Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis
Abstract
1. Introduction
2. Materials and Methods
2.1. Honey Bee Larvae Inoculated with A. apis
2.2. Enzyme Activity Analysis
2.3. Chemicals and Extraction of Metabolites
2.4. UHPLC-QTOF-MS Analysis
2.5. Statistical Analyses
3. Results
3.1. Rates of Chalkbrood Symptoms
3.2. Effects of A. apis Infection on Gut Enzyme Activities
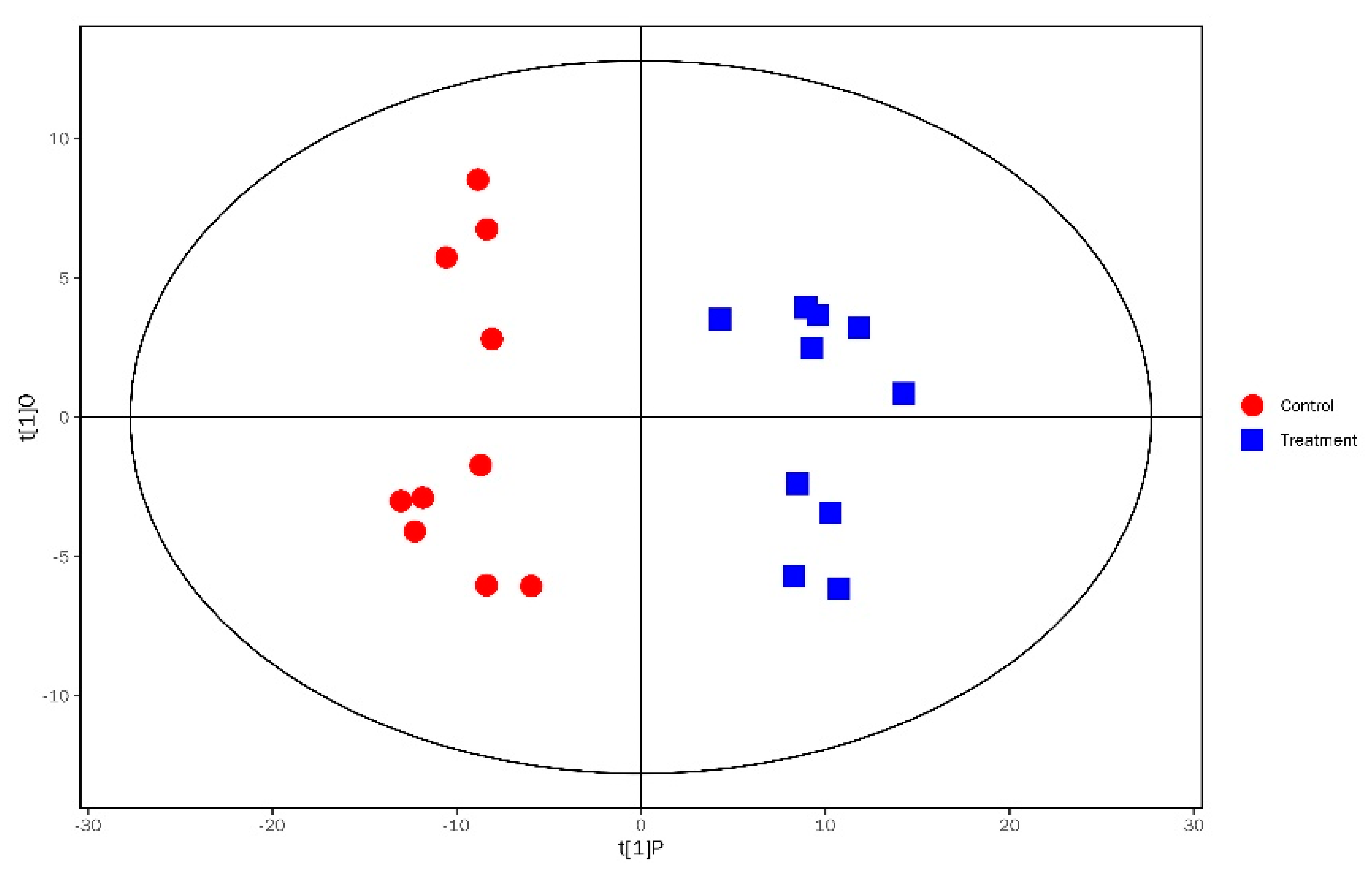
3.3. Analysis of Significantly Differential Metabolites
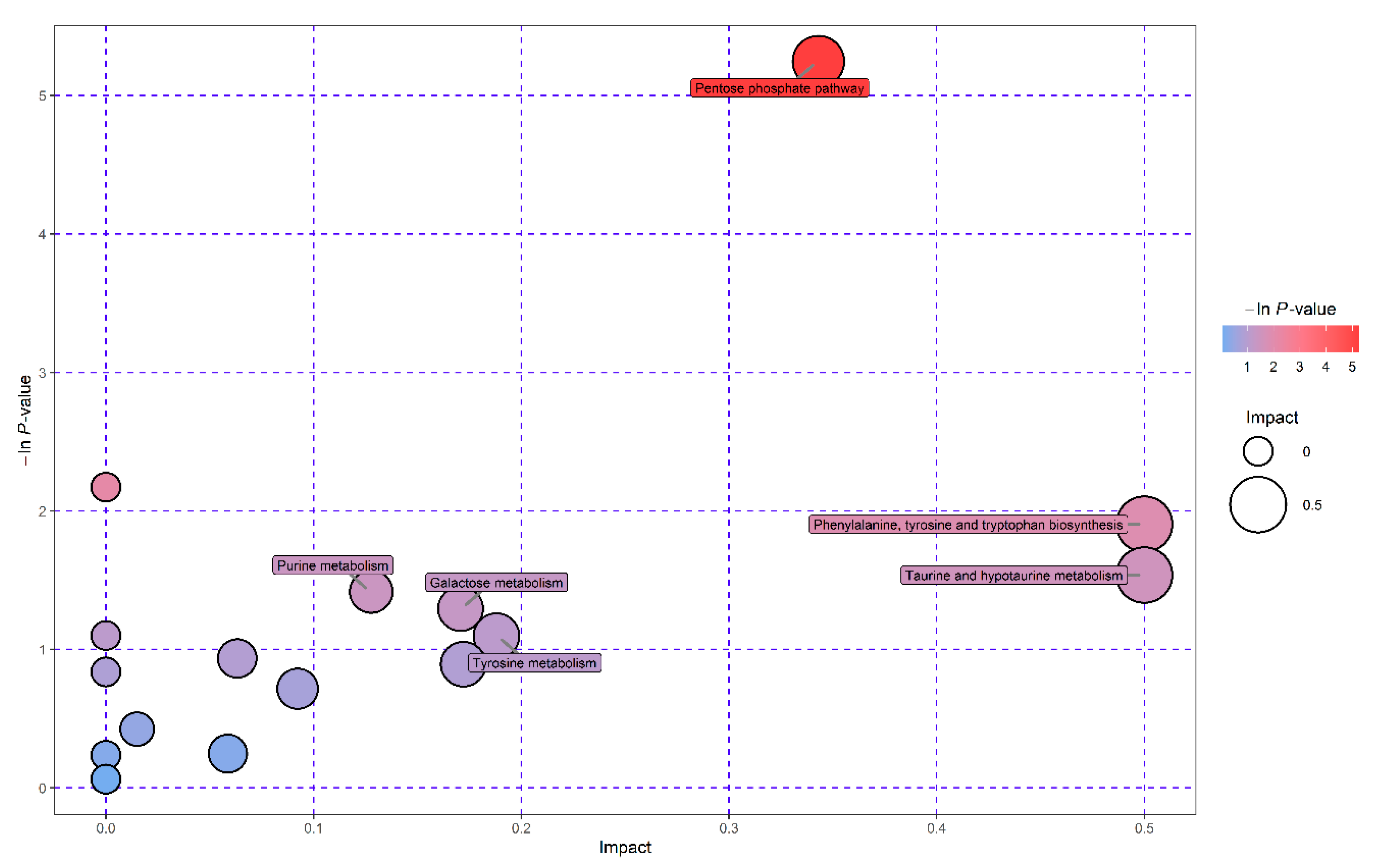
3.4. Pathway Enrichment Analysis of Significantly Differential Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, K.M.; Loh, E.H.; Rostal, M.K.; Zambrana-Torrelio, C.M.; Mendiola, L.; Daszak, P. Pathogens, pests, and economics: Drivers of honey bee colony declines and losses. EcoHealth 2013, 10, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Aronstein, K.A.; Murray, K.D. Chalkbrood disease in honey bees. J. Invertebr. Pathol. 2010, 103, S20–S29. [Google Scholar] [CrossRef] [PubMed]
- Holloway, B.; Sylvester, H.A.; Bourgeois, L.; Rinderer, T.E. Association of single nucleotide polymorphisms to resistance to chalkbrood in Apis mellifera. J. Apic. Res. 2012, 51, 154–163. [Google Scholar] [CrossRef]
- Jensen, A.B.; Aronstein, K.; Flores, J.M.; Vojvodic, S.; Palacio, M.A.; Spivak, M. Standard methods for fungal brood disease research. J. Apic. Res. 2013, 52, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Cornman, R.S.; Bennett, A.K.; Murray, K.D.; Evans, J.D.; Elsik, C.G.; Aronstein, K. Transcriptome analysis of the honey bee fungal pathogen, Ascosphaera apis: Implications for host pathogenesis. BMC Genom. 2012, 13, 285. [Google Scholar] [CrossRef]
- Matsumoto, H.; Nomura, S.; Hayakawa, Y. Changes of RNA virus infection rates and gut microbiota in young worker Apis mellifera (Hymenoptera: Apidae) of a chalkbrood-infected colony after a pollination task in a greenhouse. Appl. Entomol. Zool. 2014, 49, 395–402. [Google Scholar] [CrossRef]
- Li, Z.; Su, S.; Hamilton, M.; Yan, L.; Chen, Y. The ability to cause infection in a pathogenic fungus uncovers a new biological feature of honey bee viruses. J. Invertebr. Pathol. 2014, 120, 18–22. [Google Scholar] [CrossRef]
- Vojvodic, S.; Jensen, A.B.; James, R.; Boomsma, J.J.; Eilenberg, J. Temperature dependent virulence of obligate and facultative fungal pathogens of honeybee brood. Vet. Microbiol. 2011, 149, 200–205. [Google Scholar] [CrossRef]
- Aronstein, K.A.; Murray, K.D.; Saldivar, E. Transcriptional responses in honey bee larvae infected with chalkbrood fungus. BMC Genom. 2010, 11, 391. [Google Scholar] [CrossRef]
- Seehuus, S.-C.; Norberg, K.; Gimsa, U.; Krekling, T.; Amdam, G.V. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. USA 2006, 103, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Weirich, G.F.; Collins, A.M.; Williams, V.P. Antioxidant enzymes in the honey bee, Apis mellifera. Apidologie 2002, 33, 3–14. [Google Scholar] [CrossRef]
- Nikolić, T.V.; Purać, J.; Orčić, S.; Kojić, D.; Vujanović, D.; Stanimirović, Z.; Gržetić, I.; Ilijević, K.; Šikoparija, B.; Blagojević, D.P. Environmental effects on superoxide dismutase and catalase activity and expression in honey bee. Arch. Insect Biochem. 2015, 90, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Nikolić, T.V.; Kojić, D.; Orčić, S.; Batinić, D.; Vukašinović, E.; Blagojević, D.P.; Purać, J. The impact of sublethal concentrations of Cu, Pb and Cd on honey bee redox status, superoxide dismutase and catalase in laboratory conditions. Chemosphere 2016, 164, 98–105. [Google Scholar] [CrossRef]
- Collins, A.; Williams, V.; Evans, J. Sperm storage and antioxidative enzyme expression in the honey bee, Apis mellifera. Insect Mol. Biol. 2004, 13, 141–146. [Google Scholar] [CrossRef]
- Li, C.; Xu, B.; Wang, Y.; Yang, Z.; Yang, W. Protein content in larval diet affects adult longevity and antioxidant gene expression in honey bee workers. Entomol. Exp. Appl. 2014, 151, 19–26. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef]
- Culotta, V.C. Superoxide dismutase, oxidative stress, and cell metabolism. Curr. Top. Cell. Regul. 2001, 36, 117–132. [Google Scholar]
- Human, H.; Brodschneider, R.; Dietemann, V.; Dively, G.; Ellis, J.D.; Forsgren, E.; Fries, I.; Hatjina, F.; Hu, F.-L.; Jeffe, R.; et al. Miscellaneous standard methods for Apis mellifera research. J. Apic. Res. 2013, 52, 1–53. [Google Scholar] [CrossRef]
- James, R.; Skinner, J. PCR diagnostic methods for Ascosphaera infections in bees. J. Invertebr. Pathol. 2005, 90, 98–103. [Google Scholar] [CrossRef]
- Crailsheim, K.; Brodschneider, R.; Aupinel, P.; Behrens, D.; Genersch, E.; Vollmann, J.; Riessberger-Gallé, U. Standard methods for artificial rearing of Apis mellifera larvae. J. Apic. Res. 2013, 52, 1–16. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; He, J.; Zhao, X.; Chaimanee, V.; Huang, W.-F.; Nie, H.; Zhao, Y.; Su, S. Differential physiological effects of neonicotinoid insecticides on honey bees: A comparison between Apis mellifera and Apis cerana. Pestic. Biochem. Physiol. 2017, 140, 1–8. [Google Scholar] [CrossRef]
- Li, Z.; He, J.; Yu, T.; Chen, Y.; Huang, W.-F.; Huang, J.; Zhao, Y.; Nie, H.; Su, S. Transcriptional and physiological responses of hypopharyngeal glands in honeybees (Apis mellifera L.) infected by Nosema ceranae. Apidologie 2019, 50, 51–62. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Kuhl, C.; Tautenhahn, R.; Bottcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2011, 84, 283–289. [Google Scholar] [CrossRef]
- Sun, J.; Tower, J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol. Cell Biol. 1999, 19, 216–228. [Google Scholar] [CrossRef]
- Li, H.M.; Buczkowski, G.; Mittapalli, O.; Xie, J.; Wu, J.; Westerman, R.; Schemerhorn, B.; Murdock, L.; Pittendrigh, B. Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol. Biol. 2008, 17, 325–339. [Google Scholar] [CrossRef]
- Erlemann, K.-R.; Cossette, C.; Gravel, S.; Lesimple, A.; Lee, G.-J.; Saha, G.; Rokach, J.; Powell, W.S. Airway epithelial cells synthesize the lipid mediator 5-oxo-ETE in response to oxidative stress. Free Radic. Biol. Med. 2007, 42, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.S.; Rokach, J. Biochemistry, biology and chemistry of the 5-lipoxygenase product 5-oxo-ETE. Prog. Lipid Res. 2005, 44, 154–183. [Google Scholar] [CrossRef] [PubMed]
- Gurer, H.; Ozgunes, H.; Saygin, E.; Ercal, N. Antioxidant effect of taurine against lead-induced oxidative stress. Arch. Environ. Contam. Toxicol. 2001, 41, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Shiau, C.Y.; Chen, H.M.; Chiou, T.K. Antioxidant activities of carnosine, anserine, some free amino acids and their combination. J. Food Drug. Anal. 2003, 11, 148–153. [Google Scholar]
- Addis, P.; Shecterle, L.M.; St. Cyr, A.J. Cellular protection during oxidative stress: A potential role for D-ribose and antioxidants. J. Diet. Suppl. 2012, 9, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Gülcin, İ. Antioxidant and antiradical activities of L-carnitine. Life Sci. 2006, 78, 803–811. [Google Scholar] [CrossRef]
- Li, J.-L.; Wang, Q.-Y.; Luan, H.-Y.; Kang, Z.-C.; Wang, C.-B. Effects of L-carnitine against oxidative stress in human hepatocytes: Involvement of peroxisome proliferator-activated receptor alpha. J. Biomed. Sci. 2012, 19, 32. [Google Scholar] [CrossRef]
- Sánchez-Muros, M.-J.; Barroso, F.G.; Manzano-Agugliaro, F. Insect meal as renewable source of food for animal feeding: A review. J. Clean Prod. 2014, 65, 16–27. [Google Scholar] [CrossRef]
- Arien, Y.; Dag, A.; Zarchin, S.; Masci, T.; Shafir, S. Omega-3 deficiency impairs honey bee learning. Proc. Natl. Acad. Sci. USA 2015, 112, 15761–15766. [Google Scholar] [CrossRef]
- Rotstein, N.P.; Politi, L.E.; German, O.L.; Girotti, R. Protective effect of docosahexaenoic acid on oxidative stress-induced apoptosis of retina photoreceptors. Investig. Ophthalmol. Vis. Sci. 2003, 44, 2252–2259. [Google Scholar] [CrossRef]
- Barros, M.P.; Bechara, E.J. Luciferase and Urate may act as Antioxidant Defenses in Larval Pyrearinus termitilluminans (Elateridae: Coleoptera) During Natural Development and upon 20-Hydroxyecdysone Treatment. Photochem. Photobiol. 2000, 71, 648–654. [Google Scholar] [CrossRef]
- Wang, Y.; Kaftanoglu, O.; Brent, C.S.; Page, R.E.; Amdam, G.V. Starvation stress during larval development facilitates an adaptive response in adult worker honey bees (Apis mellifera L.). J. Exp. Biol. 2016, 219, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Read, H.W.; Balser, T.C. Reliability of muramic acid as a bacterial biomarker is influenced by methodological artifacts from streptomycin. Microb. Ecol. 2009, 57, 494–500. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Mowlds, P.; Coates, C.; Renwick, J.; Kavanagh, K. Dose-dependent cellular and humoral responses in Galleria mellonella larvae following β-glucan inoculation. Microbes Infect. 2010, 12, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Zunino, P.; Higes, M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef] [PubMed]
- Chaimanee, V.; Chantawannakul, P.; Chen, Y.; Evans, J.D.; Pettis, J.S. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J. Insect Physiol. 2012, 58, 1090–1095. [Google Scholar] [CrossRef]
- Jensen, A.B.; Pedersen, B.V.; Eilenberg, J. Differential susceptibility across honey bee colonies in larval chalkbrood resistance. Apidologie 2009, 40, 524–534. [Google Scholar] [CrossRef]
- Kuehne, A.; Emmert, H.; Soehle, J.; Winnefeld, M.; Fischer, F.; Wenck, H.; Gallinat, S.; Terstegen, L.; Lucius, R.; Hildebrand, J. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 2015, 59, 359–371. [Google Scholar] [CrossRef]
- Kumar, S.; Trivedi, P.K. Glutathione S-transferases: Role in combating abiotic stresses including arsenic detoxification in plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef]
- Slekar, K.H.; Kosman, D.J.; Culotta, V.C. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 1996, 271, 28831–28836. [Google Scholar] [CrossRef]
- Aruoma, O.; Halliwell, B.; Hoey, B.M.; Butler, J. The antioxidant action of taurine, hypotaurine and their metabolic precursors. Biochem. J. 1988, 256, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Nyhan, W.L. Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab. 2005, 86, 25–33. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A three-ring circus: Metabolism of the three proteogenic aromatic amino acids and their role in the health of plants and animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.J.; Hopkins, T.L. Tyrosine metabolism for insect cuticle tanning. Arch. Insect Biochem. Physiol. 1987, 6, 279–301. [Google Scholar] [CrossRef]
- Coelho, A.I.; Berry, G.T.; Rubio-Gozalbo, M.E. Galactose metabolism and health. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 422–427. [Google Scholar] [CrossRef] [PubMed]

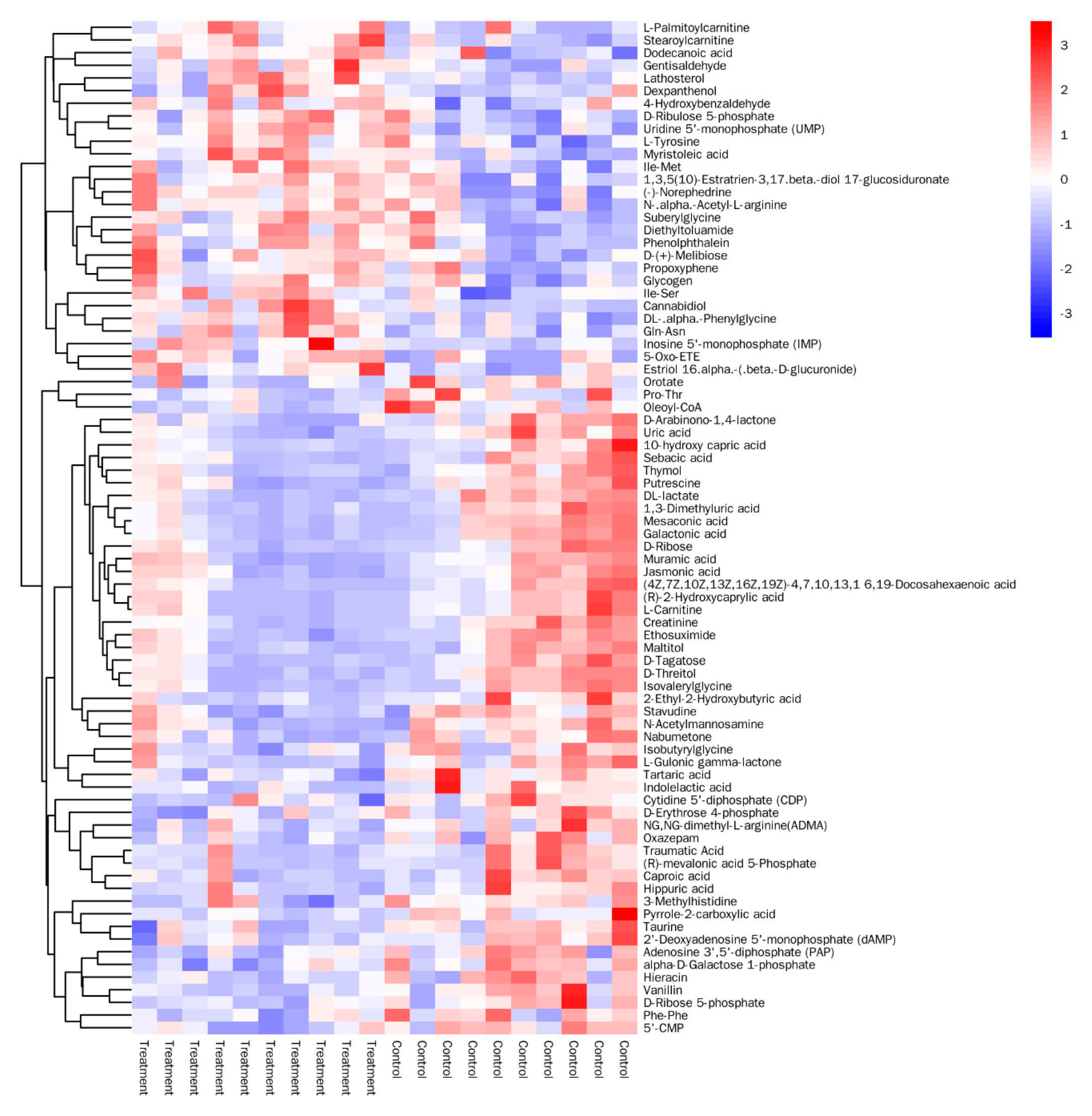
| Metabolites | HMDB ID | VIP Value | p Value | log2 Fold-Change |
|---|---|---|---|---|
| 10-hydroxy capric acid | / | 2.221866 | 0.040449 | −1.583220998 |
| Docosahexaenoic acid | HMDB02183 | 2.348188 | 0.018642 | −1.400723937 |
| DL-lactate | HMDB01311 | 2.32267 | 0.001646 | −1.383585277 |
| L-Carnitine | HMDB00062 | 2.366084 | 0.019981 | −1.249241729 |
| (R)-2-Hydroxycaprylic acid | HMDB0002264 | 2.320383 | 0.027084 | −1.238792434 |
| D-Threitol | HMDB04136 | 2.359414 | 0.001944 | −1.17578676 |
| Ethosuximide | HMDB14731 | 1.948189 | 0.007349 | −1.162173255 |
| D-Ribose | HMDB00283 | 2.181031 | 0.015077 | −1.158423407 |
| 2-Ethyl-2-Hydroxybutyric acid | HMDB01975 | 2.233223 | 0.017102 | −1.157991419 |
| Isovalerylglycine | HMDB00678 | 2.120458 | 0.008138 | −1.156552634 |
| (R)-mevalonic acid 5-Phosphate | HMDB01343 | 2.030673 | 0.02835 | −1.145439547 |
| D-Tagatose | HMDB03418 | 2.313209 | 0.009622 | −1.14421462 |
| Sebacic acid | HMDB00792 | 2.30288 | 0.017322 | −1.068341762 |
| 5-Oxo-ETE | HMDB0010217 | 1.463255 | 0.01796 | 1.059981866 |
| Traumatic Acid | HMDB00933 | 2.371702 | 0.02379 | −0.994244091 |
| Pro-Thr | HMDB0029027 | 1.823645 | 0.027625 | −0.954262735 |
| Cannabidiol | HMDB0014613 | 2.030682 | 0.008316 | 0.923946494 |
| Pyrrole-2-carboxylic acid | HMDB04230 | 2.205492 | 0.041314 | −0.900458683 |
| 1,3-Dimethyluric acid | HMDB01857 | 2.190309 | 0.008027 | −0.897637652 |
| Muramic acid | HMDB0003254 | 2.362325 | 0.019607 | −0.890529844 |
| Galactonic acid | HMDB00565 | 2.221866 | 0.003087 | −0.883735874 |
| Lathosterol | HMDB01170 | 1.710821 | 0.018031 | 0.876832978 |
| Indolelactic acid | HMDB00671 | 2.053832 | 0.01721 | −0.845847707 |
| Jasmonic acid | HMDB32797 | 2.261791 | 0.024457 | −0.822748259 |
| Hippuric acid | HMDB00714 | 2.139678 | 0.04853 | −0.813266898 |
| Creatinine | HMDB00562 | 2.372534 | 0.005277 | −0.781944734 |
| Stearoylcarnitine | HMDB00848 | 2.005948 | 0.004388 | 0.780781325 |
| Gln-Asn | HMDB0028792 | 1.732313 | 0.004701 | 0.778386048 |
| Maltitol | HMDB0002928 | 1.773791 | 0.023112 | −0.774603717 |
| 5′-CMP | HMDB0000095 | 1.934189 | 0.003266 | −0.762506922 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Hou, M.; Qiu, Y.; Zhao, B.; Nie, H.; Su, S. Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis. Insects 2020, 11, 419. https://doi.org/10.3390/insects11070419
Li Z, Hou M, Qiu Y, Zhao B, Nie H, Su S. Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis. Insects. 2020; 11(7):419. https://doi.org/10.3390/insects11070419
Chicago/Turabian StyleLi, Zhiguo, Mengshang Hou, Yuanmei Qiu, Bian Zhao, Hongyi Nie, and Songkun Su. 2020. "Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis" Insects 11, no. 7: 419. https://doi.org/10.3390/insects11070419
APA StyleLi, Z., Hou, M., Qiu, Y., Zhao, B., Nie, H., & Su, S. (2020). Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis. Insects, 11(7), 419. https://doi.org/10.3390/insects11070419




