Novel Four-Way Variant Translocation, t(1;9;22;16)(q21;q34;q11.2;q24), in a Patient with Chronic Myeloid Leukemia
Abstract
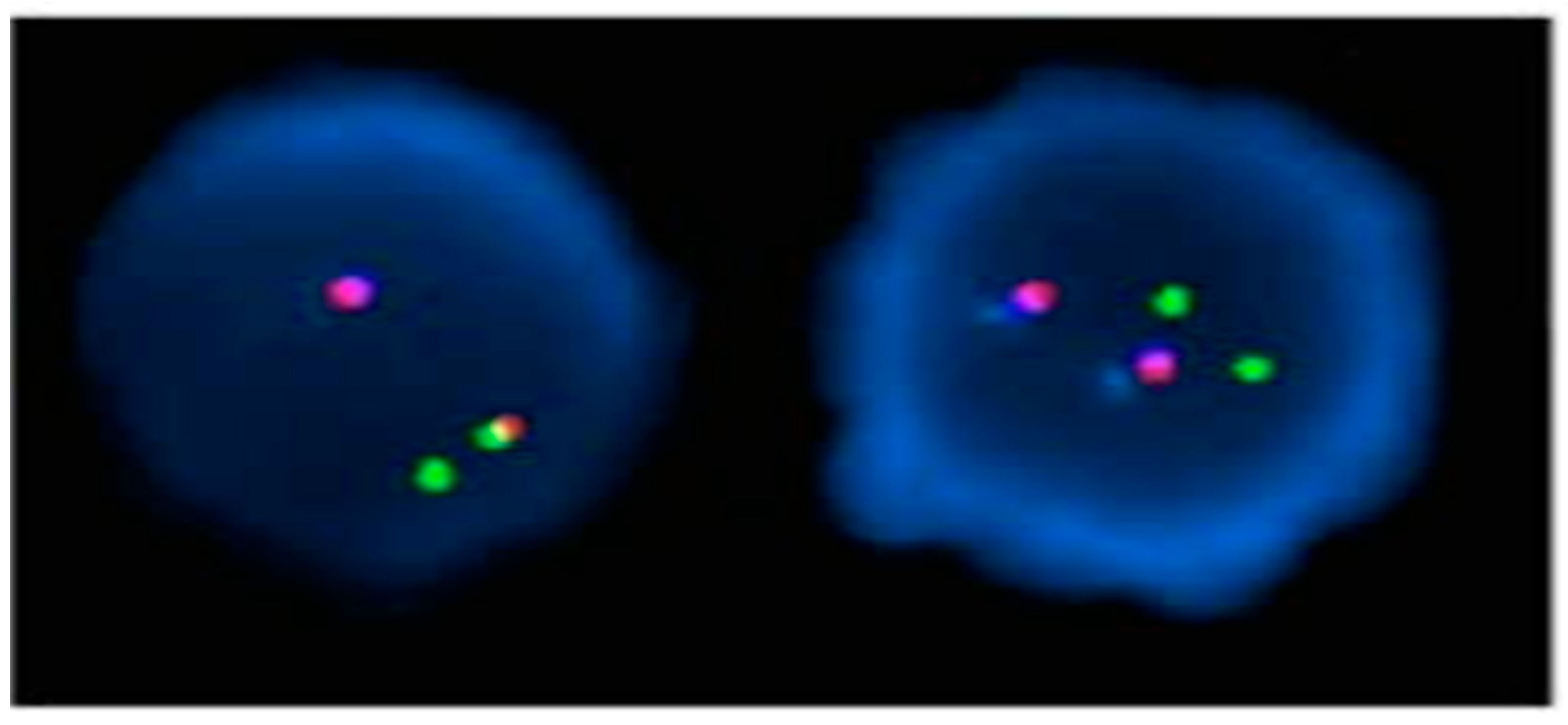
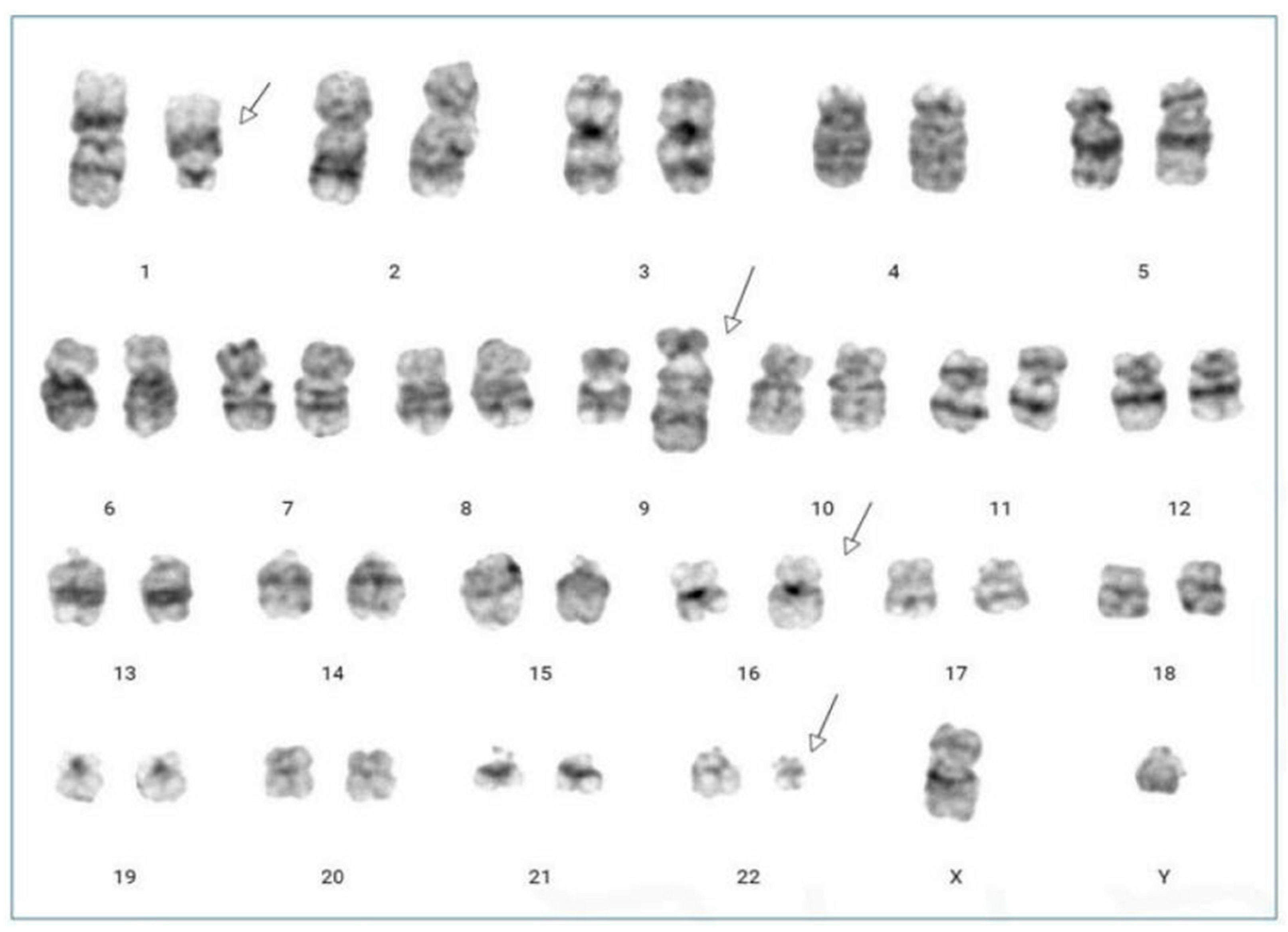
:
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowley, J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Cortes, J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood 2009, 113, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Gorusu, M.; Benn, P.; Li, Z.; Fang, M. On the genesis and prognosis of variant translocations in chronic myeloid leukemia. Cancer Genet. Cytogenet. 2007, 173, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Marzocchi, G.; Castagnetti, F.; Luatti, S.; Baldazzi, C.; Stacchini, M.; Gugliotta, G.; Amabile, M.; Specchia, G.; Sessarego, M.; Giussani, U.; et al. Variant Philadelphia translocations: Molecular-cytogenetic characterization and prognostic influence on frontline imatinib therapy, a GIMEMA Working Party on CML analysis. Blood 2011, 117, 6793–6800. [Google Scholar] [CrossRef] [PubMed]
- El-Zimaity, M.M.; Kantarjian, H.; Talpaz, M.; O’Brien, S.; Giles, F.; Garcia-Manero, G.; Verstovsek, S.; Thomas, D.; Ferrajoli, A.; Hayes, K.; et al. Results of imatinib mesylate therapy in chronic myelogenous leukaemia with variant Philadelphia chromosome. Br. J. Haematol. 2004, 125, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Reid, A.G.; Huntly, B.J.; Grace, C.; Green, A.R.; Nacheva, E.P. Survival implications of molecular heterogeneity in variant Philadelphia-positive chronic myeloid leukaemia. Br. J. Haematol. 2003, 121, 419–427. [Google Scholar] [CrossRef]
- Shetty, D.; Talker, E.; Jain, H.; Talker, J.; Patkar, N.; Subramanian, P.; Jain, H.; Bonda, A.; Punatar, S.; Gokarn, A.; et al. Evaluation of cytogenetic response in CML patients with variant Philadelphia translocation. Asia Pac. J. Clin. Oncol. 2022, 18, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Fabarius, A.; Leitner, A.; Hochhaus, A.; Müller, M.C.; Hanfstein, B.; Haferlach, C.; Göhring, G.; Schlegelberger, B.; Jotterand, M.; Reiter, A.; et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of CML: Long-term observation of 1151 patients from the randomized CML Study IV. Blood 2011, 118, 6760–6768. [Google Scholar] [CrossRef] [PubMed]
- Mitelman, F.; Johansson, B.; Mertens, F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2023. Available online: https://mitelmandatabase.isb-cgc.org (accessed on 1 November 2023).
- Johansson, B.; Fioretos, T.; Mitelman, F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002, 107, 76–94. [Google Scholar] [CrossRef] [PubMed]
- Richebourg, S.; Eclache, V.; Perot, C.; Portnoi, M.F.; Van den Akker, J.; Terré, C.; Maareck, O.; Soenen, V.; Viguié, F.; Laï, J.L.; et al. Mechanisms of genesis of variant translocation in chronic myeloid leukemia are not correlated with ABL1 or BCR deletion status or response to imatinib therapy. Cancer Genet. Cytogenet. 2008, 182, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Al Achkar, W.; Wafa, A.; Mkrtchyan, H.; Moassass, F.; Liehr, T. Novel complex translocation involving 5 different chromosomes in a chronic myeloid leukemia with Philadelphia chromosome: A case report. Mol. Cytogenet. 2009, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.J. Deletion of the derivative chromosome 9 in chronic myeloid leukemia. Methods Mol. Med. 2006, 125, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, P.B.; Nacheva, E.P.; Leversha, M.; Telford, N.; Chang, J.; Reid, A.; Bench, A.; Champion, K.; Huntly, B.; Green, A.R. Large deletions at the t(9;22) breakpoint are common and may identify a poor-prognosis subgroup of patients with chronic myeloid leukemia. Blood 2000, 95, 738–743. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, H.J.; Lee, J.H. Novel Four-Way Variant Translocation, t(1;9;22;16)(q21;q34;q11.2;q24), in a Patient with Chronic Myeloid Leukemia. Diagnostics 2024, 14, 303. https://doi.org/10.3390/diagnostics14030303
Son HJ, Lee JH. Novel Four-Way Variant Translocation, t(1;9;22;16)(q21;q34;q11.2;q24), in a Patient with Chronic Myeloid Leukemia. Diagnostics. 2024; 14(3):303. https://doi.org/10.3390/diagnostics14030303
Chicago/Turabian StyleSon, Han Joon, and Jong Ho Lee. 2024. "Novel Four-Way Variant Translocation, t(1;9;22;16)(q21;q34;q11.2;q24), in a Patient with Chronic Myeloid Leukemia" Diagnostics 14, no. 3: 303. https://doi.org/10.3390/diagnostics14030303




