Novel Domain Knowledge-Encoding Algorithm Enables Label-Efficient Deep Learning for Cardiac CT Segmentation to Guide Atrial Fibrillation Treatment in a Pilot Dataset
Abstract
:1. Introduction
2. Methods
2.1. Dataset for Training and Testing
2.2. Domain Knowledge-Encoding (DOKEN) Algorithm
- I.
- Segmentation of digital LA models: N = 6 digital LA models (publicly available) were segmented based on an iterative erosion–dilation (ED) process (Figure 2).
- II.
- Tuning ED parameter using patient LA models: The iterative ED process requires optimal iteration number as a parameter, which decides the accurate segmentation of the LA body and other substructures. To determine this parameter, 5 manually segmented LA models were used to train support vector machines (SVMs) to predict the optimal iteration in the ED process.
- I.
- Segmentation of digital LA models
- Calculate the centroid of the LA body and the centroid of each virtually dissected substructure (4 PVs and LAA).
- For each substructure centroid, create a centerline automatically by minimizing the integral of the radius of maximal inscribed spheres along the path that connects the substructure centroid to the LA body centroid.
- Replace the boundary between the left atrium and each substructure by a plane orthogonal to the corresponding centerline and close to the original boundary generated by the ED process (Figure 2(A4)).
- II.
- Tuning the ED parameter using patient LA models
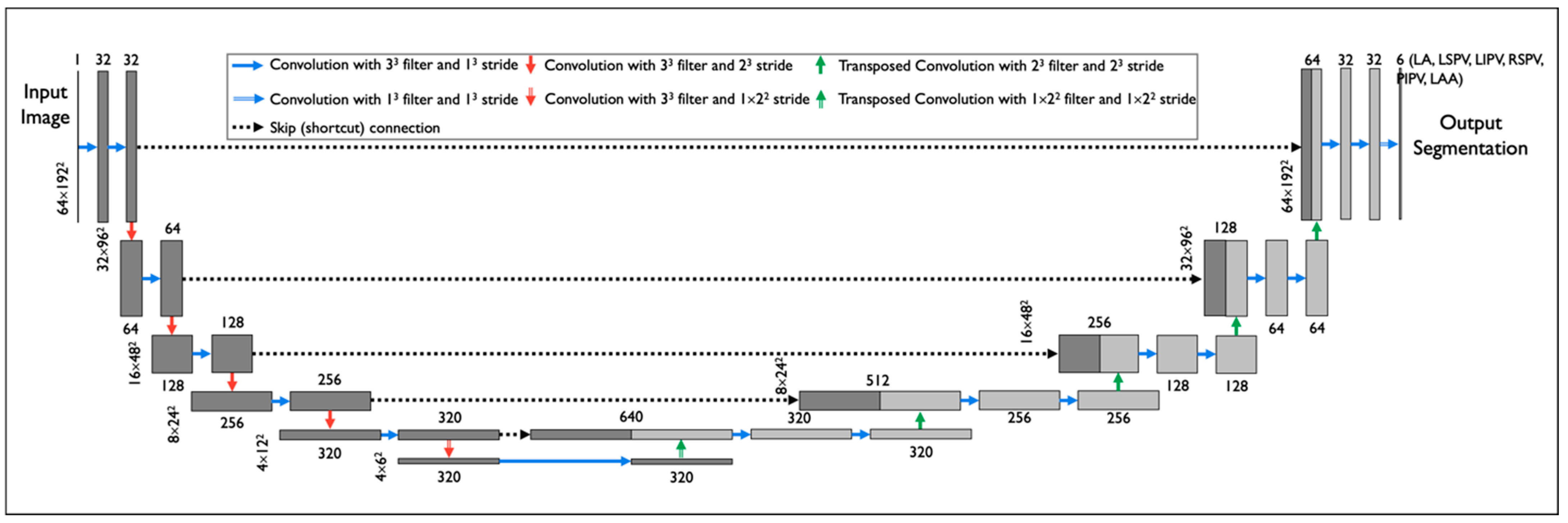
2.3. Training the DNN for CT Segmentation from a Small Training Set
2.4. Experimental Setting for Performance Evaluation
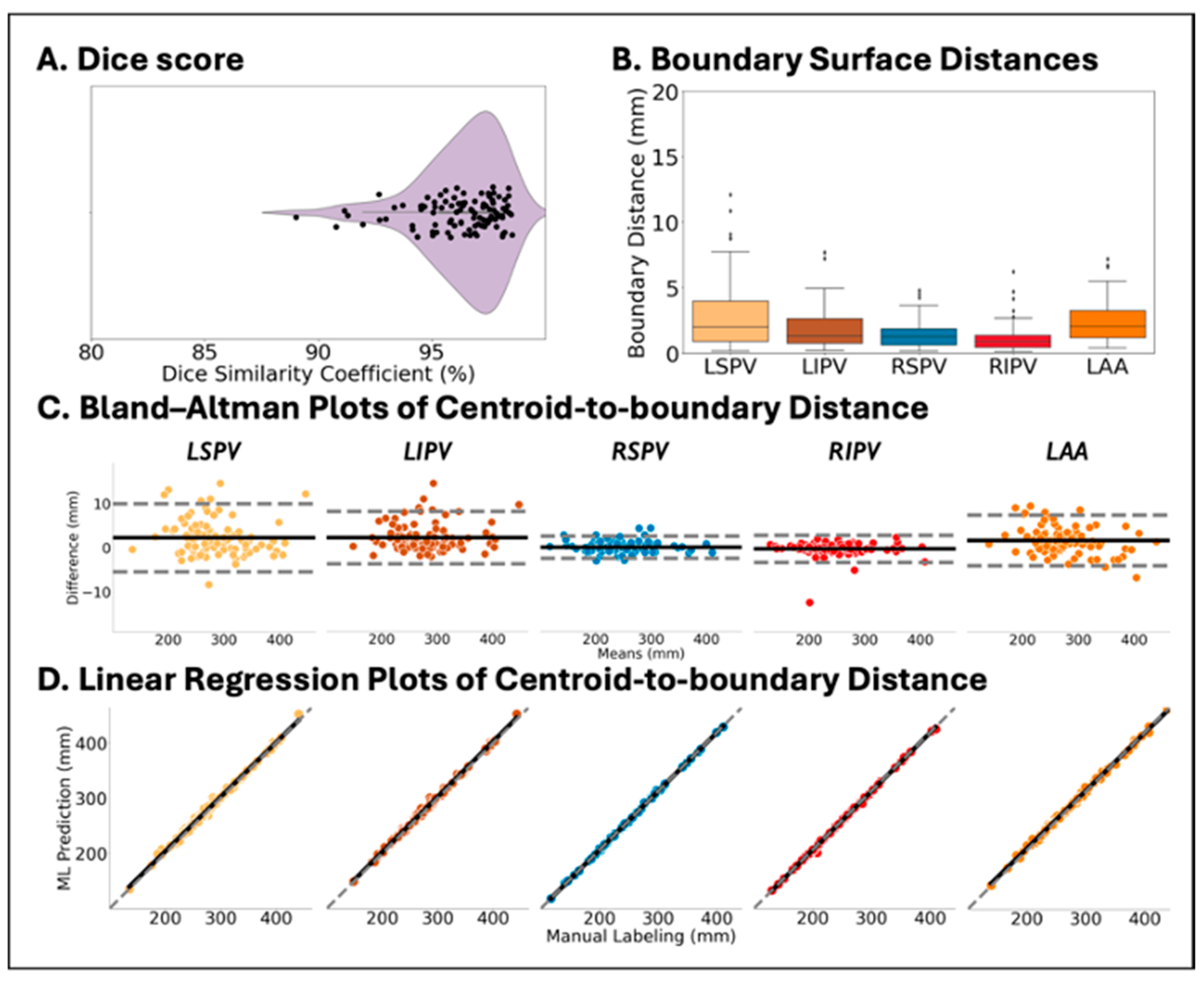
2.5. Performance Evaluation
2.6. Statistical Analysis
3. Results
3.1. DOKEN Algorithm Can Robustly Parse Cardiac Geometry
3.2. DNN Trained by DOKEN-Labeled Samples Can Accurately Segment CT
3.3. Analysis of Anatomical Variants
4. Discussion
4.1. DNN Segmentation of Cardiac CT Images
4.2. Challenges in Machine Learning
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boykov, Y.; Funka-Lea, G. Graph cuts and efficient ND image segmentation. Int. J. Comput. Vis. 2006, 70, 109–131. [Google Scholar] [CrossRef]
- Preetha, M.M.S.J.; Suresh, L.P.; Bosco, M.J. Image segmentation using seeded region growing. In Proceedings of the 2012 International Conference on Computing, Electronics and Electrical Technologies (ICCEET), Nagercoil, India, 21–22 March 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 576–583. [Google Scholar]
- Rodriguez, M.D.; Shah, M. Detecting and segmenting humans in crowded scenes. In Proceedings of the 15th ACM international conference on Multimedia, Augsburg, Germany, 24–29 September 2007; pp. 353–356. [Google Scholar]
- Rajaraman, S.; Siegelman, J.; Alderson, P.O.; Folio, L.S.; Folio, L.R.; Antani, S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access 2020, 8, 115041–115050. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, P.; Rajaraman, S.; Long, R.; Ghoraani, B.; Antani, S. Assessment of data augmentation strategies toward performance improvement of abnormality classification in chest radiographs. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Piscataway, NJ, USA; pp. 841–844. [Google Scholar]
- Tobon-Gomez, C.; Geers, A.J.; Peters, J.; Weese, J.; Pinto, K.; Karim, R.; Ammar, M.; Daoudi, A.; Margeta, J.; Sandoval, Z.; et al. Benchmark for algorithms segmenting the left atrium from 3D CT and MRI datasets. IEEE Trans. Med. Imaging 2015, 34, 1460–1473. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Li, L.; Payer, C.; Štern, D.; Urschler, M.; Heinrich, M.P.; Oster, J.; Wang, C.; Smedby, Ö.; Bian, C.; et al. Evaluation of algorithms for multi-modality whole heart segmentation: An open-access grand challenge. Med. Image Anal. 2019, 58, 101537. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, T.; Zhuang, J.; Yuan, H.; Huang, M.; Cen, J.; Jia, Q.; Dong, Y.; Shi, Y. Imagechd: A 3d computed tomography image dataset for classification of congenital heart disease. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany; pp. 77–87. [Google Scholar]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern. Med. 2018, 178, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- Futoma, J.; Simons, M.; Panch, T.; Doshi-Velez, F.; Celi, L.A. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health 2020, 2, e489–e492. [Google Scholar] [CrossRef] [PubMed]
- Markman, E.M. Categorization and Naming in Children: Problems of Induction; Mit Press: Cambridge, MA, USA, 1989. [Google Scholar]
- Van Gerven, M. Computational foundations of natural intelligence. Front. Comput. Neurosci. 2017, 11, 112. [Google Scholar] [CrossRef]
- Lake, B.M.; Salakhutdinov, R.; Tenenbaum, J.B. Human-level concept learning through probabilistic program induction. Science 2015, 350, 1332–1338. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wolterink, J.M.; Brune, C.; Veldhuis, R.N. Anatomy-aided deep learning for medical image segmentation: A review. Phys. Med. Biol. 2021, 66, 11TR01. [Google Scholar] [CrossRef]
- Xie, X.; Niu, J.; Liu, X.; Chen, Z.; Tang, S.; Yu, S. A survey on incorporating domain knowledge into deep learning for medical image analysis. Med. Image Anal. 2021, 69, 101985. [Google Scholar] [CrossRef]
- Nagel, C.; Schuler, S.; Dössel, O.; Loewe, A. A bi-atrial statistical shape model for large-scale in silico studies of human atria: Model development and application to ECG simulations. Med. Image Anal. 2021, 74, 102210. [Google Scholar] [CrossRef] [PubMed]
- Soille, P. Erosion and Dilation, Morphological Image Analysis; Springer: Berlin, Germany, 2004. [Google Scholar]
- Krissian, K.; Carreira, J.M.; Esclarin, J.; Maynar, M. Semi-automatic segmentation and detection of aorta dissection wall in MDCT angiography. Med. Image Anal. 2014, 18, 83–102. [Google Scholar] [CrossRef] [PubMed]
- Razeghi, O.; Sim, I.; Roney, C.H.; Karim, R.; Chubb, H.; Whitaker, J.; O’Neill, L.; Mukherjee, R.; Wright, M.; O’neill, M.; et al. Fully automatic atrial fibrosis assessment using a multilabel convolutional neural network. Circ. Cardiovasc. Imaging 2020, 13, e011512. [Google Scholar] [CrossRef] [PubMed]
- Piccinelli, M.; Veneziani, A.; Steinman, D.A.; Remuzzi, A.; Antiga, L. A framework for geometric analysis of vascular structures: Application to cerebral aneurysms. IEEE Trans. Med. Imaging 2009, 28, 1141–1155. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Ketkar, N. Stochastic gradient descent. In Deep Learning with Python; Springer: Berlin/Heidelberg, Germany, 2017; pp. 113–132. [Google Scholar]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [PubMed]
- Baskaran, L.; Maliakal, G.; Al’Aref, S.J.; Singh, G.; Xu, Z.; Michalak, K.; Dolan, K.; Gianni, U.; van Rosendael, A.; van den Hoogen, I.; et al. Identification and quantification of cardiovascular structures from CCTA: An end-to-end, rapid, pixel-wise, deep-learning method. Cardiovasc. Imaging 2020, 13, 1163–1171. [Google Scholar]
- Xu, H.; Niederer, S.A.; Williams, S.E.; Newby, D.E.; Williams, M.C.; Young, A.A. Whole Heart Anatomical Refinement from CCTA Using Extrapolation and Parcellation. In Proceedings of the International Conference on Functional Imaging and Modeling of the Heart, Stanford, CA, USA, 21–25 June 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 63–70. [Google Scholar]
- Chen, H.H.; Liu, C.M.; Chang, S.L.; Chang, P.Y.C.; Chen, W.S.; Pan, Y.M.; Fang, S.T.; Zhan, S.Q.; Chuang, C.M.; Lin, Y.J.; et al. Automated extraction of left atrial volumes from two-dimensional computer tomography images using a deep learning technique. Int. J. Cardiol. 2020, 316, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Yao, Z.; Ji, E.; Qiu, H.; Chen, Z.; Guo, H.; Zhuang, J.; Jia, Q.; Huang, M. Artificial intelligence–based computed tomography processing framework for surgical telementoring of congenital heart disease. ACM J. Emerg. Technol. Comput. Syst. (JETC) 2021, 17, 1–24. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; Springer: Berlin/Heidelberg, Germany, 2014; pp. 740–755. [Google Scholar]
- Sangsriwong, M.; Cismaru, G.; Puiu, M.; Simu, G.; Istratoaie, S.; Muresan, L.; Gusetu, G.; Cismaru, A.; Pop, D.; Zdrenghea, D.; et al. Formula to estimate left atrial volume using antero-posterior diameter in patients with catheter ablation of atrial fibrillation. Medicine 2021, 100, e26513. [Google Scholar] [CrossRef]
- Taubin, G. Curve and surface smoothing without shrinkage. In Proceedings of the IEEE International Conference on Computer Vision, Cambridge, MA, USA, 20–23 June 1995; IEEE: Piscataway, NJ, USA, 1995; pp. 852–857. [Google Scholar]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep learning for cardiac image segmentation: A review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, S.; Ngo, L.H.; Tugal, D.; Manning, W.J.; Nezafat, R. Incremental value of left atrial geometric remodeling in predicting late atrial fibrillation recurrence after pulmonary vein isolation: A cardiovascular magnetic resonance study. J. Am. Heart Assoc. 2018, 7, e009793. [Google Scholar] [CrossRef] [PubMed]
- Firouznia, M.; Feeny, A.K.; LaBarbera, M.A.; McHale, M.; Cantlay, C.; Kalfas, N.; Schoenhagen, P.; Saliba, W.; Tchou, P.; Barnard, J.; et al. Machine learning–derived fractal features of shape and texture of the left atrium and pulmonary veins from cardiac computed tomography scans are associated with risk of recurrence of atrial fibrillation postablation. Circ. Arrhythmia Electrophysiol. 2021, 14, e009265. [Google Scholar] [CrossRef] [PubMed]
- Tovia-Brodie, O.; Belhassen, B.; Glick, A.; Shmilovich, H.; Aviram, G.; Rosso, R.; Michowitz, Y. Use of new imaging CARTO® segmentation module software to facilitate ablation of ventricular arrhythmias. J. Cardiovasc. Electrophysiol. 2017, 28, 240–248. [Google Scholar] [CrossRef]
- Tops, L.F.; Bax, J.J.; Zeppenfeld, K.; Jongbloed, M.R.; Lamb, H.J.; van der Wall, E.E.; Schalij, M.J. Fusion of multislice computed tomography imaging with three-dimensional electroanatomic mapping to guide radiofrequency catheter ablation procedures. Heart Rhythm. 2005, 2, 1076–1081. [Google Scholar] [CrossRef]
- Feng, R.; Deb, B.; Ganesan, P.; Tjong, F.V.; Rogers, A.J.; Ruipérez-Campillo, S.; Somani, S.; Clopton, P.; Baykaner, T.; Rodrigo, M.; et al. Segmenting computed tomograms for cardiac ablation using machine learning leveraged by domain knowledge encoding. Front. Cardiovasc. Med. 2023, 10, 1189293. [Google Scholar] [CrossRef]
- Ng, A.Y. Preventing “overfitting” of cross-validation data. In Proceedings of the Fourteenth International Conference on Machine Learning (ICML), Nashville, TN, USA, 8–12 July 1997; Volume 97, pp. 245–253. [Google Scholar]
- Hu, W.; Fey, M.; Zitnik, M.; Dong, Y.; Ren, H.; Liu, B.; Catasta, M.; Leskovec, J. Open graph benchmark: Datasets for machine learning on graphs. Adv. Neural Inf. Process. Syst. 2020, 33, 22118–22133. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; Antunes, D.A.; Balaji, A.; Baraniuk, R.; Barberan, C.J.; Dannenfelser, R.; Dun, C.; Edrisi, M.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 2022, 13, 1728. [Google Scholar] [CrossRef] [PubMed]
- Vakalopoulou, M.; Chassagnon, G.; Bus, N.; Marini, R.; Zacharaki, E.I.; Revel, M.P.; Paragios, N. Atlasnet: Multi-atlas non-linear deep networks for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 658–666. [Google Scholar]
- Trutti, A.C.; Fontanesi, L.; Mulder, M.J.; Bazin, P.-L.; Hommel, B.; Forstmann, B.U. A probabilistic atlas of the human ventral tegmental area (VTA) based on 7 Tesla MRI data. Brain Struct. Funct. 2021, 226, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.; Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Bhanot, K.; Qi, M.; Erickson, J.S.; Guyon, I.; Bennett, K.P. The problem of fairness in synthetic healthcare data. Entropy 2021, 23, 1165. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Shen, J.; Dudley, J.; Kristensson, P.O. The imaginative generative adversarial network: Automatic data augmentation for dynamic skeleton-based hand gesture and human action recognition. In Proceedings of the 2021 16th IEEE International Conference on Automatic Face and Gesture Recognition (FG 2021), Jodhpur, India, 15–18 December 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–8. [Google Scholar]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesan, P.; Feng, R.; Deb, B.; Tjong, F.V.Y.; Rogers, A.J.; Ruipérez-Campillo, S.; Somani, S.; Clopton, P.; Baykaner, T.; Rodrigo, M.; et al. Novel Domain Knowledge-Encoding Algorithm Enables Label-Efficient Deep Learning for Cardiac CT Segmentation to Guide Atrial Fibrillation Treatment in a Pilot Dataset. Diagnostics 2024, 14, 1538. https://doi.org/10.3390/diagnostics14141538
Ganesan P, Feng R, Deb B, Tjong FVY, Rogers AJ, Ruipérez-Campillo S, Somani S, Clopton P, Baykaner T, Rodrigo M, et al. Novel Domain Knowledge-Encoding Algorithm Enables Label-Efficient Deep Learning for Cardiac CT Segmentation to Guide Atrial Fibrillation Treatment in a Pilot Dataset. Diagnostics. 2024; 14(14):1538. https://doi.org/10.3390/diagnostics14141538
Chicago/Turabian StyleGanesan, Prasanth, Ruibin Feng, Brototo Deb, Fleur V. Y. Tjong, Albert J. Rogers, Samuel Ruipérez-Campillo, Sulaiman Somani, Paul Clopton, Tina Baykaner, Miguel Rodrigo, and et al. 2024. "Novel Domain Knowledge-Encoding Algorithm Enables Label-Efficient Deep Learning for Cardiac CT Segmentation to Guide Atrial Fibrillation Treatment in a Pilot Dataset" Diagnostics 14, no. 14: 1538. https://doi.org/10.3390/diagnostics14141538
APA StyleGanesan, P., Feng, R., Deb, B., Tjong, F. V. Y., Rogers, A. J., Ruipérez-Campillo, S., Somani, S., Clopton, P., Baykaner, T., Rodrigo, M., Zou, J., Haddad, F., Zaharia, M., & Narayan, S. M. (2024). Novel Domain Knowledge-Encoding Algorithm Enables Label-Efficient Deep Learning for Cardiac CT Segmentation to Guide Atrial Fibrillation Treatment in a Pilot Dataset. Diagnostics, 14(14), 1538. https://doi.org/10.3390/diagnostics14141538




