Expansion of Imaginal Disc Growth Factor Gene Family in Diptera Reflects the Evolution of Novel Functions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transcriptome Preparation
2.2. Phylogenetic Analysis of IDGFs
2.3. Molecular Evolution Analysis
2.4. Gene-Level Approach Tests
2.5. Protein-Level Approach Tests
3. Results
3.1. Organization of Idgf Open Reading Frames and Genes
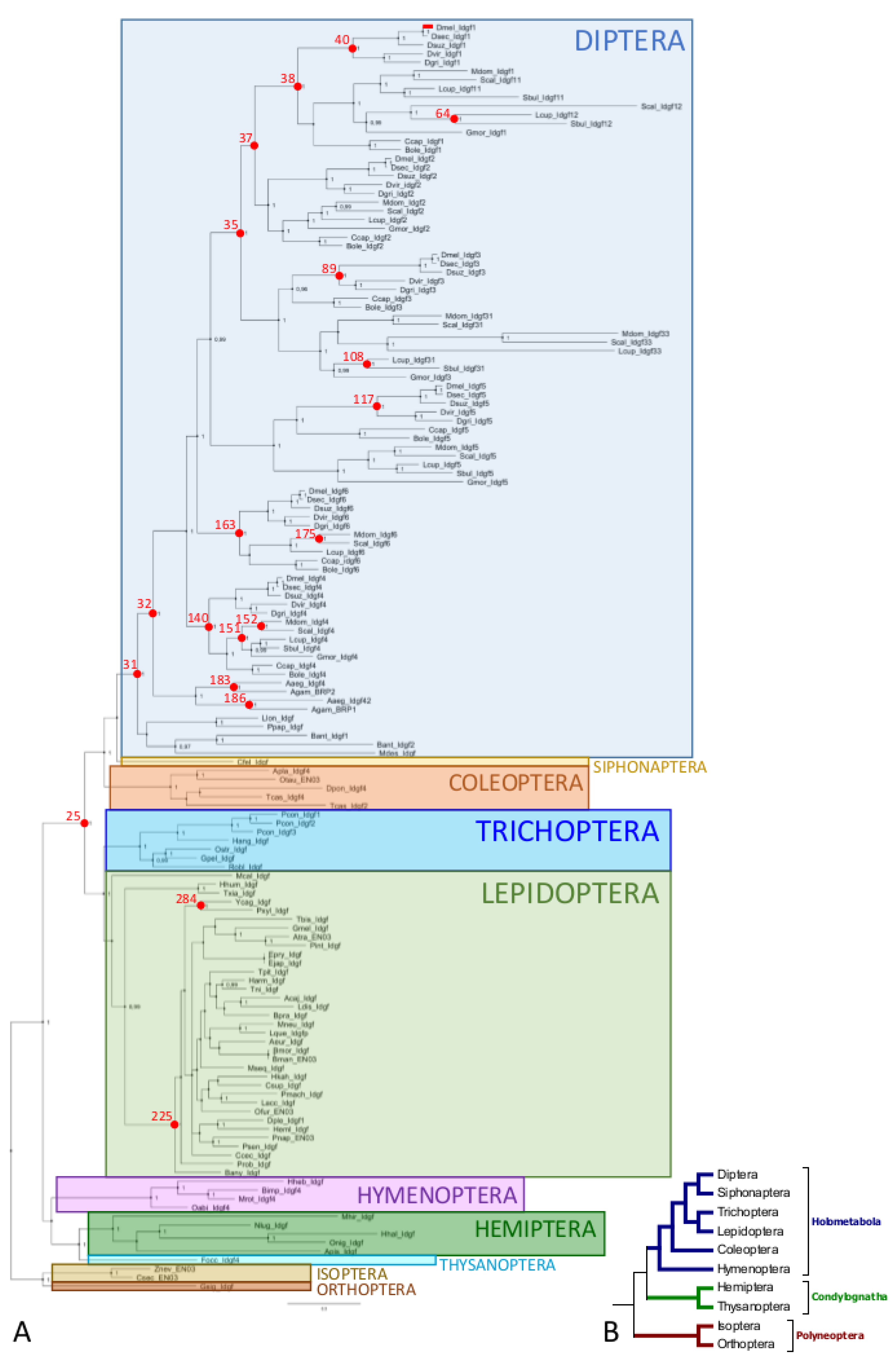
3.2. Phylogenetic Relationship of Idgfs
3.3. Diversification of Idgfs and Estimation of Evolutionary Rates
3.4. Positive Selection Participated in Radiation of the Idgf Gene Family in Diptera
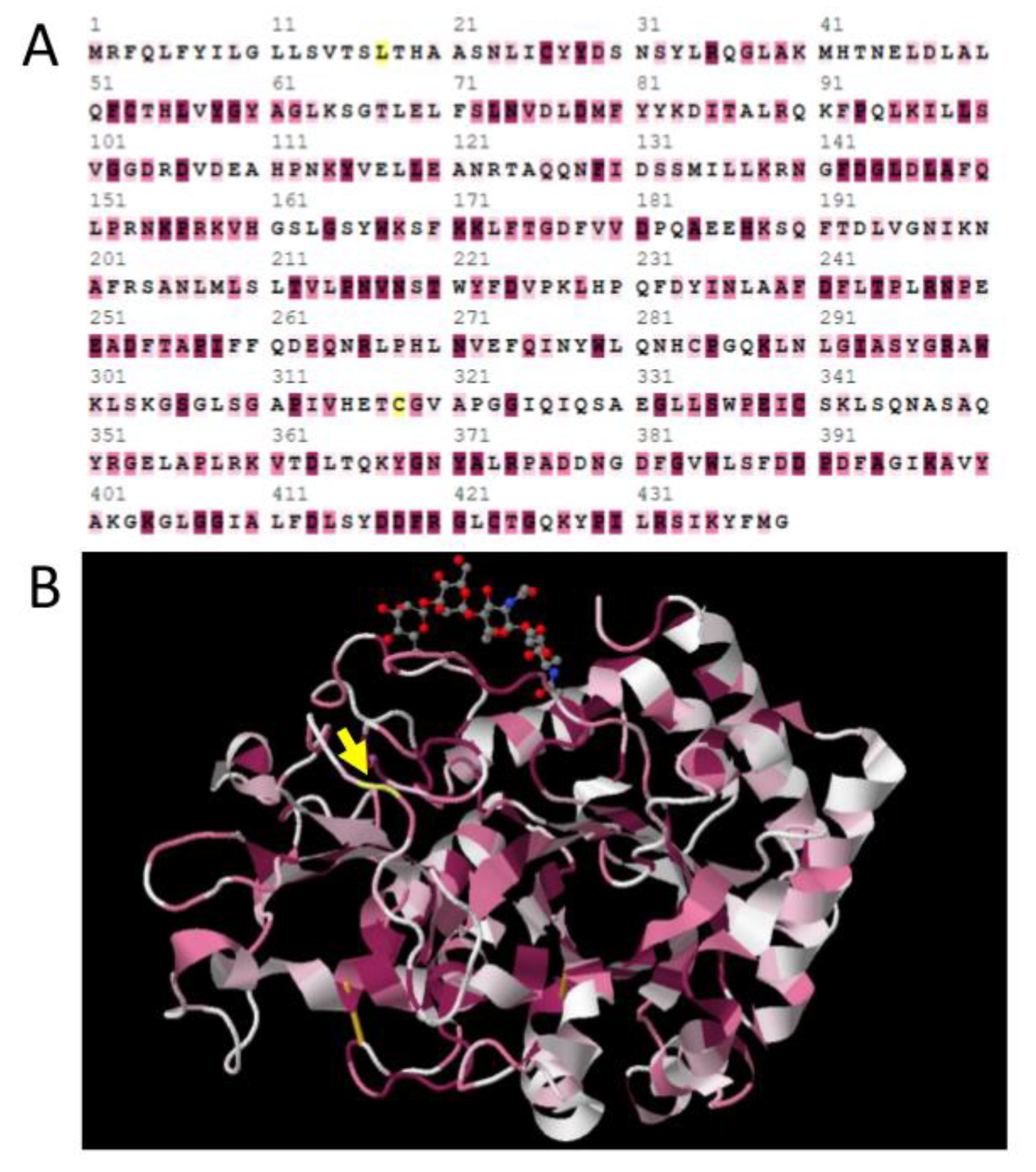
3.5. Negative Selection Preserves Conserved Structure of the 18 Glycosyl Hydrolase Domain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kawamura, K.; Shibata, T.; Saget, O.; Peel, D.; Bryant, P.J. A new family of growth factors produced by the fat body and active on Drosophila imaginal disc cells. Development 1999, 126, 211–219. [Google Scholar] [PubMed]
- Kirkpatrick, R.B.; Matico, R.E.; McNulty, D.E.; Strickler, J.E.; Rosenb, M. An abundantly secreted glycorotein from Drosophila melanogaster is related to mammalian secretory proteins produced in rheumatoid tissues and by activated macrophages. Gene 1995, 153, 147–154. [Google Scholar] [CrossRef]
- Broz, V.; Kucerova, L.; Rouhova, L.; Fleischmannova, J.; Strnad, H.; Bryant, P.J.; Zurovec, M. Drosophila imaginal disc growth factor 2 is a trophic factor involved in energy balance, detoxification, and innate immunity. Sci. Rep. 2017, 7, 43273. [Google Scholar] [CrossRef] [PubMed]
- Kucerova, L.; Broz, V.; Arefin, B.; Maaroufi, H.O.; Hurychova, J.; Strnad, H.; Zurovec, M.; Theopold, U. The Drosophila Chitinase-like protein IDGF3 is involved in protection against nematodes and in wound healing. J. Innate Immun. 2016, 8, 199–210. [Google Scholar] [CrossRef]
- Pesch, Y.Y.; Riedel, D.; Patil, K.R.; Loch, G.; Behr, M. Chitinases and imaginal disc growth factors organize the extracellular matrix formation at barrier tissues in insects. Sci. Rep. 2016, 6, 18340. [Google Scholar] [CrossRef]
- Shi, L.; Paskewitz, S.M. Identification and molecular characterization of two immune-responsive chitinase-like proteins from Anopheles gambiae. Insect Mol. Biol. 2004, 13, 387–398. [Google Scholar] [CrossRef]
- Uraki, R.; Hastings, A.K.; Brackney, D.E.; Armstrong, P.M.; Fikrig, E. AgBR1 antibodies delay lethal Aedes aegypti-borne West Nile virus infection in mice. NPJ Vaccines 2019, 4, 23. [Google Scholar] [CrossRef]
- Varela, P.F.; Llera, A.S.; Mariuzza, R.A.; Tormo, J. Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster. J. Biol. Chem. 2002, 277, 13229–13236. [Google Scholar] [CrossRef]
- Kanost, M.R.; Zepp, M.K.; Ladendorff, N.E.; Andersson, L.A. Isolation and characterization of a hemocyte aggregation inhibitor from hemolymph of Manduca sexta larvae. Arch. Insect Biochem. 1994, 27, 123–136. [Google Scholar] [CrossRef]
- Pan, Y.; Lu, P.; Wang, Y.; Yin, L.J.; Ma, H.X.; Ma, G.H.; Chen, K.P.; He, Y.Q. In silico identification of novel chitinase-like proteins in the silkworm, Bombyx mori, genome. J. Insect Sci. 2012, 12, 1–14. [Google Scholar] [CrossRef]
- Kludkiewicz, B.; Kucerova, L.; Konikova, T.; Strnad, H.; Hradilova, M.; Zaloudikova, A.; Sehadova, H.; Konik, P.; Sehnal, F.; Zurovec, M. The expansion of genes encoding soluble silk components in the greater wax moth, Galleria mellonella. Insect Biochem. Mol. Biol. 2019, 106, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Zurovcova, M.; Tatarenkov, A.; Berec, L. Differences in the pattern of evolution in six physically linked genes of Drosophila melanogaster. Gene 2006, 381, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Kucerova, L.; Zurovec, M.; Kludkiewicz, B.; Hradilova, M.; Strnad, H.; Sehnal, F. Modular structure, sequence diversification and appropriate nomenclature of seroins produced in the silk glands of Lepidoptera. Sci. Rep. 2019, 9, 3797. [Google Scholar] [CrossRef] [PubMed]
- Zurovec, M.; Yonemura, N.; Kludkiewicz, B.; Sehnal, F.; Kodrik, D.; Vieira, L.C.; Kucerova, L.; Strnad, H.; Konik, P.; Sehadova, H. Sericin composition in the silk of Antheraea yamamai. Biomacromolecules 2016, 17, 1776–1787. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- FigTree, Version 1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 4 October 2016).
- Stern, A.; Doron-Faigenboim, A.; Erez, E.; Martz, E.; Bacharach, E.; Pupko, T. Selecton 2007: Advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007, 35, W506–W511. [Google Scholar] [CrossRef] [PubMed]
- Doron-Faigenboim, A.; Stern, A.; Bacharach, E.; Pupko, T. Selecton: A server for detecting evolutionary forces at a single amino-acid site. Bioinformatics 2005, 21, 2101–2103. [Google Scholar] [CrossRef] [PubMed]
- Swanson, W.J.; Nielsen, R.; Yang, Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003, 20, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Pond, S.L.K.; Murrell, B.; Fourment, M.; Frost, S.D.W.; Delport, W.; Scheffler, K. A Random Effects Branch-Site Model for detecting episodic diversifying seection. Mol. Biol. Evol. 2011, 28, 3033–3043. [Google Scholar] [CrossRef]
- Smith, M.D.; Wertheim, J.O.; Weaver, S.; Murrell, B.; Scheffler, K.; Pond, S.L.K. Less is more: An Adaptive Branch-Site Random Effects Model for detection of episodic diversifying selection. Mol. Biol. Evol. 2015, 32, 1342–1353. [Google Scholar] [CrossRef]
- Wertheim, J.O.; Murrell, B.; Smith, M.D.; Kosakovsky Pond, S.L.; Scheffler, K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 2015, 32, 820–832. [Google Scholar] [CrossRef]
- McClellan, D.A.; McCracken, K.G. Estimating the influence of selection on the variable amino acid sites of the cytochrome B protein functional domains. Mol. Biol. Evol. 2001, 18, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Woolley, S.; Johnson, J.; Smith, M.J.; Crandall, K.A.; McClellan, D.A. TreeSAAP: Selection on amino acid properties using phylogenetic trees. Bioinformatics 2003, 19, 671–672. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernmor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- i5K Consortium. The i5K Initiative: Advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J. Hered 2013, 104, 595–600. [Google Scholar] [CrossRef]
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.W.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M.; et al. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef] [Green Version]
- Wahlberg, N.; Wheat, C.W.; Pena, C. Timing and patterns in the taxonomic diversification of Lepidoptera (butterflies and moths). PLoS ONE 2013, 8, e80875. [Google Scholar] [CrossRef]
- Nielsen, R. Statistical tests of selective neutrality in the age of genomics. Heredity 2001, 86, 641–647. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.H. Inference of selection from multiple species alignments. Curr. Opin. Genet. Dev. 2002, 12, 688–694. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Jmol: An Open-Source Java Viewer for Chemical Structures in 3D. Available online: https://www.webcitation.org/getfile?fileid=90d165802e6a28166f4628fb5502a4783e6cf685 (accessed on 23 March 2018).
- Dolezal, T.; Gazi, M.; Zurovec, M.; Bryant, P.J. Genetic analysis of the ADGF multigene family by homologous recombination and gene conversion in Drosophila. Genetics 2003, 165, 653–666. [Google Scholar] [PubMed]
- Gubb, D. Intron-Delay and the precision of expression of homeotic gene-products in Drosophila. Dev. Genet. 1986, 7, 119–131. [Google Scholar] [CrossRef]
- Troczka, B.J.; Richardson, E.; Homem, R.A.; Davies, T.G.E. An analysis of variability in genome organisation of intracellular calcium release channels across insect orders. Gene 2018, 670, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Xu, H.J.; Zhang, C.X. Chitinase-like gene family in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2015, 24, 29–40. [Google Scholar] [CrossRef]
- Omar, M.A.A.; Ao, Y.; Li, M.; He, K.; Xu, L.; Tong, H.; Jiang, M.; Li, F. The functional difference of eight chitinase genes between male and female of the cotton mealybug, Phenacoccus solenopsis. Insect Mol. Biol. 2019, 28, 550–567. [Google Scholar] [CrossRef]
- Wang, H.B.; Sakudoh, T.; Kawasaki, H.; Iwanaga, M.; Araki, K.; Fujimoto, H.; Takada, N.; Iwano, H.; Tsuchida, K. Purification and expression analysis of imaginal disc growth factor in the silkworm, Bombyx mori. J. Insect Physiol. 2009, 55, 1065–1071. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Iwami, M.; Sakurai, S. Ecdysteroid-inducible genes in the programmed cell death during insect metamorphosis. Insect Biochem. Mol. Biol. 2001, 31, 321–331. [Google Scholar] [CrossRef]
- Asgari, S.; Schmidt, O. Isolation of an imaginal disc growth factor homologue from Pieris rapae and its expression following parasitization by Cotesia rubecula. J. Insect Physiol. 2004, 50, 687–694. [Google Scholar] [CrossRef]
- Zhang, J.; Iwai, S.; Tsugehara, T.; Takeda, M. MbIDGF, a novel member of the imaginal disc growth factor family in Mamestra brassicae, stimulates cell proliferation in two lepidopteran cell lines without insulin. Insect Biochem. Mol. 2006, 36, 536–546. [Google Scholar] [CrossRef]
- Zhu, Q.S.; Arakane, Y.; Banerjee, D.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S. Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect Biochem. Mol. 2008, 38, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Li, Z.; Su, Y.; Zhao, Y.; Liu, L. Imaginal disc growth factor 4 regulates development and temperature adaptation in Bactrocera dorsalis. Sci. Rep. 2019, 9, 931. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Rubin, G.M.; Lemaitre, B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 2001, 98, 12590–12595. [Google Scholar] [CrossRef]
- Collin, M.A.; Mita, K.; Sehnal, F.; Hayashi, C.Y. Molecular evolution of lepidopteran silk proteins: Insights from the ghost moth, Hepialus californicus. J. Mol. Evol. 2010, 70, 519–529. [Google Scholar] [CrossRef]
- Zurovcova, M.; Ayala, F.J. Polymorphism patterns in two tightly linked developmental genes, Idgf1 and Idgf3, of Drosophila melanogaster. Genetics 2002, 162, 177–188. [Google Scholar]
- Li, H.; Greene, L.H. Sequence and structural analysis of the chitinase insertion domain reveals two conserved motifs involved in chitin-binding. PLoS ONE 2010, 5, e8654. [Google Scholar] [CrossRef]
- Zees, A.C.; Pyrpassopoulos, S.; Vorgias, C.E. Insights into the role of the (alpha plus beta) insertion in the TIM-barrel catalytic domain, regarding the stability and the enzymatic activity of Chitinase A from Serratia marcescens. BBA Proteins Proteom. 2009, 1794, 23–31. [Google Scholar] [CrossRef]

| Gene and Clade | Overall Mean p-Distance | S.E. |
|---|---|---|
| Schizophora * Idgf1 | 0.397 | 0.007 |
| Schizophora * Idgf2 | 0.311 | 0.007 |
| Schizophora * Idgf3 | 0.418 | 0.007 |
| Schizophora * Idgf4 | 0.261 | 0.007 |
| Schizophora * Idgf5 | 0.383 | 0.008 |
| Schizophora * Idgf6 | 0.270 | 0.007 |
| Lepidoptera ** Idgf | 0.293 | 0.007 |
| Noctuoidea *** Idgf | 0.229 | 0.008 |
| Gene and Clade | Datamonkey Selection Tests | TreeSAAP | |||||
|---|---|---|---|---|---|---|---|
| Site | Selecton | MEME | FEL | FUBAR | Property | Direction | |
| Schizophora Idgf1 | 345 | x | x | x | x | B Br p | neg pos pos |
| Schizophora Idgf5 | 31 | x | x | x | x | p Ra P | pos neg pos |
| Gene and Clade | RELAX | ||
|---|---|---|---|
| k | p | LR | |
| Schizophora Idgf1 | 1.8 | 0.085 ns | 2.97 |
| Schizophora Idgf2 | 0.38 | 1.000 ns | −4723.04 |
| 1.25 | 1.000 ns | −144.45 | |
| Schizophora Idgf3 | 1.01 | 1.000 ns | −0.04 |
| Schizophora Idgf4 | 4.36 | 1.000 ns | −3.12 |
| Schizophora Idgf5 | 1.00 | 0.953 ns | 0.00 |
| Schizophora Idgf6 | 1.74 | 0.214 ns | 1.55 |
| Lepidoptera | 1.21 | 0.004 ** | 8.11 |
| Species | Effect | Reference |
|---|---|---|
| Nilaparvata lugens | NiIdgf knockdown has no effect on morphology and survival | [48] |
| Phenacoccus solenopsis | PsIdgf knockdown has no effect on morphology and survival | [49] |
| Bombyx mori | BmIDGF is modulated in response to nutritional conditions | [50] |
| Bombyx mori | BmIDGF is induced by apoptosis or by ecdysone | [51] |
| Pieris rapae | IDGF is not affected by parasitization and polydnavirus infection | [52] |
| Mamestra brassicae | MbIDGF supports growth of two lepidopteran cell lines | [53] |
| Manduca sexta | In the presence of HAIP, cells in culture do not form aggregates | [9] |
| Tribolium castaneum | TcIdgf4 is involved in the molting process | [54] |
| Tribolium castaneum | TcIdgf2 knockdown has no phenotypic effect | [54] |
| Anopheles gambiae | AgBR1 and 2 are immune responsive proteins to bacteria | [6] |
| Aedes aegypti | AgBR1 influences mammalian host immune response | [7] |
| Bactrocera dorsalis | Idgf4 knockdown decreased larval survival under high temperature | [55] |
| Drosophila melanogaster | Idfg6 knockdown causes severe cuticular defects | [5] |
| Drosophila melanogaster | IDGF1,3,4,5, and 6 are required for chitin-ECM formation | [5] |
| Drosophila melanogaster | IDGF2 is induced by injury, supports growth of Cl.8 cells in vitro | [3] |
| Drosophila melanogaster | IDGF3 is needed for hemolymph clotting | [4] |
| Drosophila melanogaster | IDGF1 and 3 are involved in response to septic injury | [56] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zurovcova, M.; Benes, V.; Zurovec, M.; Kucerova, L. Expansion of Imaginal Disc Growth Factor Gene Family in Diptera Reflects the Evolution of Novel Functions. Insects 2019, 10, 365. https://doi.org/10.3390/insects10100365
Zurovcova M, Benes V, Zurovec M, Kucerova L. Expansion of Imaginal Disc Growth Factor Gene Family in Diptera Reflects the Evolution of Novel Functions. Insects. 2019; 10(10):365. https://doi.org/10.3390/insects10100365
Chicago/Turabian StyleZurovcova, Martina, Vladimir Benes, Michal Zurovec, and Lucie Kucerova. 2019. "Expansion of Imaginal Disc Growth Factor Gene Family in Diptera Reflects the Evolution of Novel Functions" Insects 10, no. 10: 365. https://doi.org/10.3390/insects10100365





