Adaptive Evolutionary Optimization of Deep Learning Architectures for Focused Liver Ultrasound Image Segmentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. Evolutionary Genomic Optimization
- Genome Representation: We represent each genome as a dictionary containing the following hyperparameters:
- ▪
- Dropout Rate: , where .
- ▪
- Filter Sizes: a list of integers representing the number of filters in each layer (see Table 1), .
- ▪
- Depth: d, representing the number of layers, where d ∈ [2, 5].
- ▪
- Use Skip Connections: a Boolean flag , indicating whether skip connections are included.
- Fitness Function: the fitness of each U-Net genome is evaluated based on the Dice coefficient:
- Selection: The population is sorted based on fitness scores, and the top half of the genomes is retained for the next generation. The selected genome can be represented as
- Crossover: Two parent genomes are randomly selected to produce offspring through the following rules:
- ▪
- The dropout rate and depth are averaged:
- ▪
- The filter sizes are averaged and rounded to the nearest integer:
- ▪
- The skip connection flag is randomly selected from the parents.
- Migration: Facilitates the transfer of knowledge between subpopulations:
- ▪
- After every few generations, a certain percentage of genomes are migrated between subpopulations. This can be represented as
- ▪
- This influences the fitness evaluation and crossover processes.
- Mutation: Random mutations are applied to introduce variability:
- ▪
- With a probability of 10%, the dropout rate is perturbed:
- ▪
- With a probability of 10%, each filter size is adjusted by ±8 filters, ensuring the values stay within the valid range:
- Boundary Constraints: To maintain valid parameter ranges, we apply clipping for the filter sizes and dropout rates:
- Depth Penalization: We also penalize deeper networks to prevent overfitting and manage the trade-off between model complexity and performance (avoiding extra training parameters):where d: depth and p: penalty factor.
- Convergence Criteria: We run the algorithm for a predefined number of generations (e.g., 30 epochs), gen, or until the change in average fitness across generations is below a threshold :
- Training Process
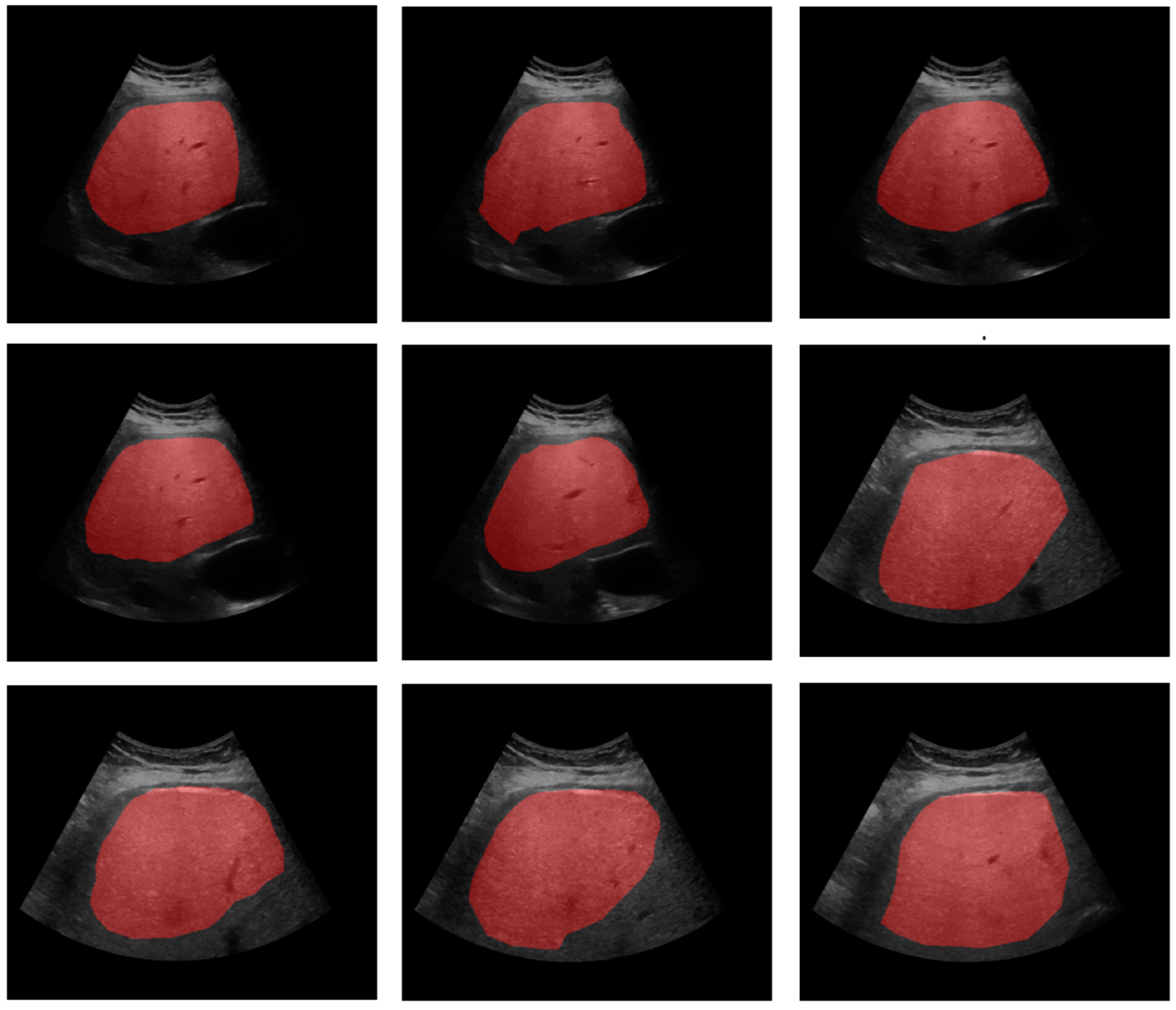
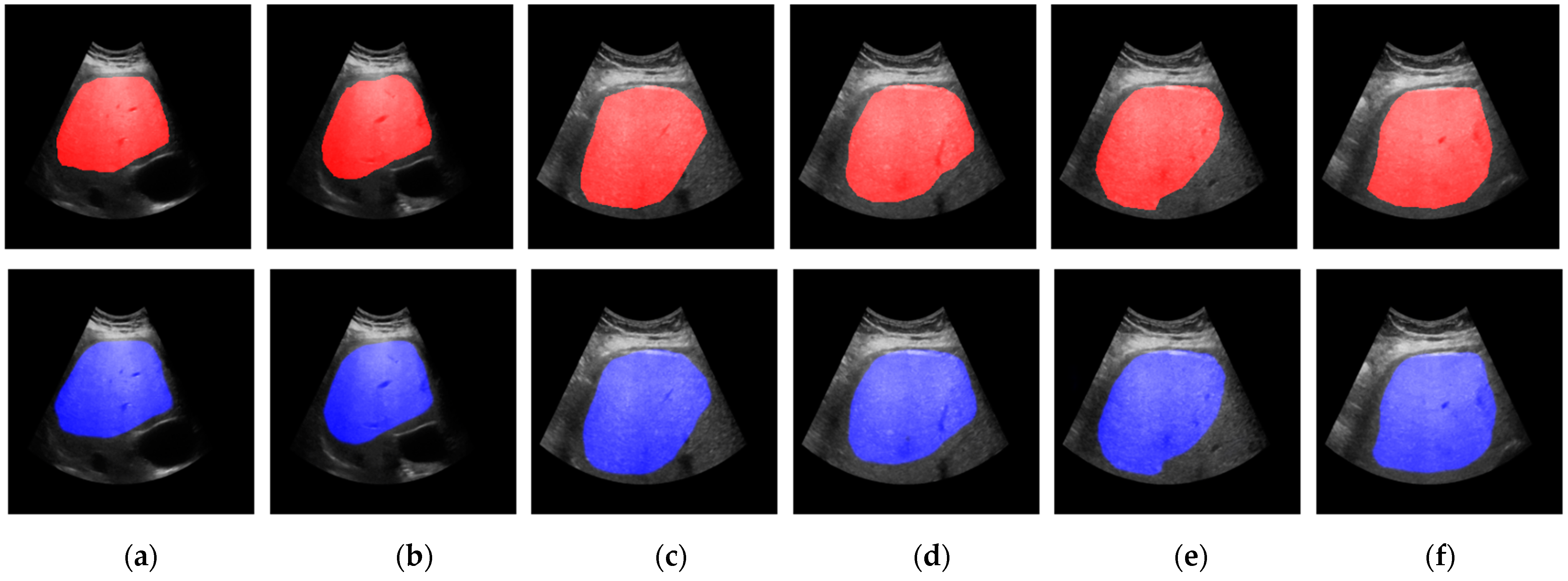
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ibrahim, M.N.; Blázquez-García, R.; Lightstone, A.; Meng, F.; Bhat, M.; El Kaffas, A.; Ukwatta, E. Automated fatty liver disease detection in point-of-care ultrasound B-mode images. J. Med. Imaging (Bellingham) 2023, 10, 034505. [Google Scholar] [CrossRef] [PubMed]
- Engin, A. Nonalcoholic Fatty Liver Disease and Staging of Hepatic Fibrosis. Adv. Exp. Med. Biol. 2024, 1460, 539–574. [Google Scholar] [CrossRef]
- Pouwels, S.; Sakran, N.; Graham, Y.; Leal, A.; Pintar, T.; Yang, W.; Kassir, R.; Singhal, R.; Mahawar, K.; Ramnarain, D. Non-alcoholic fatty liver disease (NAFLD): A review of pathophysiology, clinical management and effects of weight loss. BMC Endocr. Disord. 2022, 22, 63. [Google Scholar] [CrossRef]
- Powell, E.E.; Wong, V.W.-S.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, J.B.; Deutsch, R.; Kahen, T.; Lavine, J.E.; Stanley, C.; Behling, C. Prevalence of fatty liver in children and adolescents. Pediatrics 2006, 118, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Starekova, J.; Hernando, D.; Pickhardt, P.J.; Reeder, S.B. Quantification of Liver Fat Content with CT and MRI: State of the Art. Radiology 2021, 301, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Raptis, D.A.; Fischer, M.A.; Graf, R.; Nanz, D.; Weber, A.; Moritz, W.; Tian, Y.; Oberkofler, C.E.; Clavien, P.A. MRI: The new reference standard in quantifying hepatic steatosis? Gut 2012, 61, 117–127. [Google Scholar] [CrossRef]
- Bresnahan, R.; Duarte, R.; Mahon, J.; Beale, S.; Chaplin, M.; Bhattacharyya, D.; Houten, R.; Edwards, K.; Nevitt, S.; Maden, M.; et al. Diagnostic accuracy and clinical impact of MRI-based technologies for patients with non-alcoholic fatty liver disease: Systematic review and economic evaluation. Health Technol. Assess 2023, 27, 1–115. [Google Scholar] [CrossRef]
- Kuroda, H.; Oguri, T.; Kamiyama, N.; Toyoda, H.; Yasuda, S.; Imajo, K.; Suzuki, Y.; Sugimoto, K.; Akita, T.; Tanaka, J.; et al. Multivariable Quantitative US Parameters for Assessing Hepatic Steatosis. Radiology 2023, 309, e230341. [Google Scholar] [CrossRef]
- Paige, J.S.; Bernstein, G.S.; Heba, E.; Costa, E.A.C.; Fereirra, M.; Wolfson, T.; Gamst, A.C.; Valasek, M.A.; Lin, G.Y.; Han, A.; et al. A Pilot Comparative Study of Quantitative Ultrasound, Conventional Ultrasound, and MRI for Predicting Histology-Determined Steatosis Grade in Adult Nonalcoholic Fatty Liver Disease. AJR Am. J. Roentgenol. 2017, 208, W168–W177. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, A.; Kumar, V.; Pierce, T.T.; Li, Q.; Baikpour, M.; Rosado-Mendez, I.; Wang, M.; Guo, P.; Schoen, S.; Gu, Y.; et al. The Future Is Beyond Bright: The Evolving Role of Quantitative US for Fatty Liver Disease. Radiology 2023, 309, e223146. [Google Scholar] [CrossRef] [PubMed]
- Kadi, D.; Loomba, R.; Bashir, M.R. Diagnosis and Monitoring of Nonalcoholic Steatohepatitis: Current State and Future Directions. Radiology 2024, 310, e222695. [Google Scholar] [CrossRef] [PubMed]
- Şendur, H.N.; Özdemir Kalkan, D.; Cerit, M.N.; Kalkan, G.; Şendur, A.B.; Özhan Oktar, S. Hepatic Fat Quantification With Novel Ultrasound Based Techniques: A Diagnostic Performance Study Using Magnetic Resonance Imaging Proton Density Fat Fraction as Reference Standard. Can. Assoc. Radiol. J. 2023, 74, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.; Kim, J.-H.; Rhu, J.; Jeong, W.K.; Choi, G.-s.; Kim, J.M.; Joh, J.-W. Automated 3D liver segmentation from hepatobiliary phase MRI for enhanced preoperative planning. Sci. Rep. 2023, 13, 17605. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Abdalla, A.; Ansari, M.Y.; Ansari, M.I.; Malluhi, B.; Mohanty, S.; Mishra, S.; Singh, S.S.; Abinahed, J.; Al-Ansari, A.; et al. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med. Imaging 2022, 22, 97. [Google Scholar] [CrossRef]
- Senthilvelan, J.; Jamshidi, N. A pipeline for automated deep learning liver segmentation (PADLLS) from contrast enhanced CT exams. Sci. Rep. 2022, 12, 15794. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Zhang, H.; Cai, R.; Sui, S.Y.; Wang, R. Segmentation of liver CT images based on weighted medical transformer model. Sci. Rep. 2024, 14, 9887. [Google Scholar] [CrossRef] [PubMed]
- Gotra, A.; Sivakumaran, L.; Chartrand, G.; Vu, K.N.; Vandenbroucke-Menu, F.; Kauffmann, C.; Kadoury, S.; Gallix, B.; de Guise, J.A.; Tang, A. Liver segmentation: Indications, techniques and future directions. Insights Imaging 2017, 8, 377–392. [Google Scholar] [CrossRef]
- Rahman, H.; Bukht, T.F.N.; Imran, A.; Tariq, J.; Tu, S.; Alzahrani, A. A Deep Learning Approach for Liver and Tumor Segmentation in CT Images Using ResUNet. Bioengineering 2022, 9, 368. [Google Scholar] [CrossRef]
- Kumar, S.S.; Vinod Kumar, R.S. Literature survey on deep learning methods for liver segmentation from CT images: A comprehensive review. Multimed. Tools Appl. 2024, 83, 71833–71862. [Google Scholar] [CrossRef]
- Gross, M.; Huber, S.; Arora, S.; Ze’evi, T.; Haider, S.P.; Kucukkaya, A.S.; Iseke, S.; Kuhn, T.N.; Gebauer, B.; Michallek, F.; et al. Automated MRI liver segmentation for anatomical segmentation, liver volumetry, and the extraction of radiomics. Eur. Radiol. 2024, 34, 5056–5065. [Google Scholar] [CrossRef] [PubMed]
- Chlebus, G.; Schenk, A.; Moltz, J.H.; van Ginneken, B.; Hahn, H.K.; Meine, H. Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci. Rep. 2018, 8, 15497. [Google Scholar] [CrossRef] [PubMed]
- Gul, S.; Khan, M.S.; Bibi, A.; Khandakar, A.; Ayari, M.A.; Chowdhury, M.E.H. Deep learning techniques for liver and liver tumor segmentation: A review. Comput. Biol. Med. 2022, 147, 105620. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, F.; Sun, W.; Liu, Z.; Hou, H.; Jiang, R.; Hu, H.; Ren, P.; Zhang, R.; Zhang, X. Boundary-aware convolutional attention network for liver segmentation in ultrasound images. Sci. Rep. 2024, 14, 21529. [Google Scholar] [CrossRef]
- Ali, A.-R.; Guo, P.; Samir, A. Liver Segmentation in Ultrasound Images Using Self-Supervised Learning with Physics-inspired Augmentation and Global-Local Refinement. In Proceedings of the Canadian Conference on Artificial Intelligence, Montreal, QC, Canada, 5–9 June 2023. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Yang, Y.; Meher, P.K.; Dakua, S.P. Dense-PSP-UNet: A neural network for fast inference liver ultrasound segmentation. Comput. Biol. Med. 2023, 153, 106478. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Hijioka, S.; Hara, K.; Mizuno, N.; Imaoka, H.; Yamao, K. Endoscopic ultrasound description of liver segmentation and anatomy. Dig. Endosc. 2014, 26, 482–490. [Google Scholar] [CrossRef]
- Esneault, S.; Hraiech, N.; Delabrousse, E.; Dillenseger, J.L. Graph cut liver segmentation for interstitial ultrasound therapy. In Proceedings of the 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; Volume 2007, pp. 5247–5250. [Google Scholar] [CrossRef]
- Ji, Z.; Che, H.; Yan, Y.; Wu, J. BAG-Net: A boundary detection and multiple attention-guided network for liver ultrasound image automatic segmentation in ultrasound guided surgery. Phys. Med. Biol. 2024, 69, 035015. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.; Zhang, J.; Liu, Z.; Fan, Y.; Zheng, L.; Liu, P.; Song, H.; Lyu, G. SEG-LUS: A novel ultrasound segmentation method for liver and its accessory structures based on muti-head self-attention. Comput. Med. Imaging Graph. 2024, 113, 102338. [Google Scholar] [CrossRef]
- Gillies, D.J.; Rodgers, J.R.; Gyacskov, I.; Roy, P.; Kakani, N.; Cool, D.W.; Fenster, A. Deep learning segmentation of general interventional tools in two-dimensional ultrasound images. Med. Phys. 2020, 47, 4956–4970. [Google Scholar] [CrossRef]
- Song, K.D. Current status of deep learning applications in abdominal ultrasonography. Ultrasonography 2021, 40, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Alves, V.P.V.; Dillman, J.R.; Tkach, J.A.; Bennett, P.S.; Xanthakos, S.A.; Trout, A.T. Comparison of Quantitative Liver US and MRI in Patients with Liver Disease. Radiology 2022, 304, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Zhang, W.; Li, S.; Wang, X.; Sun, Y.; Sun, X.; Li, F.X.; Hou, C.; Lam, S.K.; Zheng, Y.P. A comprehensive benchmarking of a U-Net based model for midbrain auto-segmentation on transcranial sonography. Comput. Methods Programs Biomed. 2024, 258, 108494. [Google Scholar] [CrossRef] [PubMed]
- Cortacero, K.; McKenzie, B.; Müller, S.; Khazen, R.; Lafouresse, F.; Corsaut, G.; Van Acker, N.; Frenois, F.-X.; Lamant, L.; Meyer, N.; et al. Evolutionary design of explainable algorithms for biomedical image segmentation. Nat. Commun. 2023, 14, 7112. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. keras. 2015. Available online: https://github.com/fchollet/keras (accessed on 17 November 2024).
- Bilal; Pant, M.; Zaheer, H.; Garcia-Hernandez, L.; Abraham, A. Differential Evolution: A review of more than two decades of research. Eng. Appl. Artif. Intell. 2020, 90, 103479. [Google Scholar] [CrossRef]

| Depth |
|---|
| 3 |
| [8, 16, 128], [8, 32, 128], [16, 32, 128] |
| [8, 64, 256], [64, 128, 256], [16, 32, 64], [32, 64, 128] |
| 4 |
| [16, 32, 64, 128], [64, 128, 256, 512], [8, 32, 128, 256] |
| [8, 64, 128, 512], [16, 64, 128, 256], [32, 64, 128, 256] |
| 5 |
| [16, 32, 64, 128, 256], [32, 64, 128, 256, 512], [8, 32, 128, 256, 512], |
| [8, 64, 256, 512, 1024], [64, 128, 256, 512, 1024] |
| 6 |
| [32, 64, 128, 256, 512, 1024], [8, 16, 64, 128, 256, 512], [16, 32, 64, 128, 256, 512] |
| [8, 32, 128, 256, 512, 1024], [16, 32, 128, 256, 512, 1024] |
| Generation | Depth and Filter Sizes/Adjusted Scores |
|---|---|
| Gen 1 | 3: [8, 64, 256] (0.66874), 4: [64, 128, 256, 512] (0.85492), 5: [16, 32, 64, 128, 256] (0.86036), 6: [32, 64, 128, 256, 512, 1024] (0.85848) |
| Gen 2 | 3: [64, 128, 256] (0.49106), 4: [64, 128, 256, 512] (0.79684), 5: [16, 32, 64, 128, 256] (0.79872), 6: [32, 64, 128, 256, 512, 1024] (0.87574) |
| Gen 3 | 3: [8, 64, 256] (0.75624), 4: [64, 128, 256, 512] (0.86317), 5: [16, 32, 64, 128, 256] (0.84136), 6: [32, 64, 128, 256, 512, 1024] (0.77848) |
| Gen 4 | 3: [8, 64, 256] (0.60629), 4: [64, 128, 256, 512] (0.88488), 5: [16, 32, 64, 128, 256] (0.77838), 6: [32, 64, 128, 256, 512, 1024] (0.79681) |
| Gen 5 | 3: [64, 128, 256] (0.67891), 4: [64, 128, 256, 512] (0.85566), 5: [16, 32, 64, 128, 256] (0.79540), 6: [32, 64, 128, 256, 512, 1024] (0.78476) |
| Gen 6 | 3: [8, 64, 256] (0.85739), 4: [16, 64, 128, 256] (0.76498), 5: [16, 32, 64, 128, 256] (0.83489), 6: [32, 64, 128, 256, 512, 1024] (0.83680) |
| Gen 7 | 3: [8, 64, 256] (0.63057), 4: [16, 64, 128, 256] (0.83652), 5: [16, 32, 64, 128, 256] (0.83393), 6: [8, 32, 128, 256, 512, 1024] (0.76690) |
| Gen 8 | 3: [8, 64, 256] (0.76110), 4: [16, 64, 128, 256] (0.80789), 5: [16, 32, 64, 128, 256] (0.73106), 6: [32, 64, 128, 256, 512, 1024] (0.84866) |
| Gen 9 | 3: [16, 32, 128] (0.43480), 4: [16, 64, 128, 256] (0.81051), 5: [64, 128, 256, 512, 1024] (0.81905), 6: [8, 32, 128, 256, 512, 1024] (0.78400) |
| Gen 10 | 3: [16, 32, 128] (0.68542), 4: [16, 64, 128, 256] (0.85939), 5: [16, 32, 64, 128, 256] (0.82776), 6: [32, 64, 128, 256, 512, 1024] (0.82564) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zifan, A.; Zhao, K.; Lee, M.; Peng, Z.; Roney, L.J.; Pai, S.; Weeks, J.T.; Middleton, M.S.; Kaffas, A.E.; Schwimmer, J.B.; et al. Adaptive Evolutionary Optimization of Deep Learning Architectures for Focused Liver Ultrasound Image Segmentation. Diagnostics 2025, 15, 117. https://doi.org/10.3390/diagnostics15020117
Zifan A, Zhao K, Lee M, Peng Z, Roney LJ, Pai S, Weeks JT, Middleton MS, Kaffas AE, Schwimmer JB, et al. Adaptive Evolutionary Optimization of Deep Learning Architectures for Focused Liver Ultrasound Image Segmentation. Diagnostics. 2025; 15(2):117. https://doi.org/10.3390/diagnostics15020117
Chicago/Turabian StyleZifan, Ali, Katelyn Zhao, Madilyn Lee, Zihan Peng, Laura J. Roney, Sarayu Pai, Jake T. Weeks, Michael S. Middleton, Ahmed El Kaffas, Jeffrey B. Schwimmer, and et al. 2025. "Adaptive Evolutionary Optimization of Deep Learning Architectures for Focused Liver Ultrasound Image Segmentation" Diagnostics 15, no. 2: 117. https://doi.org/10.3390/diagnostics15020117
APA StyleZifan, A., Zhao, K., Lee, M., Peng, Z., Roney, L. J., Pai, S., Weeks, J. T., Middleton, M. S., Kaffas, A. E., Schwimmer, J. B., & Sirlin, C. B. (2025). Adaptive Evolutionary Optimization of Deep Learning Architectures for Focused Liver Ultrasound Image Segmentation. Diagnostics, 15(2), 117. https://doi.org/10.3390/diagnostics15020117






