | LCC | Kentucky bluegrass | Unknown | Silty Loam |
| (41.7445° N, 111.7949° W) | |||||
| Greenville Research Farm | GRF | Kentucky bluegrass | Unknown | Silty Loam | |
| (41.7664° N, 111.8105° W) | |||||
| Utah State University Greenhouse | USUG | Kentucky bluegrass | Unknown | Silty Loam | |
| (41.7571° N, 111.8133° W) | |||||
| Indiana | Bimel Practice Center | BPC | Bermudagrass | Patriot | Silty Clay Loam |
| (40.4376° N, 86.9178° W) | |||||
| Daniel Turfgrass Research Center | DTC | Kentucky bluegrass | Park | Silty Clay Loam | |
| (40.4411° N, 86.9317° W) | |||||
| Daniel Turfgrass Research Center | DTC | Zoysiagrass | Meyer | Silty Clay Loam | |
| (40.4415° N, 86.9325° W) |
| Species | DNA Extractions | COI | % Success | 18S | % Success | ITS2 | % Success |
|---|---|---|---|---|---|---|---|
| S. parvulus | |||||||
| Utah | 14 | 10 | 71.4 | 7 | 50.0 | 7 | 50.0 |
| Missouri | 12 | 5 | 41.6 | 5 | 41.6 | 4 | 33.3 |
| Indiana | 0 | 3 * | 0 | 3 * | |||
| S. venatus | |||||||
| Utah | 15 | 12 | 80.0 | 7 | 46.6 | 4 | 26.6 |
| Missouri | 16 | 6 | 37.5 | 5 | 31.2 | 2 | 12.5 |
| Indiana | 0 | 3 * | 0 | 3 * | |||
| S. minimus | |||||||
| Indiana | 0 | 3 * | 0 | 3 * | |||
| S. inaequalis | |||||||
| Indiana | 0 | 3 * | 0 | 3 * | |||
| S. cicatristriatus | |||||||
| Utah | 17 | 10 | 58.8 | 4 | 23.5 | 4 | 23.5 |
| S. phoeniciensis | |||||||
| Arizona | 2 | 2 | 100.0 | 2 | 100.0 | 1 | 50.0 |
| Listronotus maculicollis * | 0 | 1 * | 1 * | 1 * | |||
| Donus zoilus * | 0 | 1 * | 1 * | 1 * | |||
| Total sequences | 57 | 30 | 23 | ||||
| Avg % | 64.8 | 48.82 | 32.6 |
| Total of Larvae Identified Per Species | |||||||
| Location | Year | Grass Type | Total of Larvae Collected | Total of Larvae Sequenced | S. parvulus | S. venatus | S. minimus |
|---|---|---|---|---|---|---|---|
| Utah | 2018 | ||||||
| Cool | 24 | 17 | 15 | 2 | 0 | ||
| Warm | 0 | 0 | 0 | 0 | 0 | ||
| Indiana | 2020 | ||||||
| Cool | 63 | 41 | 28 | 5 | 8 | ||
| Warm | 51 | 36 | 33 | 3 * | |||
| Total | 138 | 94 | 76 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez-Soto, M.M.; Richmond, D.S.; Ramirez, R.A.; Xiong, X.; Enders, L.S. Characterizing Billbug (Sphenophorus spp.) Seasonal Biology Using DNA Barcodes and a Simple Morphometric Analysis. Insects 2021, 12, 930. https://doi.org/10.3390/insects12100930
Rodriguez-Soto MM, Richmond DS, Ramirez RA, Xiong X, Enders LS. Characterizing Billbug (Sphenophorus spp.) Seasonal Biology Using DNA Barcodes and a Simple Morphometric Analysis. Insects. 2021; 12(10):930. https://doi.org/10.3390/insects12100930
Chicago/Turabian StyleRodriguez-Soto, Marian M., Douglas S. Richmond, Ricardo A. Ramirez, Xi Xiong, and Laramy S. Enders. 2021. "Characterizing Billbug (Sphenophorus spp.) Seasonal Biology Using DNA Barcodes and a Simple Morphometric Analysis" Insects 12, no. 10: 930. https://doi.org/10.3390/insects12100930
APA StyleRodriguez-Soto, M. M., Richmond, D. S., Ramirez, R. A., Xiong, X., & Enders, L. S. (2021). Characterizing Billbug (Sphenophorus spp.) Seasonal Biology Using DNA Barcodes and a Simple Morphometric Analysis. Insects, 12(10), 930. https://doi.org/10.3390/insects12100930






