Federated Learning with Privacy Preserving for Multi- Institutional Three-Dimensional Brain Tumor Segmentation
Abstract
1. Introduction
2. Related Work
3. Background
3.1. Neural Network Architecture
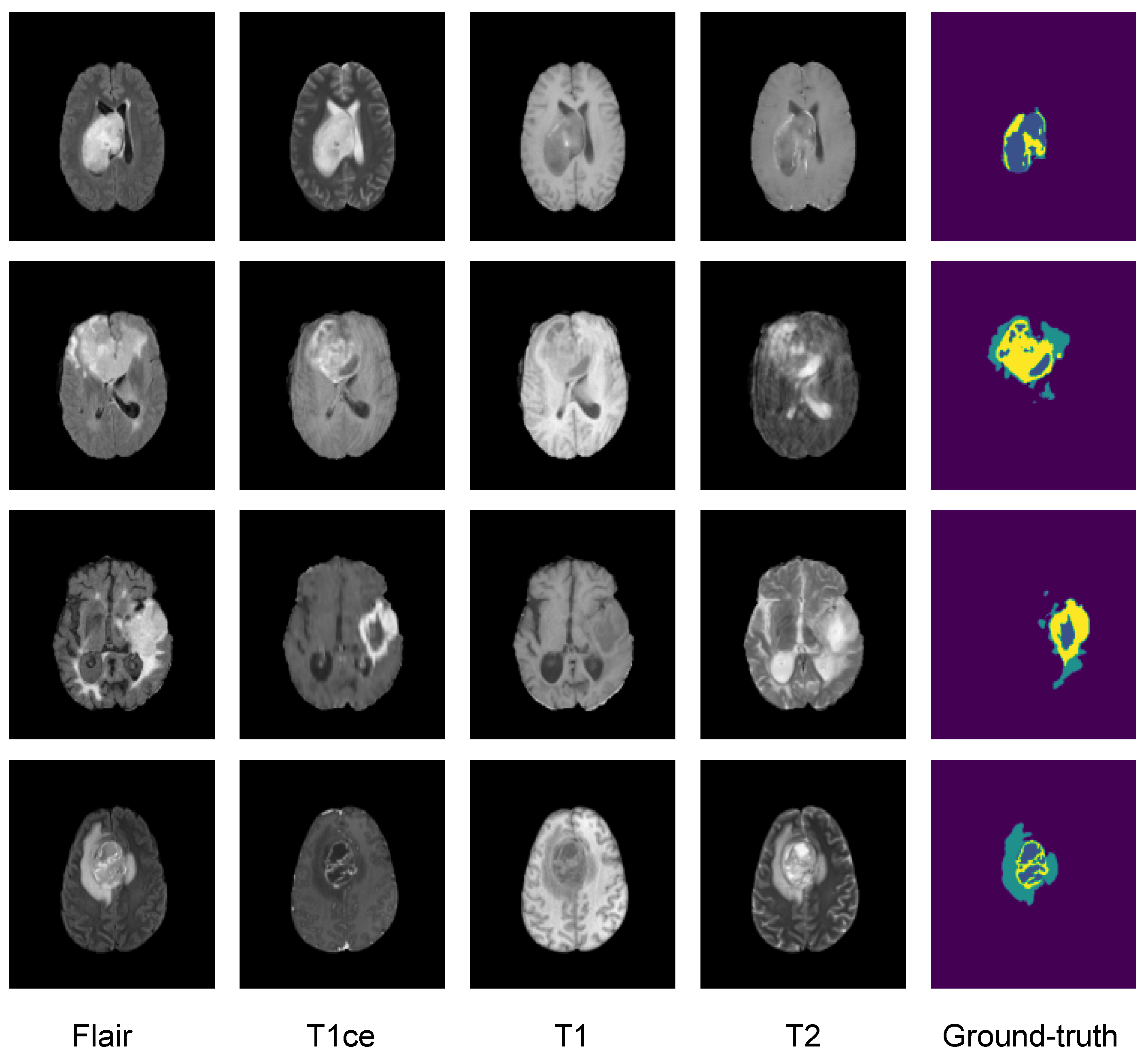
3.2. Dataset
3.3. Image Preprocessing
3.4. Data Augmentation Techniques
- Input Channel Rescaling: A factor within the range of 0.9 to 1.1 was multiplied to each voxel, with a probability of 80%.
- Input Channel Intensity Shift: A constant within the range of −0.1 to 0.1 was added to each voxel, with a probability of 10%.
- Additive Gaussian Noise: A random noise was generated using a centered normal distribution with a standard deviation of 0.1 and added to the input data.
- Input Channel Dropping: With a 16% chance, one of the input channels had all its voxel values randomly set to zero.
- Random Flip Along Each Spatial Axis: The data were subjected to a random horizontal flip, a random vertical flip, and potentially a random flip along the depth, with a probability of 80%.
3.5. Loss Function
3.6. Evaluation Metrics
4. Methodology
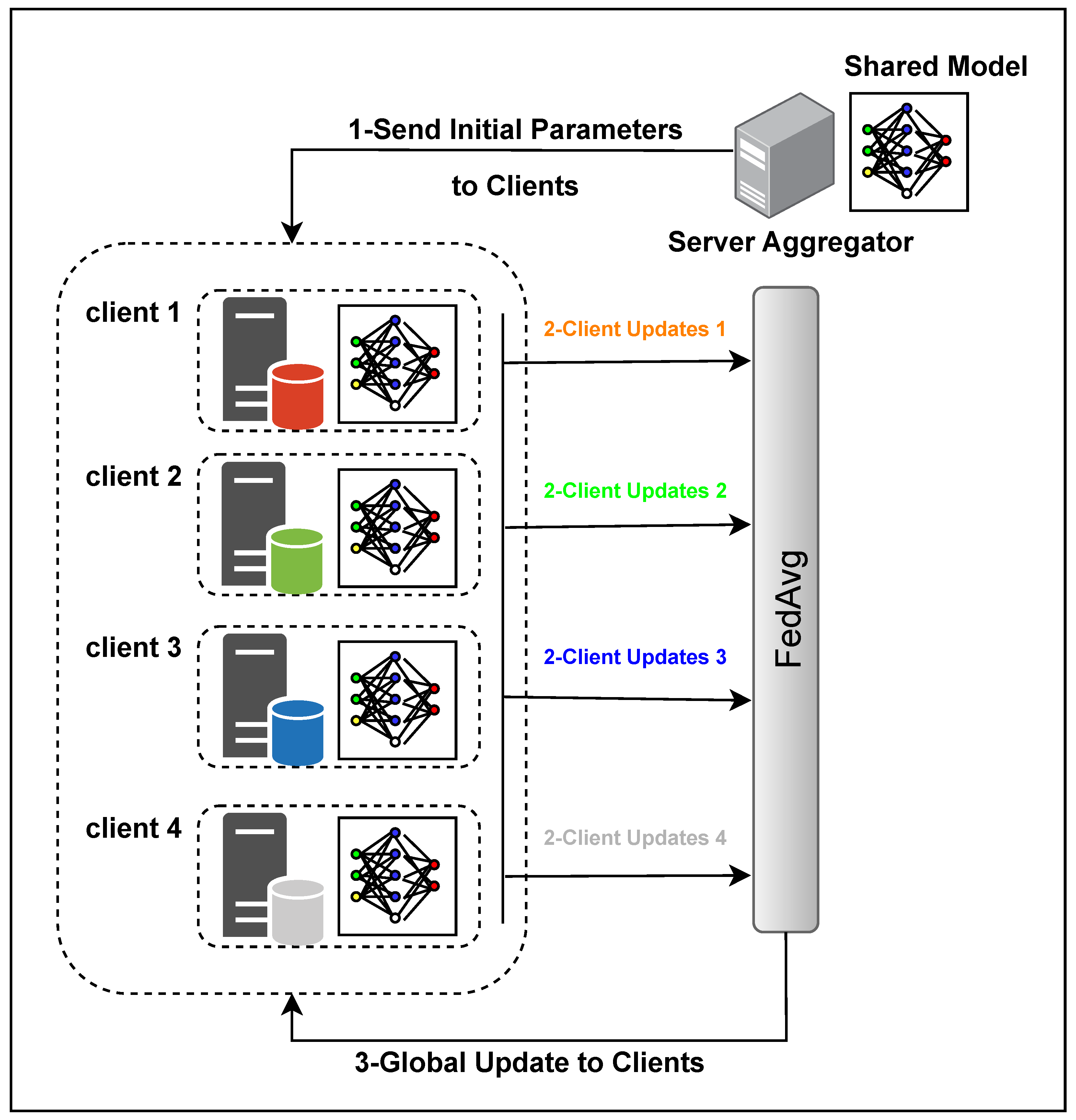
4.1. Federated Learning (FL)
4.1.1. Federated Learning Types and Conditions
- Non-IID (Non-Independently and Identically Distributed) Data: Patient data stored locally on different medical devices do not directly represent the entire population’s medical conditions.
- Unbalanced Local Data Sizes: Some medical institutions may have significantly larger datasets than others, introducing variations in the a available patient data.
- Massively Distributed: FL involves several medical institutions or clients participating in collaborative training.
- Limited Communication: When not all medical institutions are guaranteed to be online simultaneously, training may occur with a subset of devices, and the process may be asynchronous.
4.1.2. Averaging Algorithms
| Algorithm 1 The FedAvg algorithm. K clients are indexed by k, B is the local minibatch size, E is the number of local epochs, and is the learning rate. |
|
4.1.3. Overall Architecture
4.2. Privacy Preservation
- Encryption and Security
- -
- Objective: Implement robust encryption mechanisms to secure data during transmission and storage, preventing unauthorized access.
- -
- Methodology: Utilize advanced cryptographic techniques, including homomorphic encryption, and secure multiparty computation (SMPC) and blockchain to maintain the confidentiality of data.
- Differential Privacy
- -
- Objective: Ensure that the FL model does not reveal information about specific data points to protect individual data contributors.
- -
- Methodology: Introduce controlled noise or randomness to the learning process, preserving individual privacy while maintaining the utility of the model.
- Leakage Measures
- -
- Objective: Quantify and assess potential information leakage during the FL process.
- -
- Methodology: Evaluate the extent to which individual data points or model information might be unintentionally disclosed, employing leakage measures for comprehensive analysis.
- Communication Overhead
- -
- Objective: Minimize the amount of communication between the central server and participating clients to enhance privacy.
- -
- Methodology: Optimize communication protocols and reduce unnecessary data exchange, balancing the need for information transfer with privacy considerations.
- Security Analysis
- -
- Objective: Conduct a thorough security analysis to identify vulnerabilities and threats to the FL system.
- -
- Methodology: Assess the system’s robustness against potential attacks, ensuring the implementation of effective countermeasures.
- Threat Modeling
- -
- Objective: Anticipate potential threats to the privacy of the FL system.
- -
- Methodology: Develop models to understand and mitigate identified threats, aligning the privacy measures with the anticipated risks.
- User Perception
- -
- Objective: Consider end-user perceptions and expectations regarding privacy protection measures.
- -
- Methodology: Align PP mechanisms with user expectations, ensuring transparency and user acceptance.
- Trade-offs
- -
- Objective: Acknowledge and discuss trade-offs between privacy preservation and model performance.
- -
- Methodology: Evaluate the impact of privacy measures on model utility and find a balance that aligns with the overarching goals of the FL system.
- Evolving Attack Techniques
- -
- Objective: Stay informed about emerging privacy attack techniques.
- -
- Methodology: Continuously update FL system defenses to adapt to evolving threats, ensuring the system’s resilience against new attack vectors.
- Reward-Driven Approaches
- -
- Objective: Encourage participants to contribute data while protecting their privacy.
- -
- Methodology: Implement incentive structures that reward data contributors, striking a balance between participation encouragement and privacy preservation.
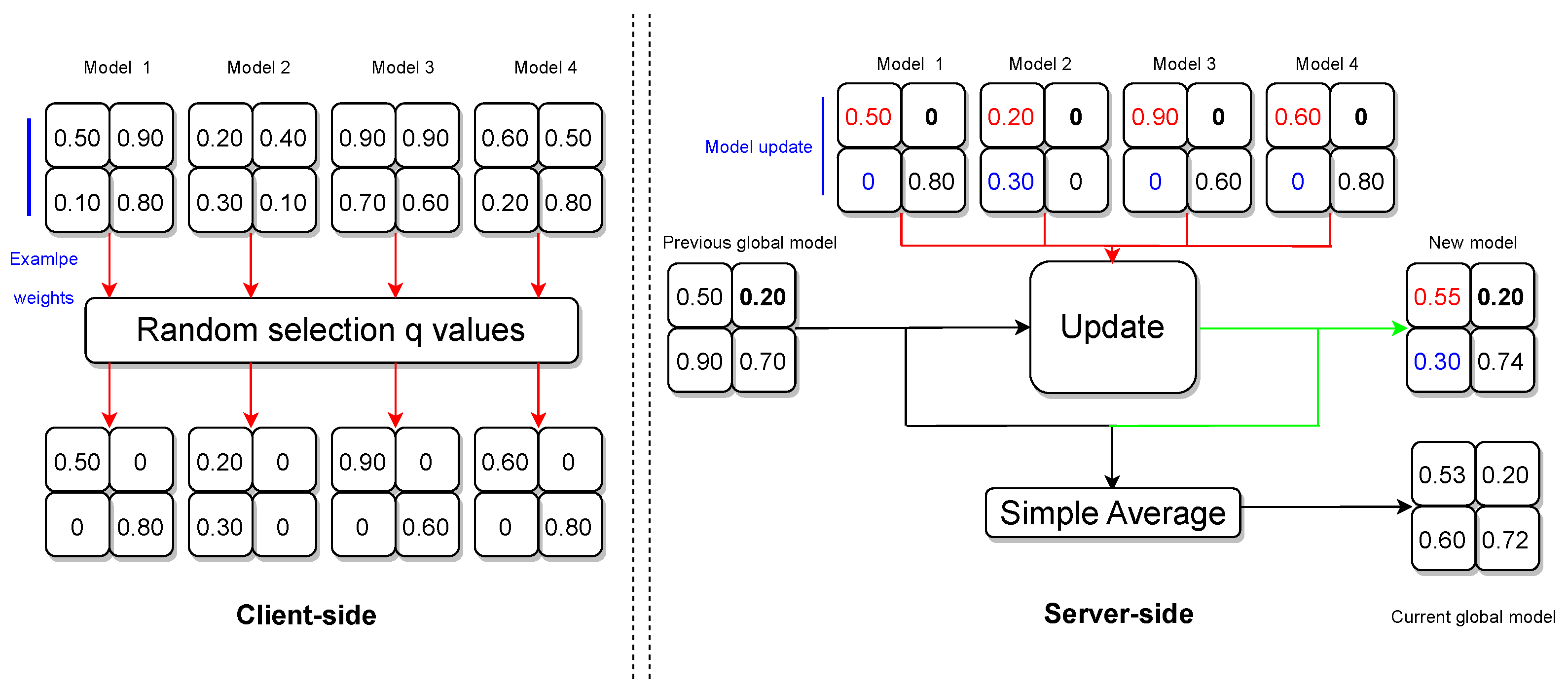
4.3. Privacy-Preserving Algorithm
- Handling Zero Weights
- -
- If a weight is 0 for all clients, the algorithm retains the previous global model weight for that position in the new model.
- -
- If a weight is 0 for all clients except one, the algorithm incorporates the nonzero weight from the single client into the new model.
- -
- If weights are nonzero for a subset of clients, the algorithm computes the average of those nonzero weights and disregards clients with 0 weights.
- -
- If weights are nonzero for all clients, the algorithm computes the average of those weights.
- Global Model Update
- -
- The algorithm concludes by computing a simple average between the previous global model and the new model weights.
- -
- The adjusted server model is shared with all participating clients in preparation for the forthcoming federation round.
4.4. Training and Validation Details
5. Results and Discussion
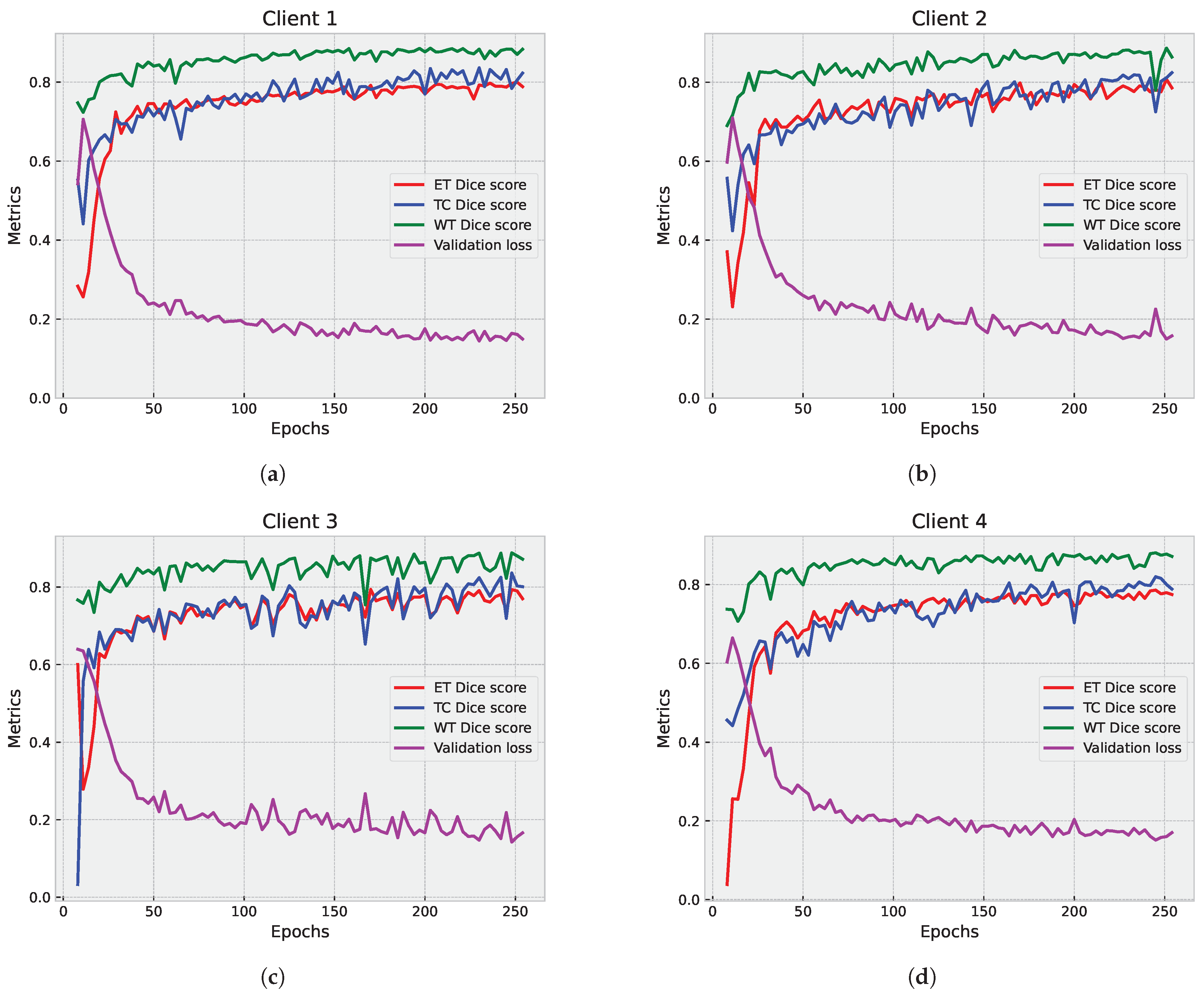
5.1. First Step Validation
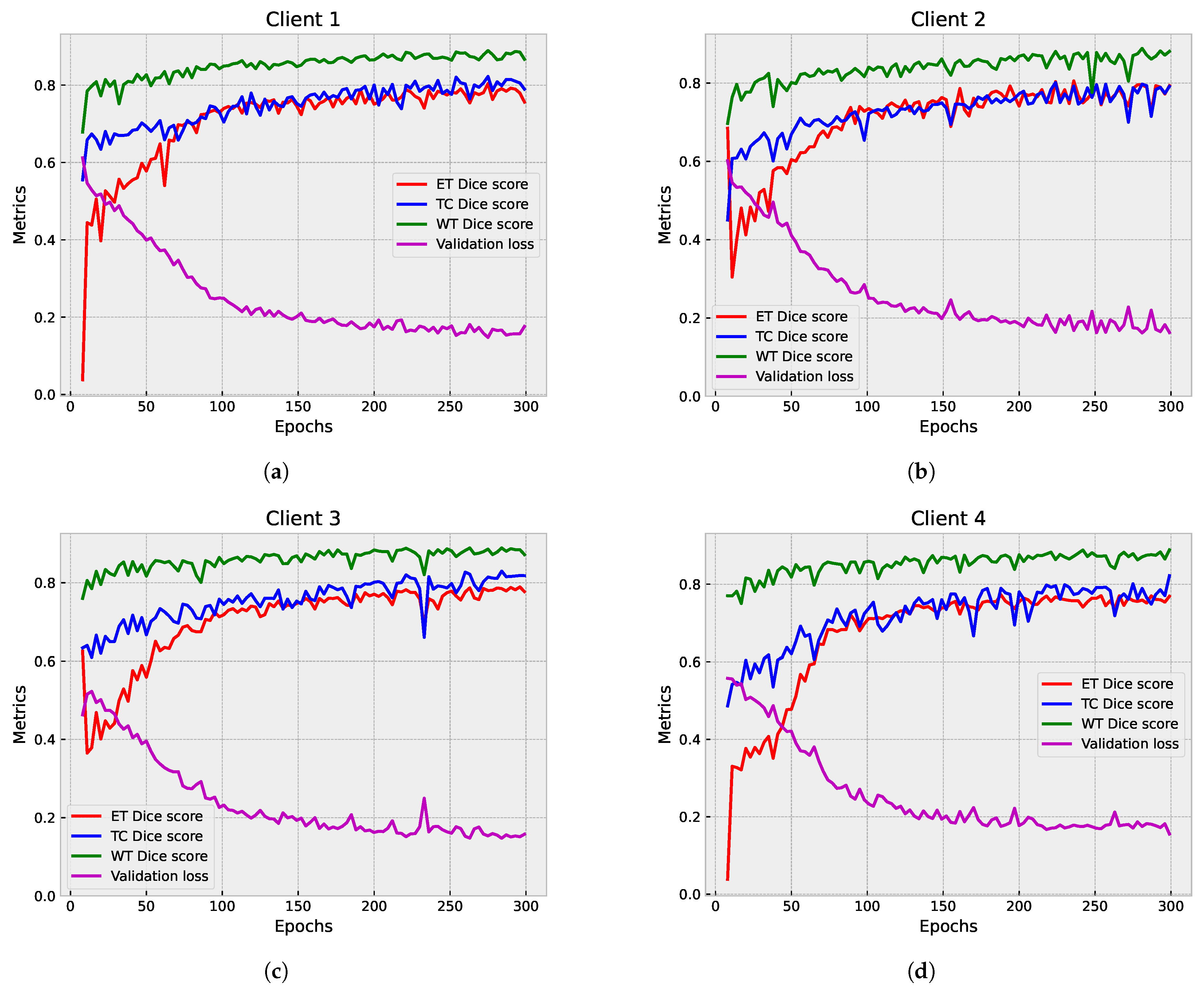
5.1.1. Partial Federated Deep Model
5.1.2. Full Federated Deep Model
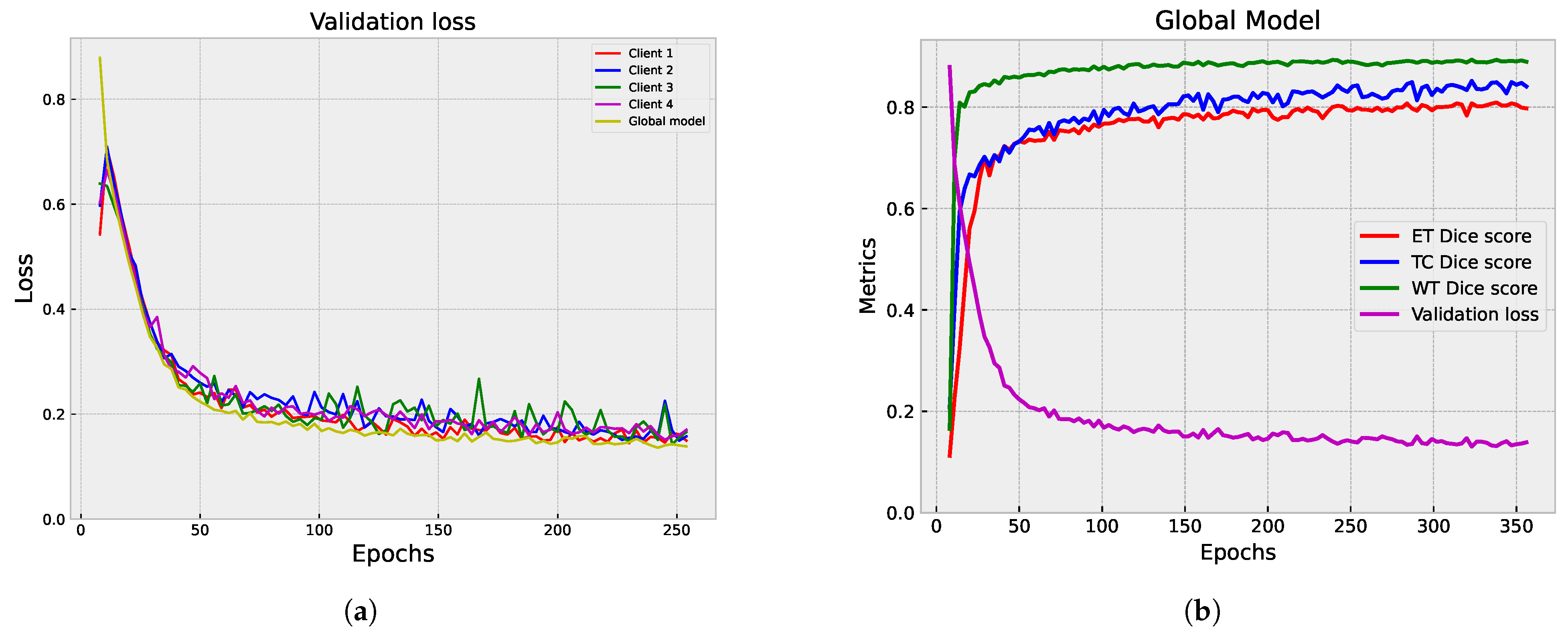
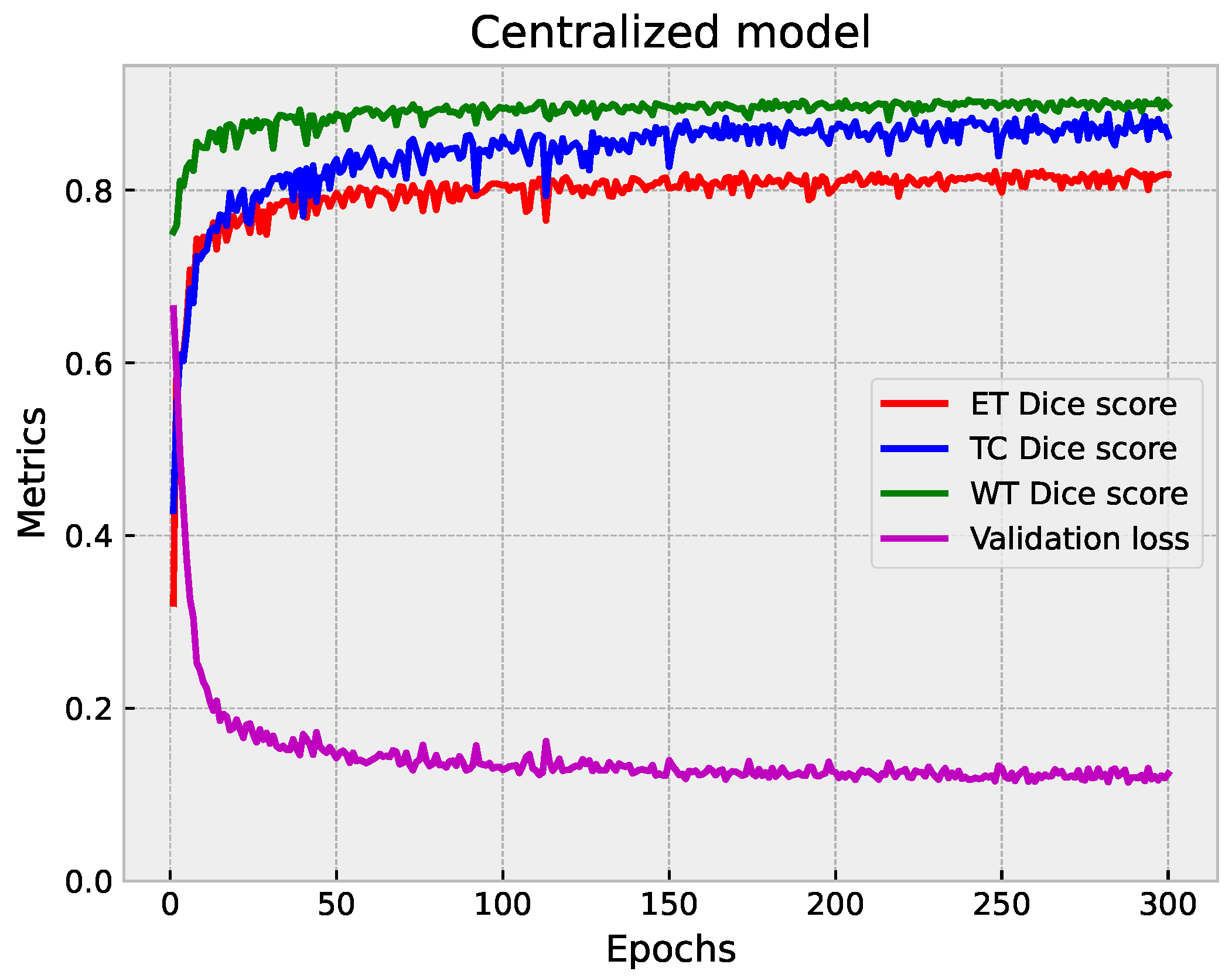
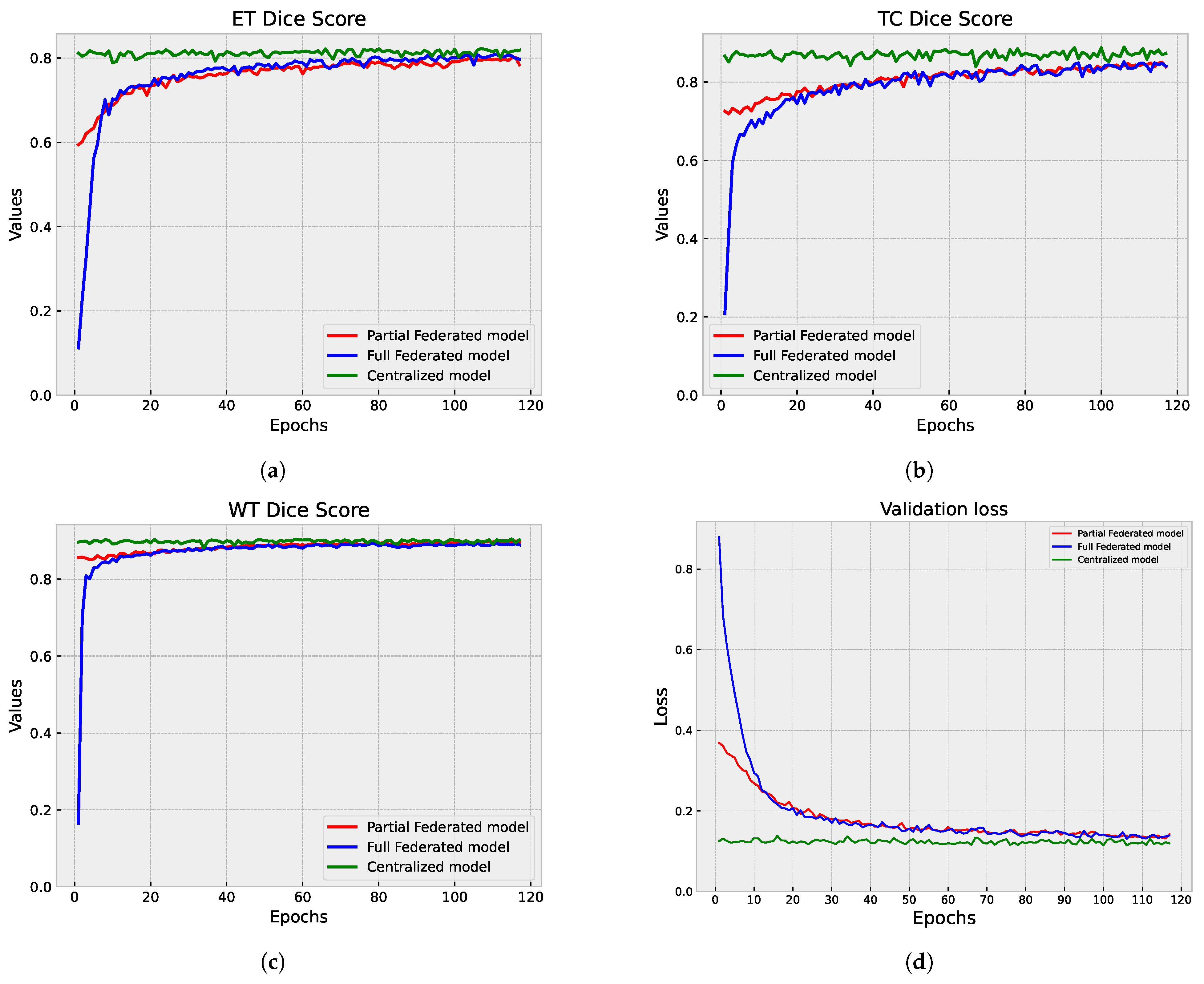
5.1.3. Centralized vs. Federated Approach: Convergence and Performance Analysis
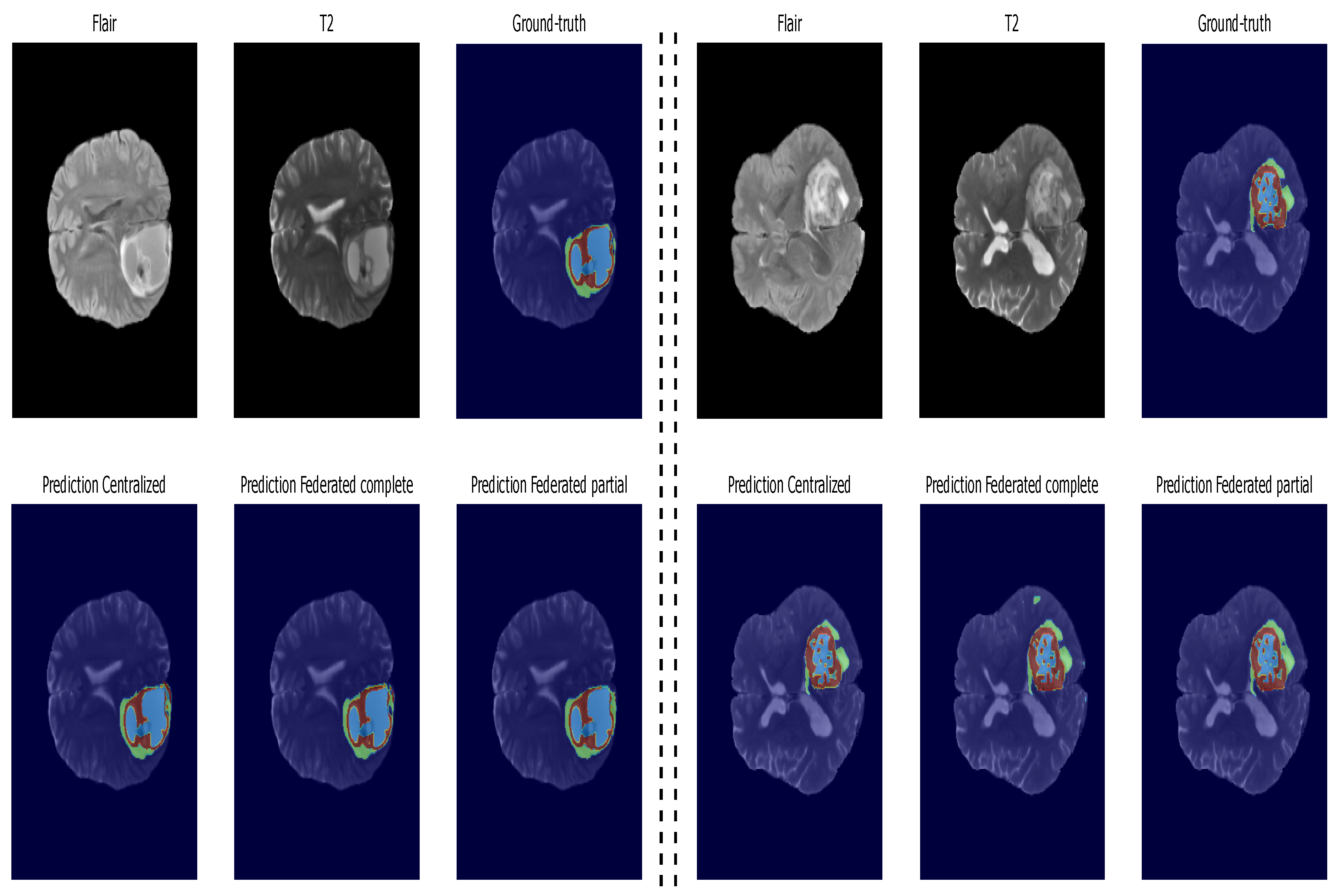
5.2. Second Step of Validation: Whole-Image Validation
5.3. Inference: Testing on Unseen Data
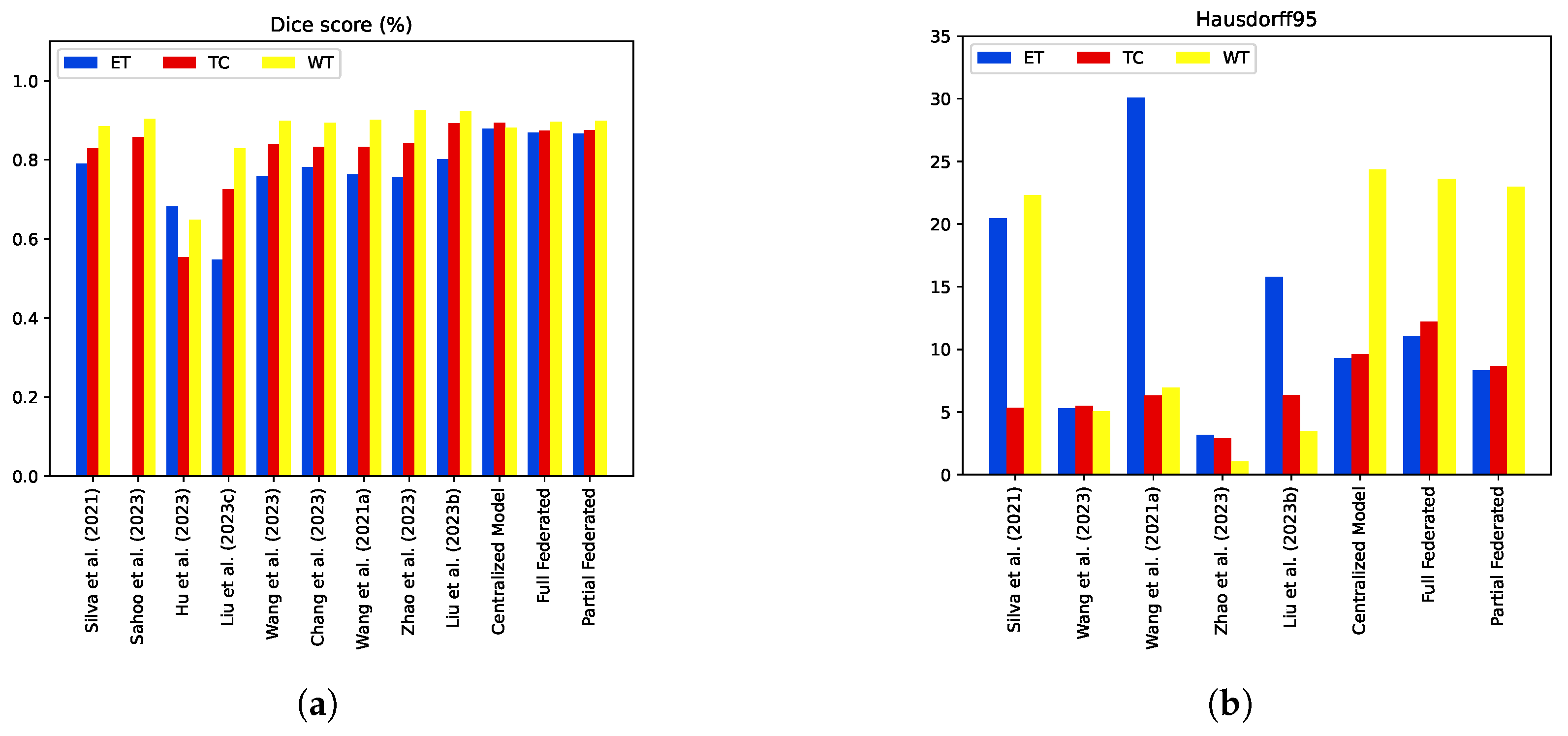
5.4. Model Comparisons with the State-of-the-Art Methods
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Bouamrane, A.; Derdour, M. Enhancing Lung Cancer Detection and Classification Using Machine Learning and Deep Learning Techniques: A Comparative Study. In Proceedings of the 2023 International Conference on Networking and Advanced Systems (ICNAS), Algiers, Algeria, 21–23 October 2023; IEEE: New York, NY, USA, 2023; pp. 1–6. [Google Scholar]
- Gasmi, M.; Derdour, M.; Gahmous, A. Transfer learning for the classification of small-cell and non-small-cell lung cancer. In Proceedings of the International Conference on Intelligent Systems and Pattern Recognition, Hammamet, Tunisia, 24–26 March 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 341–348. [Google Scholar]
- Gasmi, M.; Derdour, M.; Gahmousse, A.; Amroune, M.; Bendjenna, H.; Sahraoui, B. Multi-Input CNN for molecular classification in breast cancer. In Proceedings of the 2021 International Conference on Recent Advances in Mathematics and Informatics (ICRAMI), Tebessa, Algeria, 21–22 September 2021; IEEE: New York, NY, USA, 2021; pp. 1–5. [Google Scholar]
- Menaceur, S.; Derdour, M.; Bouramoul, A. Using Query Expansion Techniques and Content-Based Filtering for Personalizing Analysis in Big Data. Int. J. Inf. Technol. Web Eng. (IJITWE) 2020, 15, 77–101. [Google Scholar] [CrossRef]
- Mounir, A.; Adel, A.; Makhlouf, D.; Sébastien, L.; Philippe, R. A New Two-Level Clustering Approach for Situations Management in Distributed Smart Environments. Int. J. Ambient. Comput. Intell. (IJACI) 2019, 10, 91–111. [Google Scholar] [CrossRef]
- Kahil, M.S.; Bouramoul, A.; Derdour, M. GreedyBigVis–A greedy approach for preparing large datasets to multidimensional visualization. Int. J. Comput. Appl. 2022, 44, 760–769. [Google Scholar] [CrossRef]
- Kahil, M.S.; Bouramoul, A.; Derdour, M. Multi Criteria-Based Community Detection and Visualization in Large-scale Networks Using Label Propagation Algorithm. In Proceedings of the 2021 International Conference on Recent Advances in Mathematics and Informatics (ICRAMI), Tebessa, Algeria, 21–22 September 2021; IEEE: New York, NY, USA, 2021; pp. 1–6. [Google Scholar]
- Silva, C.A.; Pinto, A.; Pereira, S.; Lopes, A. Multi-stage Deep Layer Aggregation for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Eds.; Springer: Cham, Switzerland, 2021; pp. 179–188. [Google Scholar]
- Sahoo, A.K.; Parida, P.; Muralibabu, K.; Dash, S. An improved DNN with FFCM method for multimodal brain tumor segmentation. Intell. Syst. Appl. 2023, 18, 200245. [Google Scholar] [CrossRef]
- Hu, J.; Gu, X.; Wang, Z.; Gu, X. Mixture of calibrated networks for domain generalization in brain tumor segmentation. Knowl. -Based Syst. 2023, 270, 110520. [Google Scholar] [CrossRef]
- Liu, Z.; Wei, J.; Li, R.; Zhou, J. Learning multi-modal brain tumor segmentation from privileged semi-paired MRI images with curriculum disentanglement learning. Comput. Biol. Med. 2023, 159, 106927. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Bai, X. Gradient-assisted deep model for brain tumor segmentation by multi-modality MRI volumes. Biomed. Signal Process. Control 2023, 85, 105066. [Google Scholar] [CrossRef]
- Chang, Y.; Zheng, Z.; Sun, Y.; Zhao, M.; Lu, Y.; Zhang, Y. DPAFNet: A Residual Dual-Path Attention-Fusion Convolutional Neural Network for Multimodal Brain Tumor Segmentation. Biomed. Signal Process. Control 2023, 79, 104037. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, Y.; Li, J.; Wu, H.; Wang, S.; Dong, X.; Yu, H. A lightweight hierarchical convolution network for brain tumor segmentation. BMC Bioinform. 2021, 22, 636. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xing, Z.; Chen, Z.; Wan, L.; Han, T.; Fu, H.; Zhu, L. Uncertainty-aware multi-dimensional mutual learning for brain and brain tumor segmentation. IEEE J. Biomed. Health Inform. 2023, 27, 4362–4372. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ma, C.; She, W.; Wang, X. TransMVU: Multi-view 2D U-Nets with transformer for brain tumour segmentation. IET Image Process. 2023, 17, 1874–1882. [Google Scholar] [CrossRef]
- Mohammed, B.A.; Senan, E.M.; Alshammari, T.S.; Alreshidi, A.; Alayba, A.M.; Alazmi, M.; Alsagri, A.N. Hybrid techniques of analyzing mri images for early diagnosis of brain tumours based on hybrid features. Processes 2023, 11, 212. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E.; Rassem, T.H.; Aljaloud, A.S.; Mohammed, B.A.; Al-Mekhlafi, Z.G. Early diagnosis of brain tumour mri images using hybrid techniques between deep and machine learning. Comput. Math. Methods Med. 2022, 2022, 8330833. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, B.A.; Senan, E.M.; Al-Mekhlafi, Z.G.; Rassem, T.H.; Makbol, N.M.; Alanazi, A.A.; Almurayziq, T.S.; Ghaleb, F.A.; Sallam, A.A. Multi-method diagnosis of CT images for rapid detection of intracranial hemorrhages based on deep and hybrid learning. Electronics 2022, 11, 2460. [Google Scholar] [CrossRef]
- Sheller, M.J.; Reina, G.A.; Edwards, B.; Martin, J.; Bakas, S. Multi-institutional deep learning modeling without sharing patient data: A feasibility study on brain tumor segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers, Part I 4. Springer: Berlin/Heidelberg, Germany, 2019; pp. 92–104. [Google Scholar]
- Qiu, L.; Cheng, J.; Gao, H.; Xiong, W.; Ren, H. Federated semi-supervised learning for medical image segmentation via pseudo-label denoising. IEEE J. Biomed. Health Inform. 2023, 27, 4672–4683. [Google Scholar] [CrossRef]
- Elbachir, Y.M.; Makhlouf, D.; Mohamed, G.; Bouhamed, M.M.; Abdellah, K. Federated Learning for Multi-institutional on 3D Brain Tumor Segmentation. In Proceedings of the 2024 6th International Conference on Pattern Analysis and Intelligent Systems (PAIS), El Oued, Algeria, 24–25 April 2024; IEEE: New York, NY, USA, 2024; pp. 1–8. [Google Scholar]
- Xu, X.; Deng, H.H.; Gateno, J.; Yan, P. Federated multi-organ segmentation with inconsistent labels. IEEE Trans. Med. Imaging 2023, 42, 2948–2960. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, C.; Qin, J.; Dou, Q.; Heng, P.A. Feddg: Federated domain generalization on medical image segmentation via episodic learning in continuous frequency space. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 1013–1023. [Google Scholar]
- Agrawal, S.; Sarkar, S.; Aouedi, O.; Yenduri, G.; Piamrat, K.; Alazab, M.; Bhattacharya, S.; Maddikunta, P.K.R.; Gadekallu, T.R. Federated Learning for intrusion detection system: Concepts, challenges and future directions. Comput. Commun. 2022, 195, 346–361. [Google Scholar] [CrossRef]
- Lazzarini, R.; Tianfield, H.; Charissis, V. Federated learning for IoT intrusion detection. AI 2023, 4, 509–530. [Google Scholar] [CrossRef]
- Wang, W.; He, F.; Li, Y.; Tang, S.; Li, X.; Xia, J.; Lv, Z. Data information processing of traffic digital twins in smart cities using edge intelligent federation learning. Inf. Process. Manag. 2023, 60, 103171. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, S.; Fujita, H.; Hu, X.; Zhang, Y.; Wang, B.; Wang, K. Fault diagnosis based on federated learning driven by dynamic expansion for model layers of imbalanced client. Expert Syst. Appl. 2024, 238, 121982. [Google Scholar] [CrossRef]
- Chen, J.; Xue, J.; Wang, Y.; Huang, L.; Baker, T.; Zhou, Z. Privacy-Preserving and Traceable Federated Learning for data sharing in industrial IoT applications. Expert Syst. Appl. 2023, 213, 119036. [Google Scholar] [CrossRef]
- Qi, T.; Wu, F.; Wu, C.; He, L.; Huang, Y.; Xie, X. Differentially private knowledge transfer for federated learning. Nat. Commun. 2023, 14, 3785. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Wu, F.; Zhang, J. Selective knowledge sharing for privacy-preserving federated distillation without a good teacher. Nat. Commun. 2024, 15, 349. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Milletarì, F.; Xu, D.; Rieke, N.; Hancox, J.; Zhu, W.; Baust, M.; Cheng, Y.; Ourselin, S.; Cardoso, M.J.; et al. Privacy-preserving federated brain tumour segmentation. In Proceedings of the Machine Learning in Medical Imaging: 10th International Workshop, MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, 13 October 2019; Proceedings 10. Springer: Berlin/Heidelberg, Germany, 2019; pp. 133–141. [Google Scholar]
- Ziller, A.; Passerat-Palmbach, J.; Ryffel, T.; Usynin, D.; Trask, A.; Junior, I.D.L.C.; Mancuso, J.; Makowski, M.; Rueckert, D.; Braren, R.; et al. Privacy-preserving medical image analysis. arXiv 2020, arXiv:2012.06354. [Google Scholar]
- Lu, Y.; Huang, X.; Zhang, K.; Maharjan, S.; Zhang, Y. Blockchain and federated learning for 5G beyond. IEEE Netw. 2020, 35, 219–225. [Google Scholar] [CrossRef]
- Xu, G.; Zhou, Z.; Dong, J.; Zhang, L.; Song, X. A blockchain-based federated learning scheme for data sharing in industrial internet of things. IEEE Internet Things J. 2023, 10, 21467–21478. [Google Scholar] [CrossRef]
- Sameera, K.M.; Nicolazzo, S.; Arazzi, M.; Nocera, A.; KA, R.R.; Vinod, P.; Conti, M. Privacy-Preserving in Blockchain-based Federated Learning Systems. arXiv 2024, arXiv:2401.03552. [Google Scholar]
- Kaissis, G.A.; Makowski, M.R.; Rückert, D.; Braren, R.F. Secure, privacy-preserving and federated machine learning in medical imaging. Nat. Mach. Intell. 2020, 2, 305–311. [Google Scholar] [CrossRef]
- Wang, Y.; Su, Z.; Zhang, N.; Benslimane, A. Learning in the air: Secure federated learning for UAV-assisted crowdsensing. IEEE Trans. Netw. Sci. Eng. 2020, 8, 1055–1069. [Google Scholar] [CrossRef]
- Zhang, C.; Li, S.; Xia, J.; Wang, W.; Yan, F.; Liu, Y. {BatchCrypt}: Efficient homomorphic encryption for {Cross-Silo} federated learning. In Proceedings of the 2020 USENIX Annual Technical Conference (USENIX ATC 20), Online, 15–17 July 2020; pp. 493–506. [Google Scholar]
- Zhang, L.; Xu, J.; Vijayakumar, P.; Sharma, P.K.; Ghosh, U. Homomorphic encryption-based privacy-preserving federated learning in iot-enabled healthcare system. IEEE Trans. Netw. Sci. Eng. 2022, 10, 2864–2880. [Google Scholar] [CrossRef]
- Henry, T.; Carré, A.; Lerousseau, M.; Estienne, T.; Robert, C.; Paragios, N.; Deutsch, E. Brain Tumor Segmentation with Self-ensembled, Deeply-Supervised 3D U-Net Neural Networks: A BraTS 2020 Challenge Solution. Lect. Notes Comput. Sci. (Incl. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2021, 12658 LNCS, 327–339. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Brendan McMahan, H.; Moore, E.; Ramage, D.; Hampson, S.; Agüera y Arcas, B. Communication-efficient learning of deep networks from decentralized data. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, AISTATS 2017, Lauderdale, FL, USA, 20–22 April 2017; Volume 54. [Google Scholar]
- Konečný, J.; McMahan, B.; Ramage, D. Federated Optimization:Distributed Optimization Beyond the Datacenter. arXiv 2015, arXiv:1511.03575. [Google Scholar]
- Hitaj, B.; Ateniese, G.; Perez-Cruz, F. Deep models under the GAN: Information leakage from collaborative deep learning. In Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security, Dallas, TX, USA, 30 October–3 November 2017; pp. 603–618. [Google Scholar]




| Method | Dice | Sensitivity | Hausdorff95 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | ET | WT | TC | ET | WT | TC | |
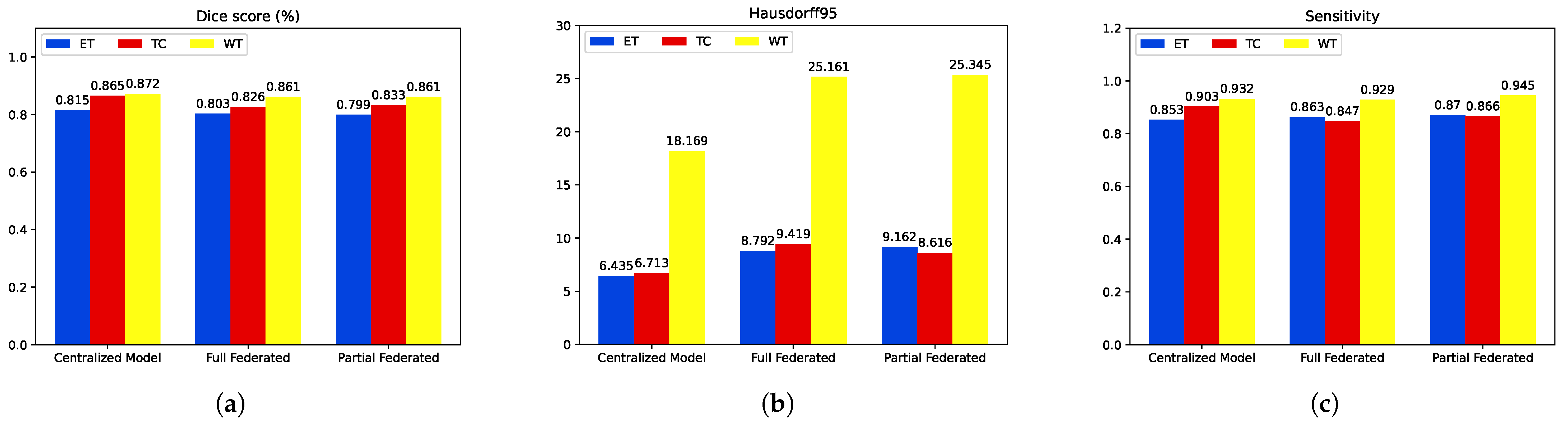
| Proposed Centralized Model | 0.815 | 0.865 | 0.872 | 0.853 | 0.932 | 0.903 | 6.435 | 18.169 | 6.713 |
| Full Federated | 0.803 | 0.826 | 0.861 | 0.863 | 0.929 | 0.847 | 8.792 | 25.161 | 9.419 |
| Partial Federated | 0.798 | 0.833 | 0.861 | 0.870 | 0.945 | 0.866 | 9.162 | 25.345 | 8.616 |
| Method | Dice | Sensitivity | Hausdorff95 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | ET | WT | TC | ET | WT | TC | |
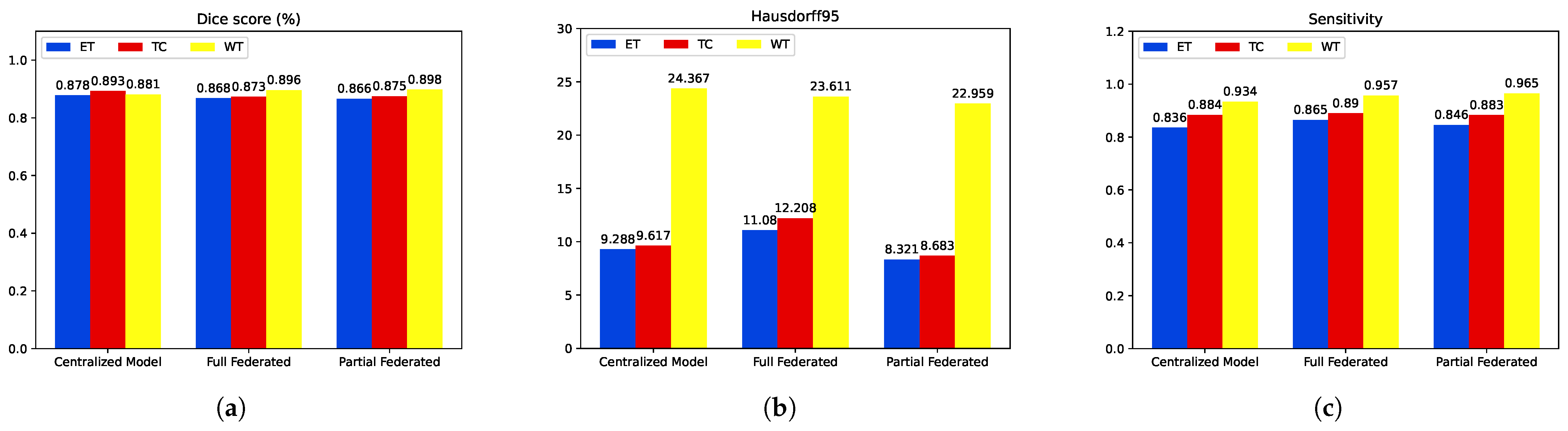
| Proposed Centralized Model | 0.878 | 0.893 | 0.881 | 0.836 | 0.934 | 0.884 | 9.28 | 24.36 | 9.61 |
| Full Federated | 0.868 | 0.873 | 0.896 | 0.865 | 0.957 | 0.890 | 11.088 | 23.611 | 12.208 |
| Partial Federated | 0.866 | 0.875 | 0.898 | 0.846 | 0.965 | 0.883 | 8.321 | 22.959 | 8.683 |
| Method | Dice | Sensitivity | Specificity | Hausdorff95 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | ET | WT | TC | ET | WT | TC | ET | WT | TC | |
| [10] | 0.79000 | 0.82970 | 0.88580 | - | - | - | - | - | - | 20.44 | 22.32 | 5.32 |
| [11] | - | 0.85750 | 0.90360 | - | 0.892 | 0.841 | - | 0.995 | 0.994 | - | - | - |
| [12] | 0.68250 | 0.55480 | 0.64820 | - | - | - | - | - | - | - | - | - |
| [13] | 0.54800 | 0.72620 | 0.82910 | - | - | - | - | - | - | - | - | - |
| [14] | 0.75800 | 0.84020 | 0.89910 | - | - | - | - | - | - | 5.29 | 5.07 | 5.51 |
| [15] | 0.78100 | 0.83200 | 0.89400 | - | - | - | - | - | - | - | - | - |
| [16] | 0.76380 | 0.83320 | 0.90100 | - | - | - | - | - | - | 30.09 | 6.96 | 6.30 |
| [17] | 0.75600 | 0.84300 | 0.9240 | - | - | - | - | - | - | 3.19 | 1.04 | 2.88 |
| [18] | 0.80200 | 0.8920 | 0.9230 | - | - | - | - | - | - | 15.80 | 3.44 | 6.35 |
| Proposed Centralized Model | 0.878 | 0.893 | 0.881 | 0.836 | 0.934 | 0.884 | 0.999 | 0.998 | 0.999 | 9.28 | 24.36 | 9.61 |
| Full Federated | 0.868 | 0.873 | 0.896 | 0.865 | 0.957 | 0.890 | 0.999 | 0.997 | 0.998 | 11.088 | 23.611 | 12.208 |
| Partial Federated | 0.866 | 0.875 | 0.898 | 0.846 | 0.965 | 0.883 | 0.999 | 0.998 | 0.999 | 8.321 | 22.959 | 8.683 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yahiaoui, M.E.; Derdour, M.; Abdulghafor, R.; Turaev, S.; Gasmi, M.; Bennour, A.; Aborujilah, A.; Sarem, M.A. Federated Learning with Privacy Preserving for Multi- Institutional Three-Dimensional Brain Tumor Segmentation. Diagnostics 2024, 14, 2891. https://doi.org/10.3390/diagnostics14242891
Yahiaoui ME, Derdour M, Abdulghafor R, Turaev S, Gasmi M, Bennour A, Aborujilah A, Sarem MA. Federated Learning with Privacy Preserving for Multi- Institutional Three-Dimensional Brain Tumor Segmentation. Diagnostics. 2024; 14(24):2891. https://doi.org/10.3390/diagnostics14242891
Chicago/Turabian StyleYahiaoui, Mohammed Elbachir, Makhlouf Derdour, Rawad Abdulghafor, Sherzod Turaev, Mohamed Gasmi, Akram Bennour, Abdulaziz Aborujilah, and Mohamed Al Sarem. 2024. "Federated Learning with Privacy Preserving for Multi- Institutional Three-Dimensional Brain Tumor Segmentation" Diagnostics 14, no. 24: 2891. https://doi.org/10.3390/diagnostics14242891
APA StyleYahiaoui, M. E., Derdour, M., Abdulghafor, R., Turaev, S., Gasmi, M., Bennour, A., Aborujilah, A., & Sarem, M. A. (2024). Federated Learning with Privacy Preserving for Multi- Institutional Three-Dimensional Brain Tumor Segmentation. Diagnostics, 14(24), 2891. https://doi.org/10.3390/diagnostics14242891










