Postmortem Minimally Invasive Autopsy in Critically Ill COVID-19 Patients at the Bedside: A Proof-of-Concept Study at the ICU
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design
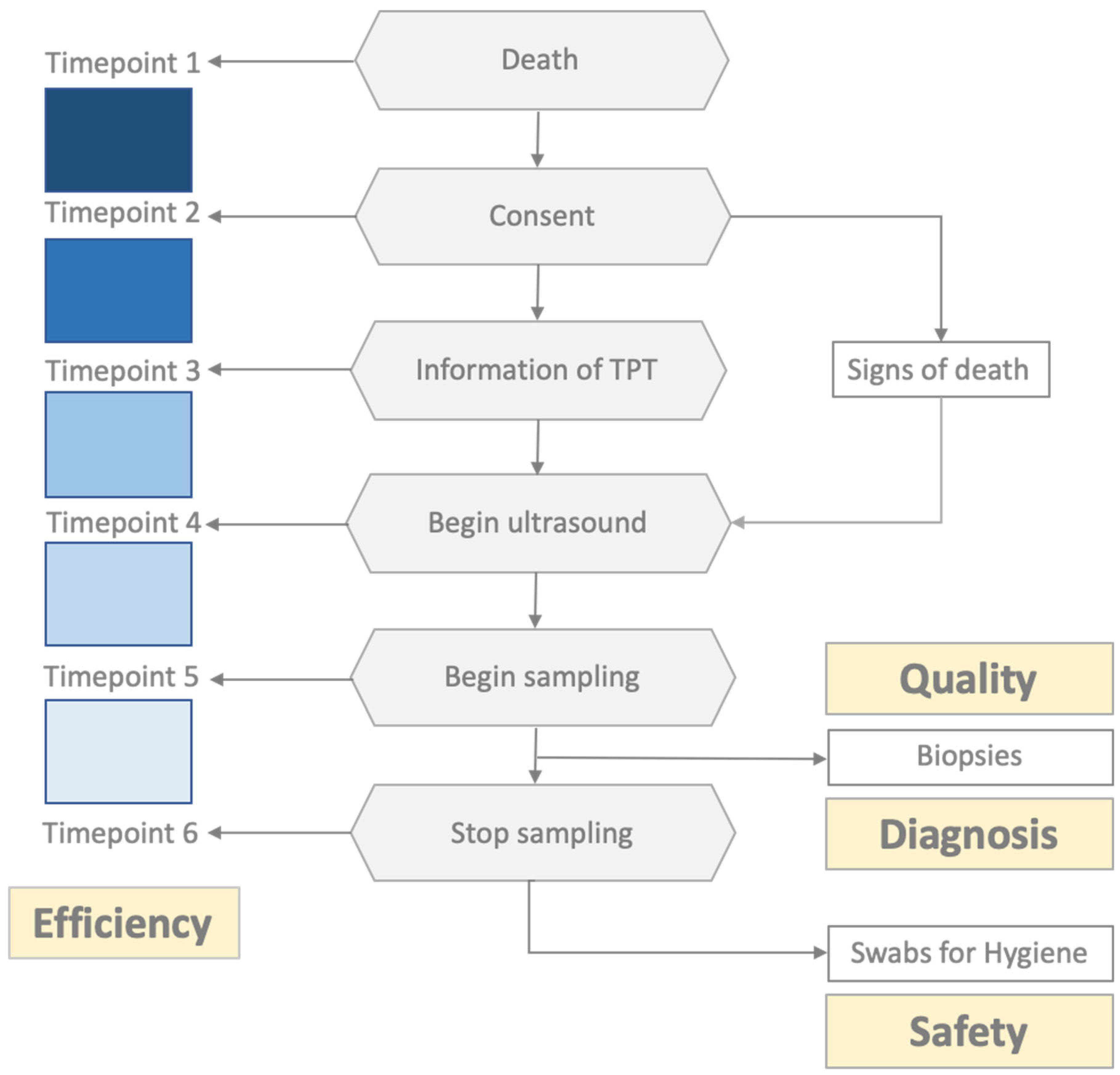
2.2. Logistical Aspects and Standard Operating Procedures
2.3. Assessment Tools for Time Efficiency and Sample Quality
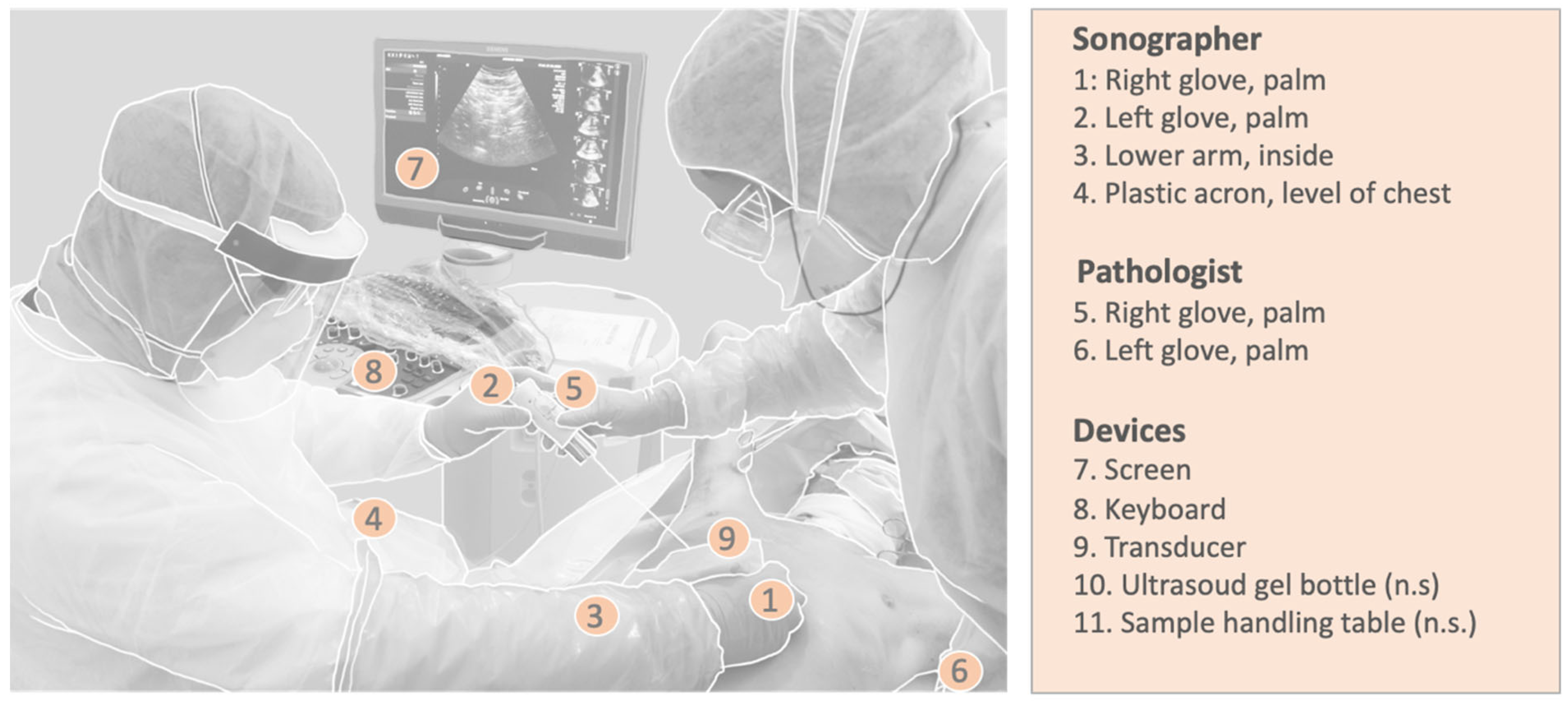
2.4. Safety/Hygiene
3. Results
3.1. Quality Assessment
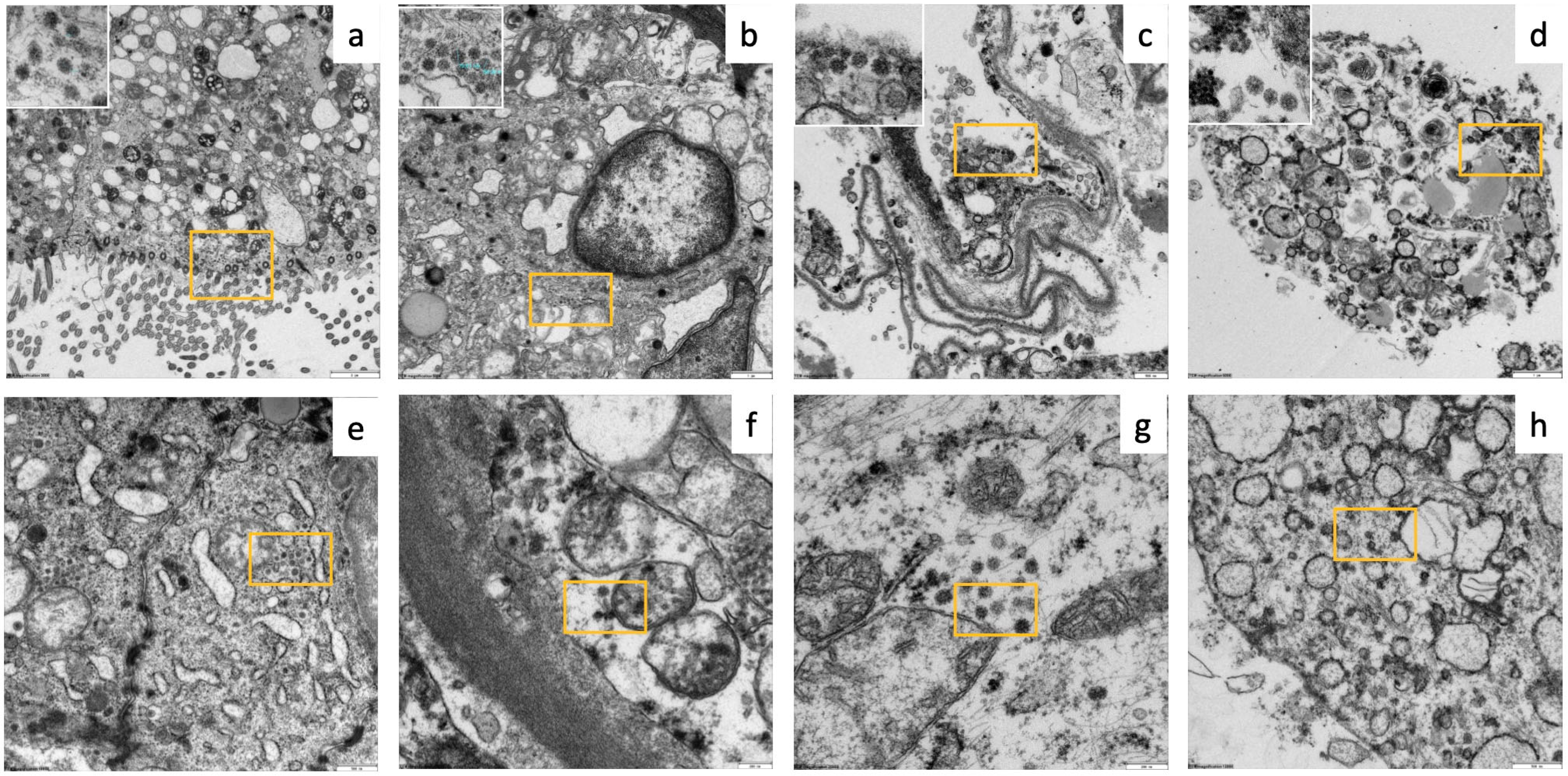
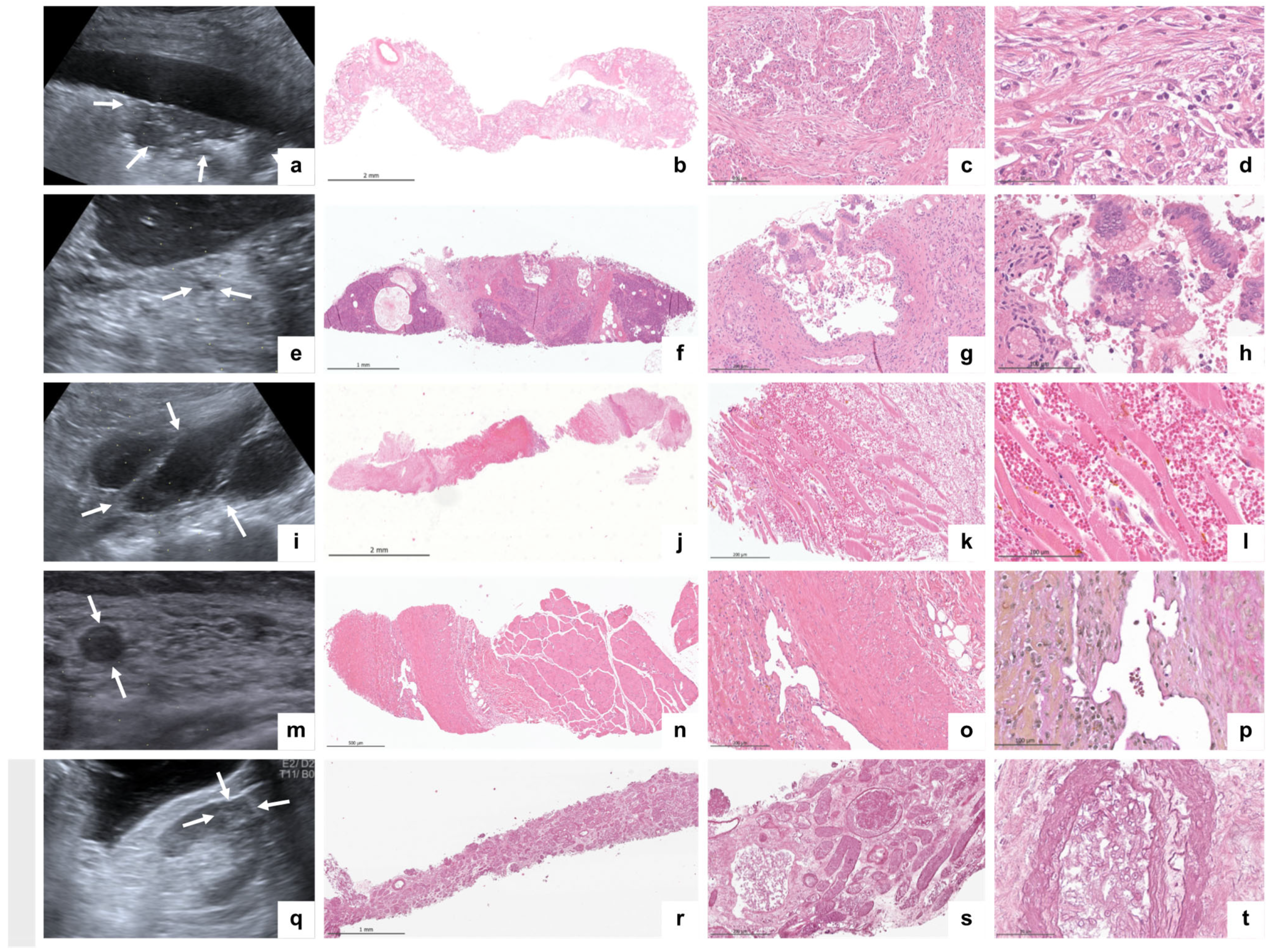
3.2. Imaging and Histopathological Findings
3.3. Safety (Hygiene Control)
4. Discussion
5. Conclusions
6. Research in Context
6.1. Evidence before This Study
6.2. Added Value of This Study
6.3. Implications of All the Available Evidence
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| US-MIA | Ultrasound-guided minimally invasive autopsy |
| MIA | Minimal invasive autopsy |
| VOC | Variants of concern |
| PMI | Postmortem interval |
| SOPs | Standard operating procedures |
| NGS | Next-generation sequencing |
| CT | Computed tomography |
| MITS | Minimally invasive tissue sampling |
| ICU | Intensive care unit |
| TUM | Technical University of Munich |
| RT-PCR | Reverse transcription polymerase chain reaction |
| PPE | Personal protective equipment |
| MRI | München rechts der Isar |
| TPT | Tissue procurement team |
| FFPE | Paraffin embedded |
| H&E | Hematoxylin and Eosin |
| PAS | Periodic Acid–Schiff |
| EvG | Elastica van Gieson’s stain |
| RAP | Rapid research autopsy programs |
| EM | Electron microscopy |
| WHO | World Health Organization |
References
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Khan, A.; Wani, M.Y.; Ahmad, A.; Duse, A. COVID-19: A state of art on immunological responses, mutations, and treatment modalities in riposte. J. Infect. Public Health 2023, 16, 233–249. [Google Scholar] [CrossRef]
- Arish, M.; Qian, W.; Narasimhan, H.; Sun, J. COVID-19 immunopathology: From acute diseases to chronic sequelae. J. Med. Virol. 2023, 95, e28122. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Huerne, K.; Filion, K.B.; Grad, R.; Ernst, P.; Gershon, A.S.; Eisenberg, M.J. Epidemiological and clinical perspectives of long COVID syndrome. Am. J. Med. Open 2023, 9, 100033. [Google Scholar] [CrossRef] [PubMed]
- Fricke-Galindo, I.; Falfán-Valencia, R. Genetics Insight for COVID-19 Susceptibility and Severity: A Review. Front. Immunol. 2021, 12, 622176. [Google Scholar] [CrossRef] [PubMed]
- Zuin, M.; Engelen, M.M.; Bilato, C.; Vanassche, T.; Rigatelli, G.; Verhamme, P.; Vandenbriele, C.; Zuliani, G.; Roncon, L. Prevalence of Acute Pulmonary Embolism at Autopsy in Patients With COVID-19. Am. J. Cardiol. 2022, 171, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Valdivia-Mazeyra, M.F.; Salas, C.; Nieves-Alonso, J.M.; Martín-Fragueiro, L.; Bárcena, C.; Muñoz-Hernández, P.; Villar-Zarra, K.; Martín-López, J.; Ramasco-Rueda, F.; Fraga, J.; et al. Increased number of pulmonary megakaryocytes in COVID-19 patients with diffuse alveolar damage: An autopsy study with clinical correlation and review of the literature. Virchows Arch. 2021, 478, 487–496. [Google Scholar] [CrossRef]
- Remmelink, M.; De Mendonça, R.; D’haene, N.; De Clercq, S.; Verocq, C.; Lebrun, L.; Lavis, P.; Racu, M.-L.; Trépant, A.-L.; Maris, C.; et al. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Crit. Care 2020, 24, 495. [Google Scholar] [CrossRef]
- Jeican, I.I.; Inișca, P.; Gheban, D.; Anton, V.; Lazăr, M.; Vică, M.L.; Mironescu, D.; Rebeleanu, C.; Crivii, C.B.; Aluaș, M.; et al. Histopathological Lung Findings in COVID-19 B.1.617.2 SARS-CoV-2 Delta Variant. J. Pers. Med. 2023, 13, 279. [Google Scholar] [CrossRef]
- Wichmann, D.; Sperhake, J.P.; Lütgehetmann, M.; Steurer, S.; Edler, C.; Heinemann, A.; Heinrich, F.; Mushumba, H.; Kniep, I.; Schröder, A.S.; et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann. Intern. Med. 2020, 173, 268–277. [Google Scholar] [CrossRef]
- Ghosh, A.; Colling, R. An overview of COVID-19 for diagnostic pathologists: Clinicopathological correlation and diagnostic techniques. Diagn. Histopathol. 2020, 26, 529–536. [Google Scholar] [CrossRef]
- Stempsey, W.E. The Penetrating Gaze and the Decline of the Autopsy. AMA J. Ethics 2016, 18, 833–838. [Google Scholar]
- Rusu, S.; Lavis, P.; Salgado, V.D.; Van Craynest, M.-P.; Creteur, J.; Salmon, I.; Brasseur, A.; Remmelink, M. Comparison of antemortem clinical diagnosis and post-mortem findings in intensive care unit patients. Virchows Arch. 2021, 479, 385–392. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Massoud, T.F.; Gambhir, S.S. Molecular imaging in living subjects: Seeing fundamental biological processes in a new light. Genes Dev. 2003, 17, 545–580. [Google Scholar] [CrossRef] [PubMed]
- Ziv, E.; Durack, J.C.; Solomon, S.B. The Importance of Biopsy in the Era of Molecular Medicine. Cancer J. 2016, 22, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.G.; Munoz-Aguirre, M.; Reverter, F.; Sa Godinho, C.P.; Sousa, A.; Amadoz, A.; Sodaei, R.; Hidalgo, M.R.; Pervouchine, D.; Carbonell-Caballero, J.; et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 2018, 9, 490. [Google Scholar] [CrossRef]
- Barner, A.; Burian, E.; Simon, A.; Castillo, K.; Waschulzik, B.; Braren, R.; Heemann, U.; Osterwalder, J.; Spiel, A.; Heim, M.; et al. Pulmonary Findings in Hospitalized COVID-19 Patients Assessed by Lung Ultrasonography (LUS)—A Prospective Registry Study. Ultraschall. Med. 2023, 44, e248–e256. [Google Scholar] [CrossRef] [PubMed]
- Welsh, T.S.; Kaplan, J. The role of postmortem examination in medical education. Mayo Clin. Proc. 1998, 73, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Juan, H.F.; Huang, H.C. Quantitative analysis of high-throughput biological data. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2023, 13, e1658. [Google Scholar] [CrossRef]
- Yao, X.-H.; Luo, T.; Shi, Y.; He, Z.-C.; Tang, R.; Zhang, P.-P.; Cai, J.; Zhou, X.-D.; Jiang, D.-P.; Fei, X.-C.; et al. A cohort autopsy study defines COVID-19 systemic pathogenesis. Cell Res. 2021, 31, 836–846. [Google Scholar] [CrossRef]
- Layne, S.P.; Walters, K.-A.; Kash, J.C.; Taubenberger, J.K. More autopsy studies are needed to understand the pathogenesis of severe COVID-19. Nat. Med. 2022, 28, 427–428. [Google Scholar] [CrossRef]
- Krook, M.A.; Chen, H.-Z.; Bonneville, R.; Allenby, P.; Roychowdhury, S. Rapid Research Autopsy: Piecing the Puzzle of Tumor Heterogeneity. Trends Cancer 2019, 5, 1–5. [Google Scholar] [CrossRef]
- A Cox, J.; Lukande, R.L.; Kalungi, S.; Van de Vijver, K.; Van Marck, E.; Nelson, A.M.; Munema, A.; Manabe, Y.C.; Colebunders, R. Practice of percutaneous needle autopsy; a descriptive study reporting experiences from Uganda. BMC Clin. Pathol. 2014, 14, 44. [Google Scholar] [CrossRef]
- Rakislova, N.; Marimon, L.; Ismail, M.R.; Carrilho, C.; Fernandes, F.; Ferrando, M.; Castillo, P.; Rodrigo-Calvo, M.T.; Guerrero, J.; Ortiz, E.; et al. Minimally Invasive Autopsy Practice in COVID-19 Cases: Biosafety and Findings. Pathogens 2021, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Bassat, Q.; Varo, R.; Hurtado, J.C.; Marimon, L.; Ferrando, M.; Ismail, M.R.; Carrilho, C.; Fernandes, F.; Castro, P.; Maixenchs, M.; et al. Minimally Invasive Tissue Sampling as an Alternative to Complete Diagnostic Autopsies in the Context of Epidemic Outbreaks and Pandemics: The Example of Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2021, 73 (Suppl. S5), S472–S479. [Google Scholar] [CrossRef]
- Wagensveld, I.M.; Weustink, A.C.; Kors, J.A.; Blokker, B.M.; Hunink, M.G.M.; Oosterhuis, J.W. Effect of minimally invasive autopsy and ethnic background on acceptance of clinical postmortem investigation in adults. PLoS ONE 2020, 15, e0232944. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, J.; Wang, S.; Li, X.; Zhou, J.; Huang, B.; Luo, D.; Cao, Q.; Chen, Y.; Chen, S.; et al. Progression to fibrosing diffuse alveolar damage in a series of 30 minimally invasive autopsies with COVID-19 pneumonia in Wuhan, China. Histopathology 2021, 78, 542–555. [Google Scholar] [CrossRef]
- D’onofrio, V.; Donders, E.; Abeele, M.-E.V.; Dubois, J.; Cartuyvels, R.; Achten, R.; Lammens, M.; Dendooven, A.; Driessen, A.; Augsburg, L.; et al. The clinical value of minimal invasive autopsy in COVID-19 patients. PLoS ONE 2020, 15, e0242300. [Google Scholar] [CrossRef] [PubMed]
- Krasemann, S.; Dittmayer, C.; von Stillfried, S.; Meinhardt, J.; Heinrich, F.; Hartmann, K.; Pfefferle, S.; Thies, E.; von Manitius, R.; Aschman, T.A.D.; et al. Assessing and improving the validity of COVID-19 autopsy studies—A multicentre approach to establish essential standards for immunohistochemical and ultrastructural analyses. EBioMedicine 2022, 83, 104193. [Google Scholar] [CrossRef] [PubMed]
- Brandner, J.M.; Boor, P.; Borcherding, L.; Edler, C.; Gerber, S.; Heinemann, A.; Hilsenbeck, J.; Kasajima, A.; Lohner, L.; Märkl, B.; et al. Contamination of personal protective equipment during COVID-19 autopsies. Virchows Arch. 2022, 480, 519–528. [Google Scholar] [CrossRef]

| Case #1 | Case #2 | Case #3 | Case #4 | Case #5 | |
|---|---|---|---|---|---|
| Age (years), sex | 75, male | 65, male | 78, male | 73, female | 87, male |
| BMI | 27.1 | 29 | 25.5 | 24.8 | 24.5 |
| ICU stay (days) | 24 | 2 | 26 | 9 | 32 |
| Hospital stay (days) | 24 | 43 | 26 | 17 | 34 |
| SOFA score | 12 | 8 | 13 | 8 | 16 |
| Mechanical ventilation (days) | 24 | 2 | 26 | 9 | 32 |
| Vaccination status until time of death | 2×, BioNTech/Pfizer | 2×, BioNTech/Pfizer | 2×, BioNTech/Pfizer | 2×, BioNTech/Pfizer | 3×, BioNTech/Pfizer |
| COVID-19-specific medication | Dexamethasone and casirivimab/Imdevimab | Dexamethasone and casirivimab/Imdevimab | Dexamethasone and casirivimab/Imdevimab | Dexamethasone and casirivimab/Imdevimab | Dexamethasone |
| Medical history | AH, T2DM, and FSGS (under therapy with ciclosorin and rituximab) | AH, CHD, and diverticuosis history of DLBCL (RD negative) and tMDS/AML (under therapy, RD negative) | T2DM, CHD, atherosclerosis, and history of prostate cancer (RD negative) | AH, CHD, idiopathic lung fibrosis, cardiac enlargement, and idiopathic MDS (high risk) | AH, COPD, thrombosis of left saphenous vein and intramuscular veins, and pulmonary artery embolism |
| Imaging findings (CT) | CO-RADS 6 | CO-RADS 6 (left lung); renal and splenic infarctions | CO-RADS 6 | CO-RADS 6 | Suspicious for COVID-19 (CO-RADS 4) |
| Clinical diagnosis leading to death | COVID-19 pneumonia, ARDS, and hypoxaemia | COVID-19 pneumonia and suspected fungal infection | COVID-19 pneumonia, ARDS, respiratory global insufficiency, suspected HSV, and fungal infection | COVID-19 pneumonia and septic multiorgan failure | COVID-19 pneumonia, ARDS, suspected bacterial superinfection, and hypoxaemia |
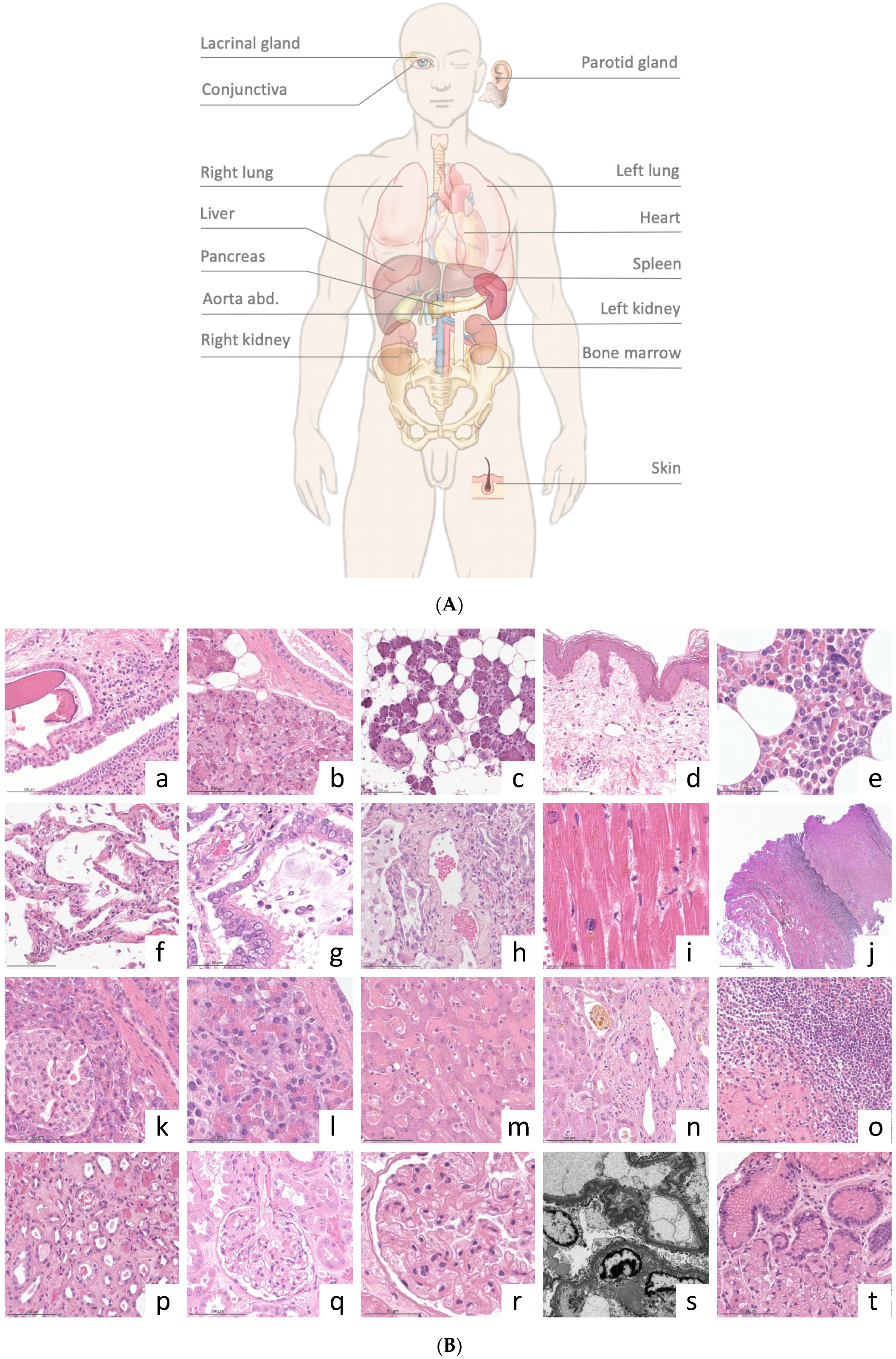
| Organ/Target | Representativity (%) | Mean Length of Biopsy (mm) |
|---|---|---|
| Right lung | 21/21 (100) | 14 (8–20) |
| Left lung | 15/16 (94) | 10 (4–15) |
| Liver | 15/15 (100) | 14 (11–17) |
| Right kidney | 8/8 (100) | 11 (5–15) |
| Left kidney | 5/5 (100) | 14 (3–15) |
| Heart | 9/9 (100) | 13 (5–25) |
| Pancreas | 5/6 (83) | 10 (8–12) |
| Abdominal aorta | 4/5 (80) | 8 (4–10) |
| Spleen | 5/5 (100) | 14 (9–17) |
| Lacrimal gland/conjunctiva | 5/5 (100) | 8.8 (6–11) |
| Parotid gland | 5/5 (80) | 9.6 (7–12) |
| Skin | 5/5 (100) | 10 (8–14) |
| Bone marrow | 4/5 (80) | 12 (9–16) |
| Total | 106/110 (96) | 11 (4–25) |
| Ultrasound | Pulmonary Histomorphology | Extrapulmonary Histomorphology | |
|---|---|---|---|
| Case #1 | Bilateral pleural effusions. Bilateral multiple peripheral lung consolidation. Hepatomegaly. Hepatic steatosis grade 1. Hydrops of the gallbladder. | Extensive DAD (mainly proliferative; partly exudative) and microthrombi. | Subacute and acute hepatic congestion; non-alcoholic fatty liver disease (5%). Interstitial pancreatic fibrosis; pancreatic acinar atrophy. Pancreatic intraepithelial neoplasia (low grade). Splenic red pulp expansion; white pulp hypoplasia. Acute renal tubular injury, microthrombi, and minimal focal segmental glomerulosclerosis. Myocardial interstitial fibrosis. Chronic conjunctivitis. Parotid lipomatosis. Dermal perivascular lymphocytic inflammation. |
| Case #2 | Pleural effusion (right); small lung consolidations (left). Ascites. Hepatomegaly and aerobilia. Liver cyst (9 mm). Bilateral kidney and spleen infarctions. | Beginning DAD (exudative). Microthrombi. Mild interstitial fibrosis. Anthracosis. Emphysema. | Fungal sepsis (Aspergillus spp.) with the colonization of renal infarctions. Hepatocellular and canalicular cholestasis, chronic portal inflammation, hemosiderosis, bile cast nephropathy, acute renal tubular injury, splenic hemosiderosis, white pulp hypoplasia, myocardial interstitial fibrosis (infarct-like) bone marrow hypoplasia, edema, and hemosiderosis. |
| Case #3 | Pleural effusion (right); bilateral multiple peripheral lung consolidation Aerobilia. Pancreatic lipomatosis. Aortic sclerosis. Retroperitoneal hematoma. | Extensive DAD (exudative and proliferative). Interstitial fibrosis. Bacterial superinfection. Microthrombi. No HSV or fungi. | Hepatocellular single cell necrosis, cholestasis, sinusoidal neutrophils, and non-alcoholic fatty liver disease (5%). Splenic red pulp expansion; white pulp hypoplasia. Acute renal tubular injury. Renal interstitial fibrosis; focal global glomerulosclerosis. Pancreatic lipomatosis. Retroperitoneal hematoma. Parotid lipomatosis. Lymphocytic inflammation of conjunctiva and lacrimal gland dermal perivascular lymphocytic inflammation. |
| Case #4 | Bilateral pleural effusions. Bilateral multiple peripheral lung consolidation. Ascites. Hepatomegaly; aerobilia; splenomegaly. Pancreatic lipomatosis. Pancreatic cyst (5 mm). | Extensive DAD (exudative and proliferative); microthrombi pulmonary congestion. | Acute hepatic congestion; sinusoidal neutrophils. Non-alcoholic fatty liver disease (5%). Pancreatic intraepithelial neoplasia (low grade). Splenic red pulp expansion; white pulp hypoplasia. Acute renal tubular injury. Myocardial hypertrophy. Myelodysplastic syndrome with blast excess 2 (18% blasts). Chronic conjunctivitis. Dermal perivascular lymphocytic inflammation. Parotid lipomatosis. |
| Case #5 | Bilateral multiple peripheral lung consolidation. Hepatic steatosis grade 1. Aortic sclerosis. Muscle vein thrombosis (M. soleus). | DAD (exudative and proliferative); microthrombi; infarct-like hemorrhage; bacterial superinfection | Severe acute hepatic congestion, sinusoidal neutrophils, and hepatocellular cholestasis. Non-alcoholic fatty liver disease (2%). Acute renal tubular injury, focal global glomerulosclerosis, and renal microthrombi. Pancreatic lipomatosis. Splenic red pulp expansion; white pulp hypoplasia. Chronic and acute conjunctivitis. Dermal perivascular lymphocytic inflammation. Aortic sclerosis. Muscle vein thrombosis in organization. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lahmer, T.; Weirich, G.; Porubsky, S.; Rasch, S.; Kammerstetter, F.A.; Schustetter, C.; Schüffler, P.; Erber, J.; Dibos, M.; Delbridge, C.; et al. Postmortem Minimally Invasive Autopsy in Critically Ill COVID-19 Patients at the Bedside: A Proof-of-Concept Study at the ICU. Diagnostics 2024, 14, 294. https://doi.org/10.3390/diagnostics14030294
Lahmer T, Weirich G, Porubsky S, Rasch S, Kammerstetter FA, Schustetter C, Schüffler P, Erber J, Dibos M, Delbridge C, et al. Postmortem Minimally Invasive Autopsy in Critically Ill COVID-19 Patients at the Bedside: A Proof-of-Concept Study at the ICU. Diagnostics. 2024; 14(3):294. https://doi.org/10.3390/diagnostics14030294
Chicago/Turabian StyleLahmer, Tobias, Gregor Weirich, Stefan Porubsky, Sebastian Rasch, Florian A. Kammerstetter, Christian Schustetter, Peter Schüffler, Johanna Erber, Miriam Dibos, Claire Delbridge, and et al. 2024. "Postmortem Minimally Invasive Autopsy in Critically Ill COVID-19 Patients at the Bedside: A Proof-of-Concept Study at the ICU" Diagnostics 14, no. 3: 294. https://doi.org/10.3390/diagnostics14030294
APA StyleLahmer, T., Weirich, G., Porubsky, S., Rasch, S., Kammerstetter, F. A., Schustetter, C., Schüffler, P., Erber, J., Dibos, M., Delbridge, C., Kuhn, P. H., Jeske, S., Steinhardt, M., Chaker, A., Heim, M., Heemann, U., Schmid, R. M., Weichert, W., Stock, K. F., & Slotta-Huspenina, J. (2024). Postmortem Minimally Invasive Autopsy in Critically Ill COVID-19 Patients at the Bedside: A Proof-of-Concept Study at the ICU. Diagnostics, 14(3), 294. https://doi.org/10.3390/diagnostics14030294







