Aneuploid CTC and CEC
Abstract
:1. Background
2. Aneuploidy
3. CTC and CEC
4. Conventional Strategies and Current Progress in CTC Detection
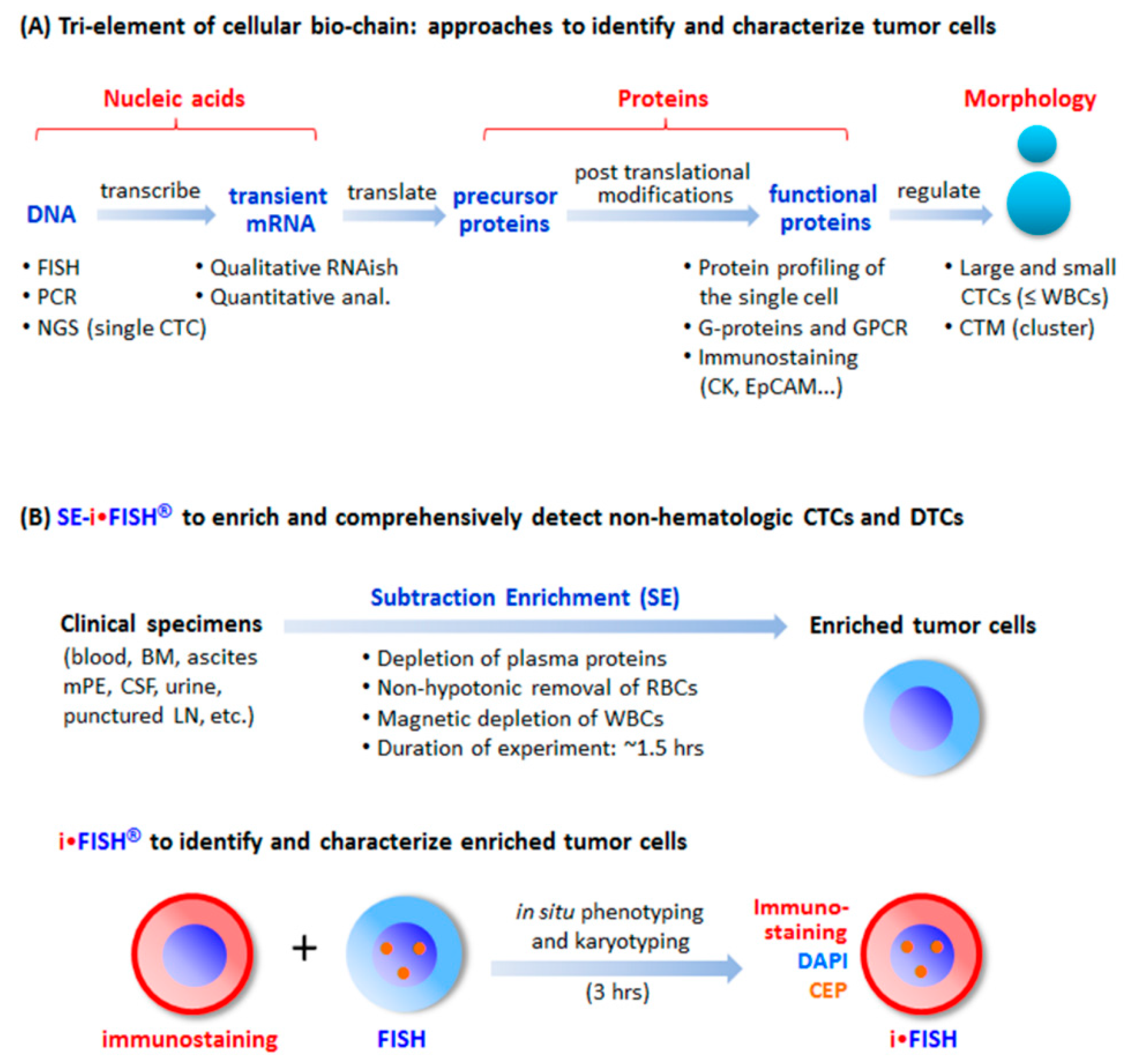
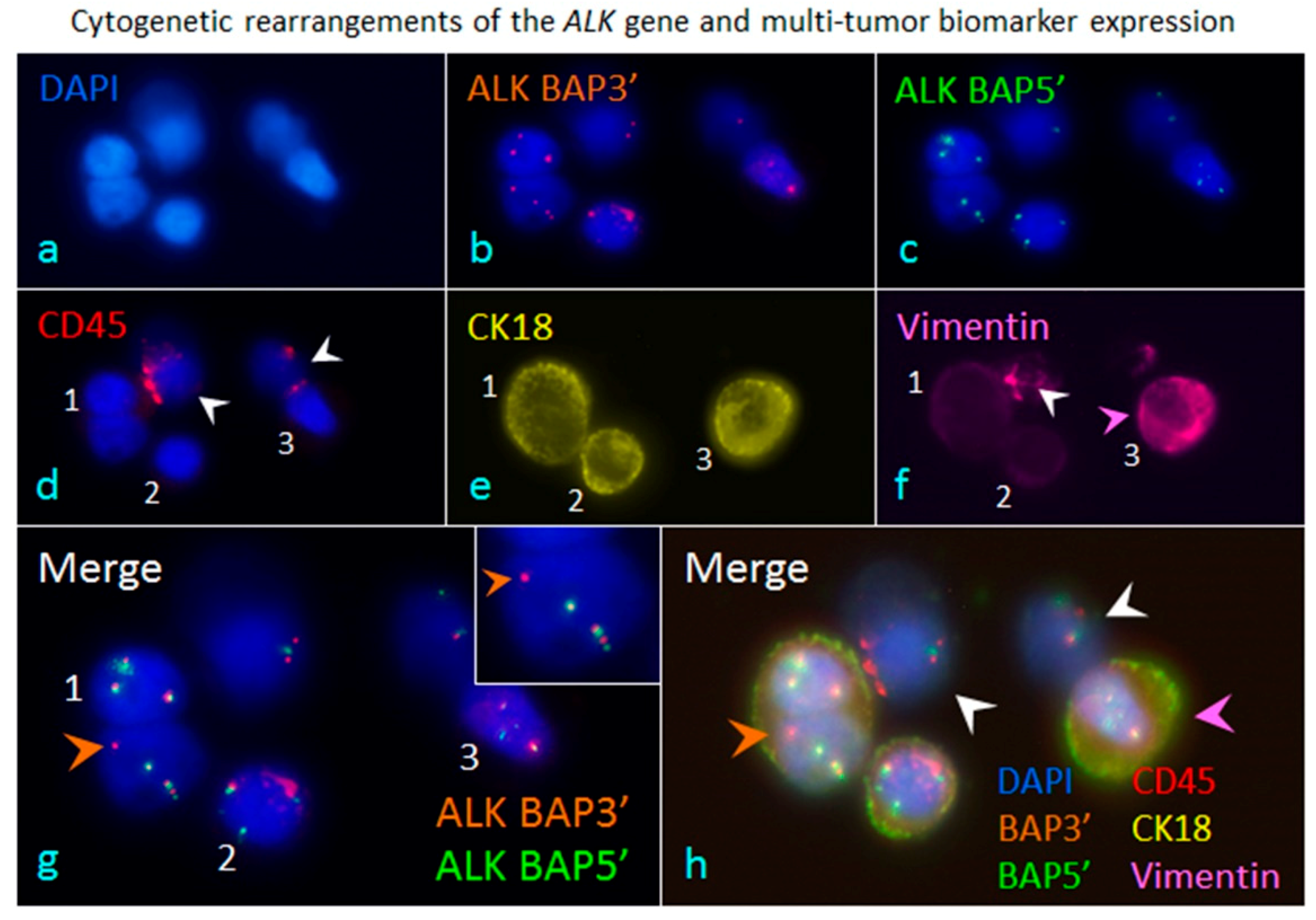
5. Comprehensive in Situ Phenotypic, Karyotypic, and Cytogenetic characterization as well as Classification of CTCs by SE-iFISH
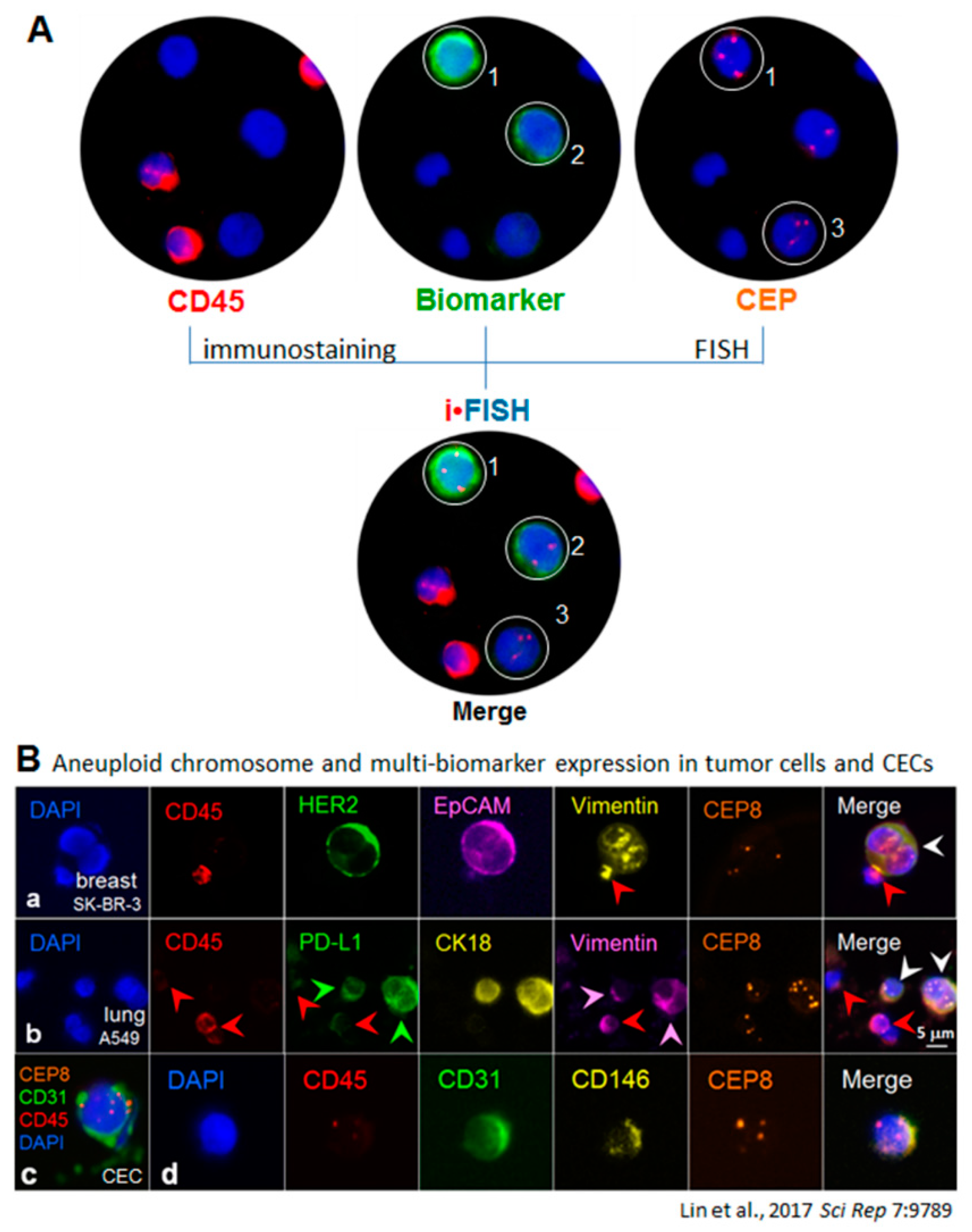
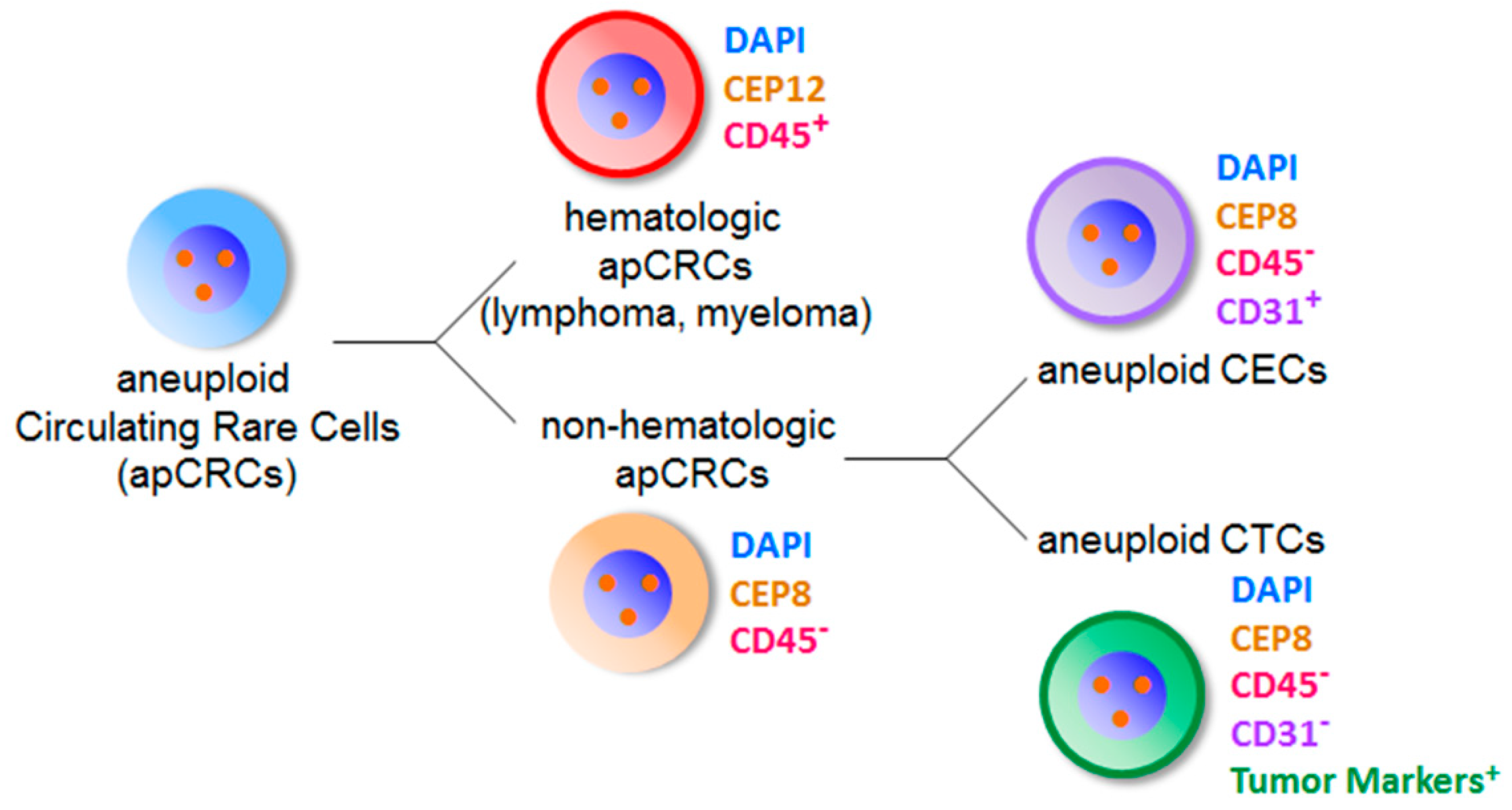
6. Co-Detection of Aneuploid CTCs and CECs
7. Proteomic and Genomic Profiling of Single Aneuploid CTCs and CECs
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Heller, G.; McCormack, R.; Kheoh, T.; Molina, A.; Smith, M.R.; Dreicer, R.; Saad, F.; de Wit, R.; Aftab, D.T.; Hirmand, M.; et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J. Clin. Oncol. 2018, 36, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Stoecklein, N.H.; Lin, P.P.; Gires, O. Circulating and disseminated tumor cells: Diagnostic tools and therapeutic targets in motion. Oncotarget 2017, 8, 1884–1912. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Shaked, Y.; Mancuso, P.; Kerbel, R.S. The multifaceted circulating endothelial cell in cancer: Towards marker and target identification. Nat. Rev. Cancer 2006, 6, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Cima, I.; Kong, S.L.; Sengupta, D.; Tan, I.B.; Phyo, W.M.; Lee, D.; Hu, M.; Iliescu, C.; Alexander, I.; Goh, W.L.; et al. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci. Transl. Med. 2016, 8, 345ra389. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.J.; Resio, B.; Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012, 13, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.; Weaver, B.A.; Cleveland, D.W. On the road to cancer: Aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 2005, 5, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.P.; Gires, O.; Wang, D.D.; Li, L.; Wang, H. Comprehensive in situ co-detection of aneuploid circulating endothelial and tumor cells. Sci. Rep. 2017, 7, 9789. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015, 16, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.A.; Cleveland, D.W. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006, 18, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.R.; Amon, A. Aneuploidy: Cancer’s fatal flaw? Cancer Res. 2009, 69, 5289–5291. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.P. Integrated EpCAM-independent subtraction enrichment and iFISH strategies to detect and classify disseminated and circulating tumors cells. Clin. Transl. Med. 2015, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Akino, T.; Hida, K.; Hida, Y.; Tsuchiya, K.; Freedman, D.; Muraki, C.; Ohga, N.; Matsuda, K.; Akiyama, K.; Harabayashi, T.; et al. Cytogenetic abnormalities of tumor-associated endothelial cells in human malignant tumors. Am. J. Pathol. 2009, 175, 2657–2667. [Google Scholar] [CrossRef] [PubMed]
- Hida, K.; Klagsbrun, M. A new perspective on tumor endothelial cells: Unexpected chromosome and centrosome abnormalities. Cancer Res. 2005, 65, 2507–2510. [Google Scholar] [CrossRef] [PubMed]
- Sen, S. Aneuploidy and cancer. Curr. Opin. Oncol. 2000, 12, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, L.; Swanton, C. The Role of Aneuploidy in Cancer Evolution. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Passerini, V.; Ozeri-Galai, E.; de Pagter, M.S.; Donnelly, N.; Schmalbrock, S.; Kloosterman, W.P.; Kerem, B.; Storchova, Z. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016, 7, 10754. [Google Scholar] [CrossRef] [PubMed]
- Durrbaum, M.; Storchova, Z. Effects of aneuploidy on gene expression: Implications for cancer. FEBS J. 2016, 283, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Manchado, E.; Malumbres, M. Targeting aneuploidy for cancer therapy. Cell 2011, 144, 465–466. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.C.; Williams, B.R.; Siegel, J.J.; Amon, A. Identification of aneuploidy-selective antiproliferation compounds. Cell 2011, 144, 499–512. [Google Scholar] [CrossRef]
- Pantel, K.; Brakenhoff, R.H.; Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer 2008, 8, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Zhang, X.T.; Ge, S.; Gao, J.; Gong, J.F.; Lu, M.; Zhang, Q.Y.; Cao, Y.S.; Wang, D.D.; Lin, P.P.; et al. Clinical significance of phenotyping and karyotyping of circulating tumor cells in patients with advanced gastric cancer. Oncotarget 2014, 5, 6594–6602. [Google Scholar] [CrossRef] [PubMed]
- Bayarri-Lara, C.; Ortega, F.G.; Cueto Ladron de Guevara, A.; Puche, J.L.; Ruiz Zafra, J.; de Miguel-Perez, D.; Ramos, A.S.; Giraldo-Ospina, C.F.; Navajas Gomez, J.A.; Delgado-Rodriguez, M.; et al. Circulating Tumor Cells Identify Early Recurrence in Patients with Non-Small Cell Lung Cancer Undergoing Radical Resection. PLoS ONE 2016, 11, e0148659. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Xu, J.; Zhang, A.; Wang, X.; Tang, R.; Zhang, X.; Yin, H.; Liu, M.; Wang, D.D.; et al. Quantified postsurgical small cell size CTCs and EpCAM(+) circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018, 412, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Gradilone, A.; Naso, G.; Raimondi, C.; Cortesi, E.; Gandini, O.; Vincenzi, B.; Saltarelli, R.; Chiapparino, E.; Spremberg, F.; Cristofanilli, M.; et al. Circulating tumor cells (CTCs) in metastatic breast cancer (MBC): Prognosis, drug resistance and phenotypic characterization. Ann. Oncol. 2011, 22, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, D.D.; Yang, M.; Chen, D.; Pang, L.; Guo, S.; Cai, J.; Wery, J.P.; Li, L.; Li, H.; et al. Comprehensive characterization of chemotherapeutic efficacy on metastases in the established gastric neuroendocrine cancer patient derived xenograft model. Oncotarget 2015, 6, 15639–15651. [Google Scholar] [CrossRef] [PubMed]
- Boos, C.J.; Lip, G.Y.; Blann, A.D. Circulating endothelial cells in cardiovascular disease. J. Am. Coll. Cardiol. 2006, 48, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, H.D.; Namdarian, B.; Corcoran, N.M.; Costello, A.J.; Hovens, C.M. Circulating endothelial cells as biomarkers of prostate cancer. Nat. Rev. Urol. 2008, 5, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 2002, 29, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, P.; Burlini, A.; Pruneri, G.; Goldhirsch, A.; Martinelli, G.; Bertolini, F. Resting and activated endothelial cells are increased in the peripheral blood of cancer patients. Blood 2001, 97, 3658–3661. [Google Scholar] [CrossRef] [PubMed]
- Beerepoot, L.V.; Mehra, N.; Vermaat, J.S.; Zonnenberg, B.A.; Gebbink, M.F.; Voest, E.E. Increased levels of viable circulating endothelial cells are an indicator of progressive disease in cancer patients. Ann. Oncol. 2004, 15, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Kawaishi, M.; Fujiwara, Y.; Fukui, T.; Kato, T.; Yamada, K.; Ohe, Y.; Kunitoh, H.; Sekine, I.; Yamamoto, N.; Nokihara, H.; et al. Circulating endothelial cells in non-small cell lung cancer patients treated with carboplatin and paclitaxel. J. Thorac. Oncol. 2009, 4, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.K.; Lee, H.J.; Choi, H.N.; Ahn, J.H.; Choi, J.Y.; Song, H.S.; Lee, K.H.; Yoon, Y.; Yi, L.S.; Kim, J.S.; et al. Characterization of CD45-/CD31+/CD105+ circulating cells in the peripheral blood of patients with gynecologic malignancies. Clin. Cancer Res. 2013, 19, 5340–5350. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Went, P.T.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam protein expression in human carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Stoecklein, N.H. Dynamic EpCAM expression on circulating and disseminating tumor cells: Causes and consequences. Cell Mol. Life Sci. 2014, 71, 4393–4402. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Zhang, H.; Wang, D.D.; Li, L.; Lin, P.P. Enhanced detection and comprehensive in situ phenotypic characterization of circulating and disseminated heteroploid epithelial and glioma tumor cells. Oncotarget 2015, 6, 27049–27064. [Google Scholar] [CrossRef] [PubMed]
- Driemel, C.; Kremling, H.; Schumacher, S.; Will, D.; Wolters, J.; Lindenlauf, N.; Mack, B.; Baldus, S.A.; Hoya, V.; Pietsch, J.M.; et al. Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene 2014, 33, 4904–4915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The Identification and Characterization of Breast Cancer CTCs Competent for Brain Metastasis. Sci. Transl. Med. 2013, 5, 189er185. [Google Scholar] [CrossRef] [PubMed]
- Grover, P.K.; Cummins, A.G.; Price, T.J.; Roberts-Thomson, I.C.; Hardingham, J.E. Circulating tumour cells: The evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann. Oncol. 2014, 25, 1506–1516. [Google Scholar] [CrossRef] [PubMed]
- Mikolajczyk, S.D.; Millar, L.S.; Tsinberg, P.; Coutts, S.M.; Zomorrodi, M.; Pham, T.; Bischoff, F.Z.; Pircher, T.J. Detection of EpCAM-Negative and Cytokeratin-Negative Circulating Tumor Cells in Peripheral Blood. J. Oncol. 2011, 2011, 252361. [Google Scholar] [CrossRef] [PubMed]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.; Schutze, K.; Capron, F.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef]
- Ito, H.; Inoue, H.; Kimura, S.; Ohmori, T.; Ishikawa, F.; Gohda, K.; Sato, J. Prognostic impact of the number of viable circulating cells with high telomerase activity in gastric cancer patients: A prospective study. Int. J. Oncol. 2014, 45, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Alunni-Fabbroni, M.; Sandri, M.T. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods 2010, 50, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.; van Dalum, G.; Beck, M.; Terstappen, L.W. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS ONE 2013, 8, e61770. [Google Scholar] [CrossRef] [PubMed]
- Willipinski-Stapelfeldt, B.; Riethdorf, S.; Assmann, V.; Woelfle, U.; Rau, T.; Sauter, G.; Heukeshoven, J.; Pantel, K. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin. Cancer Res. 2005, 11, 8006–8014. [Google Scholar] [CrossRef] [PubMed]
- Pfitzner, C.; Schroder, I.; Scheungraber, C.; Dogan, A.; Runnebaum, I.B.; Durst, M.; Hafner, N. Digital-Direct-RT-PCR: A sensitive and specific method for quantification of CTC in patients with cervical carcinoma. Sci. Rep. 2014, 4, 3970. [Google Scholar] [CrossRef] [PubMed]
- Palmirotta, R.; Lovero, D.; Silvestris, E.; Felici, C.; Quaresmini, D.; Cafforio, P.; Silvestris, F. Next-generation Sequencing (NGS) Analysis on Single Circulating Tumor Cells (CTCs) with No Need of Whole-genome Amplification (WGA). Cancer Genom. Proteom. 2017, 14, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Yee, S.S.; Lieberman, D.B.; Blanchard, T.; Rader, J.; Zhao, J.; Troxel, A.B.; DeSloover, D.; Fox, A.J.; Daber, R.D.; Kakrecha, B.; et al. A novel approach for next-generation sequencing of circulating tumor cells. Mol. Genet. Genom. Med. 2016, 4, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Katz, R.L.; He, W.; Khanna, A.; Fernandez, R.L.; Zaidi, T.M.; Krebs, M.; Caraway, N.P.; Zhang, H.Z.; Jiang, F.; Spitz, M.R.; et al. Genetically abnormal circulating cells in lung cancer patients: An antigen-independent fluorescence in situ hybridization-based case-control study. Clin. Cancer Res. 2010, 16, 3976–3987. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Bischoff, F.Z.; Mayer, J.A.; Wong, K.L.; Pham, T.; Bottsford-Miller, J.; Stone, R.L.; Lin, Y.G.; Jaladurgam, P.; Roh, J.W.; et al. A novel platform for detection of CK+ and CK− CTCs. Cancer Discov. 2011, 1, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Barbazan, J.; Dunkel, Y.; Li, H.; Nitsche, U.; Janssen, K.P.; Messer, K.; Ghosh, P. Prognostic Impact of Modulators of G proteins in Circulating Tumor Cells from Patients with Metastatic Colorectal Cancer. Sci. Rep. 2016, 6, 22112. [Google Scholar] [CrossRef] [PubMed]
- Payne, R.E.; Wang, F.; Su, N.; Krell, J.; Zebrowski, A.; Yague, E.; Ma, X.J.; Luo, Y.; Coombes, R.C. Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br. J. Cancer 2012, 106, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Sinkala, E.; Sollier-Christen, E.; Renier, C.; Rosas-Canyelles, E.; Che, J.; Heirich, K.; Duncombe, T.A.; Vlassakis, J.; Yamauchi, K.A.; Huang, H.; et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 2017, 8, 14622. [Google Scholar] [CrossRef] [PubMed]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef] [PubMed]
- O’Hayre, M.; Vazquez-Prado, J.; Kufareva, I.; Stawiski, E.W.; Handel, T.M.; Seshagiri, S.; Gutkind, J.S. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 2013, 13, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Fischer, T.; Lavoie, C.; Huang, H.; Farquhar, M.G. Calnuc plays a role in dynamic distribution of Gαi but not Gβ subunits and modulates ACTH secretion in AtT-20 neuroendocrine secretory cells. Mol. Neurodegener. 2009, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Yao, Y.; Hofmeister, R.; Tsien, R.Y.; Farquhar, M.G. Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J. Cell Biol. 1999, 145, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Hao, H.; Li, L.; Zhou, X.; Guo, Z.; Zhang, L.; Zhang, X.; Zhong, W.; Guo, H.; Bremner, R.M.; et al. Preliminary investigation of the clinical significance of detecting circulating tumor cells enriched from lung cancer patients. J. Thorac. Oncol. 2009, 4, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Ran, R.; Li, L.; Wang, M.; Wang, S.; Zheng, Z.; Lin, P.P. Determination of EGFR mutations in single cells microdissected from enriched lung tumor cells in peripheral blood. Anal. Bioanal. Chem. 2013, 405, 7377–7382. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Ding, J.; Wang, M.; Li, N.; Yang, H.; Wang, K.; Wang, D.; Lin, P.P.; Li, M.; et al. Clinical significance of detecting CSF-derived tumor cells in breast cancer patients with leptomeningeal metastasis. Oncotarget 2018, 9, 2705–2714. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Fischer, T.; Weiss, T.; Farquhar, M.G. Calnuc, an EF-hand Ca2+ binding protein, specifically interacts with the C-terminal α5-helix of Gαi3. Proc. Natl. Acad. Sci. USA 2000, 97, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, I.; Planchard, D. ALK inhibitors in non-small cell lung cancer: The latest evidence and developments. Ther. Adv. Med. Oncol. 2016, 8, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Swennenhuis, J.F.; Tibbe, A.G.; Levink, R.; Sipkema, R.C.; Terstappen, L.W. Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytom. Part A 2009, 75, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Hida, K.; Hida, Y.; Amin, D.N.; Flint, A.F.; Panigrahy, D.; Morton, C.C.; Klagsbrun, M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004, 64, 8249–8255. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, A.; Achatz, M.I.; Borresen-Dale, A.L.; Hainaut, P.; Olivier, M. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 2007, 26, 2157–2165. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, K.J.; McDonald, E.R., 3rd; Schlabach, M.R.; Billy, E.; Hoffman, G.R.; deWeck, A.; Ruddy, D.A.; Venkatesan, K.; Yu, J.; McAllister, G.; et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 2016, 351, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Kerbel, R.S.; Yu, J.; Tran, J.; Man, S.; Viloria-Petit, A.; Klement, G.; Coomber, B.L.; Rak, J. Possible mechanisms of acquired resistance to anti-angiogenic drugs: Implications for the use of combination therapy approaches. Cancer Metast. Rev. 2001, 20, 79–86. [Google Scholar] [CrossRef]
- Bussolati, B.; Deambrosis, I.; Russo, S.; Deregibus, M.C.; Camussi, G. Altered angiogenesis and survival in human tumor-derived endothelial cells. FASEB J. 2003, 17, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, P.P. Aneuploid CTC and CEC. Diagnostics 2018, 8, 26. https://doi.org/10.3390/diagnostics8020026
Lin PP. Aneuploid CTC and CEC. Diagnostics. 2018; 8(2):26. https://doi.org/10.3390/diagnostics8020026
Chicago/Turabian StyleLin, Peter Ping. 2018. "Aneuploid CTC and CEC" Diagnostics 8, no. 2: 26. https://doi.org/10.3390/diagnostics8020026
APA StyleLin, P. P. (2018). Aneuploid CTC and CEC. Diagnostics, 8(2), 26. https://doi.org/10.3390/diagnostics8020026




