1. Introduction
Two-dimensional to three-dimensional registration is the process that matches a 3D model of an object with 2D image pairs and recovers the 3D pose of the object. High-speed biplanar videoradiography (HSBV) or dual fluoroscopy (DF) is an X-ray-based imaging system that can capture a series of 2D radiographic image pairs. It uses a low-dose X-ray to provide dynamic imagery of the bones. The 3D bone models can be obtained from magnetic resonance imaging (MRI) or computed tomography (CT) scans. Registering a 3D bone model to sequential 2D image pairs allows the determination of dynamic bony poses and provides valuable 3D kinematics information. Accurately estimating these kinematic parameters is critical for calculating joint cartilage contact mechanics that can provide insights into the mechanical processes and mechanisms of joint degeneration or pathology. For example, with the registered 3D bone models for the tibia (shank bone) and the femur (thigh bone), the tibiofemoral soft tissue model overlap can be quantified to estimate the cartilage deformation or contact regions [
1,
2,
3,
4]. An increased cartilage deformation rate under loading could be an early sign of osteoarthritis [
2,
5]. The cartilage thickness for early osteoarthritic or healthy knees under loading has been found to range from 0.3 mm to 1.2 mm [
2]. Therefore, the estimation accuracy of the cartilage contact thickness should be sub-millimeter for an early osteoarthritis diagnosis, and the same accuracy is required for the estimation at registration.
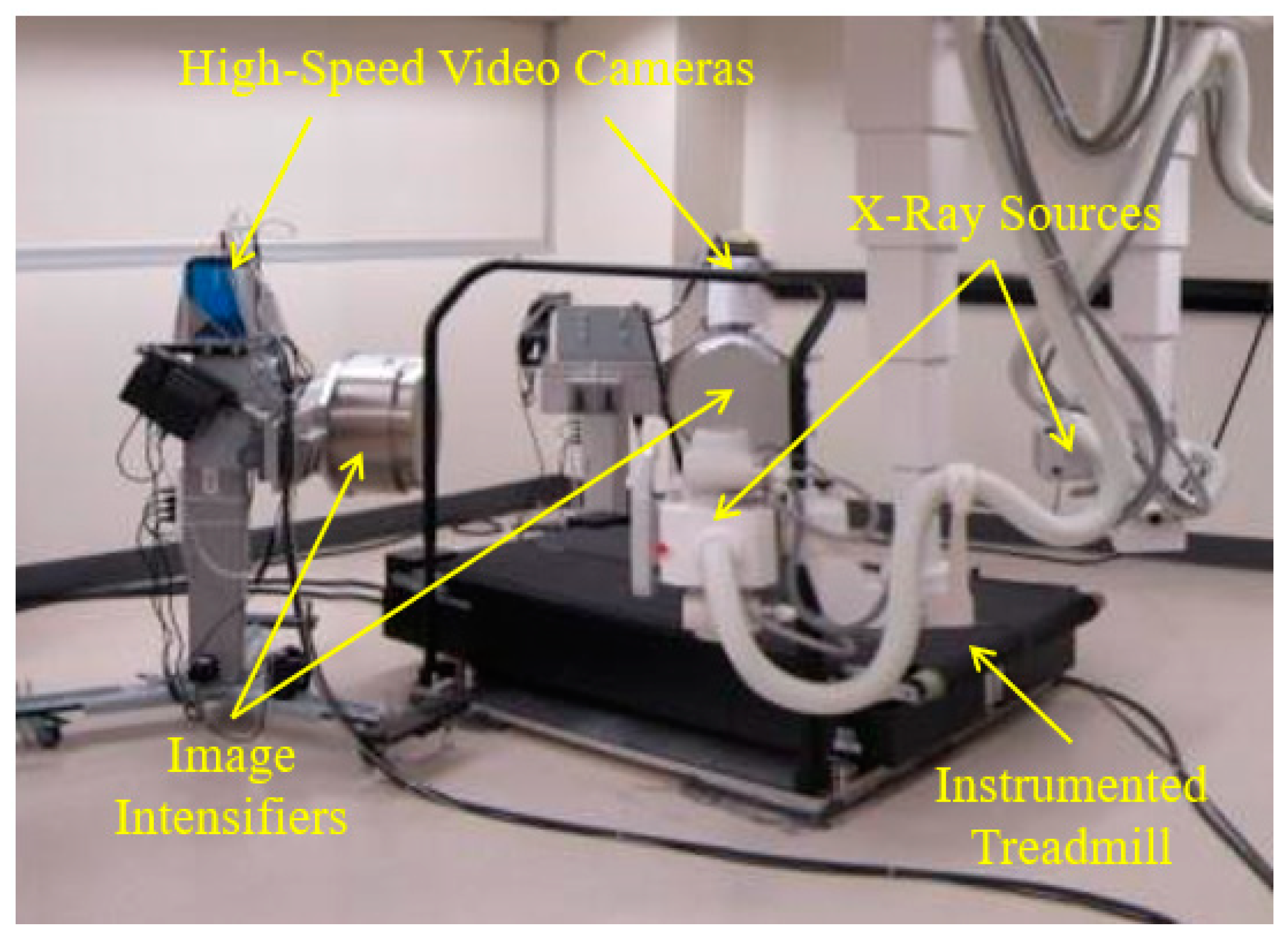
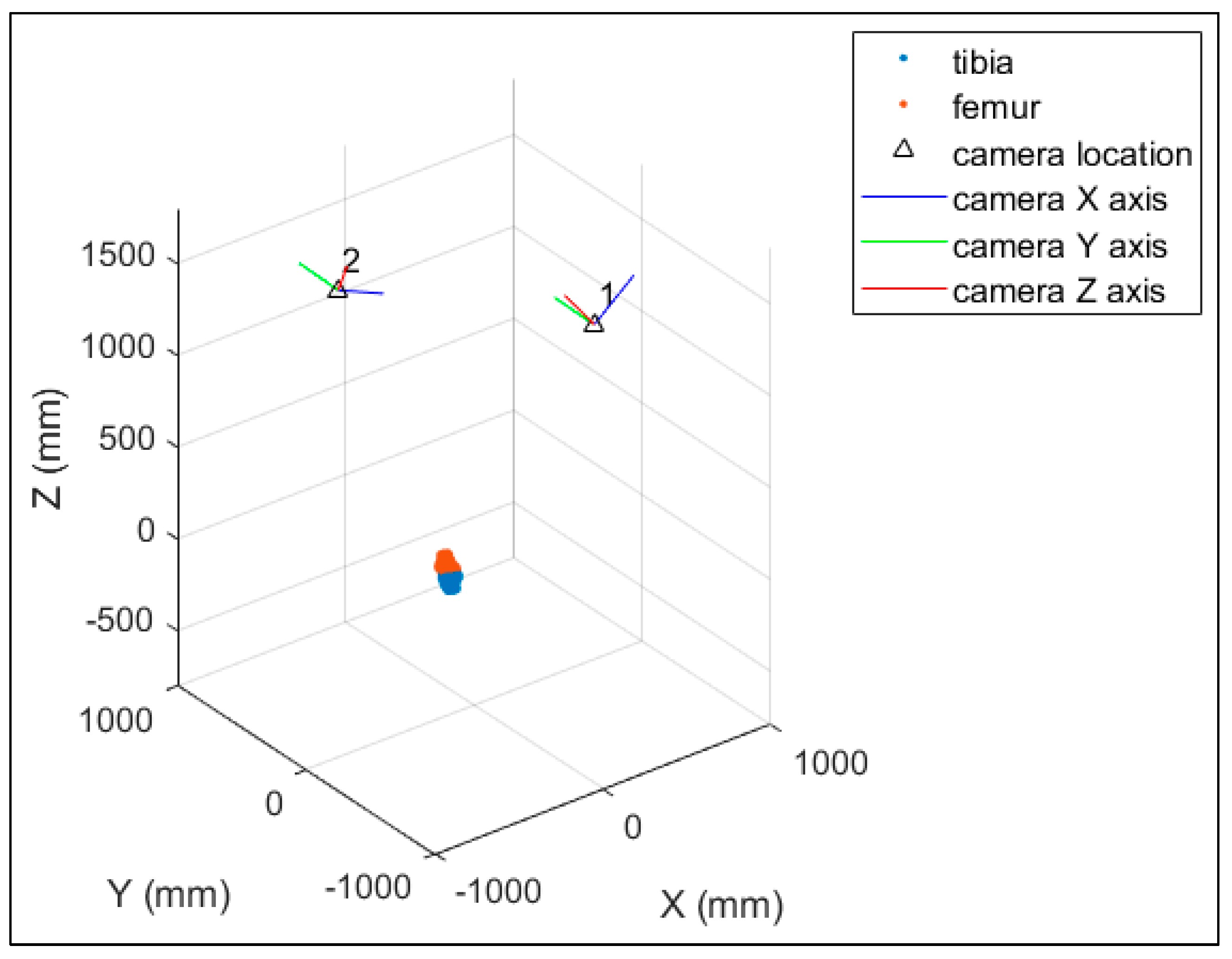
The HSBV system in the current study (
Figure 1) is located at the Clinical Movement Assessment Laboratory, University of Calgary, Canada. It is composed of two X-ray sources (time-synchronized, G-1086, Varian, Palo Alto, CA, USA), two X-ray image intensifiers (406 mm diameter, E5876SD-P2A, Toshiba, Tokyo, Japan), and two high-frame-rate video cameras (DIMAX, PCO, Kelheim, Germany). Dynamic movement, such as walking or running, is possible with an instrumented treadmill (Bertec, Columbus, OH, USA) positioned within the equipment’s field of view.
The HSBV motion reconstruction procedure has the following four steps: 3D bone model reconstruction, HSBV system photogrammetric self-calibration, biplane videoradiograph series acquisition, and 3D motion reconstruction by 2D–3D registration. As this research focuses on determining cartilage contact, MRI-derived 3D reference bone models are used because they can acquire detailed soft tissue contrast that is difficult to obtain from CT scans. The HSBV system in the current study was calibrated using a bundle adjustment method [
6] to obtain the system’s geometry and distortion correction terms. This is a rigorous method and is more flexible than the commonly used direct linear transformation (DLT) technique [
6,
7].
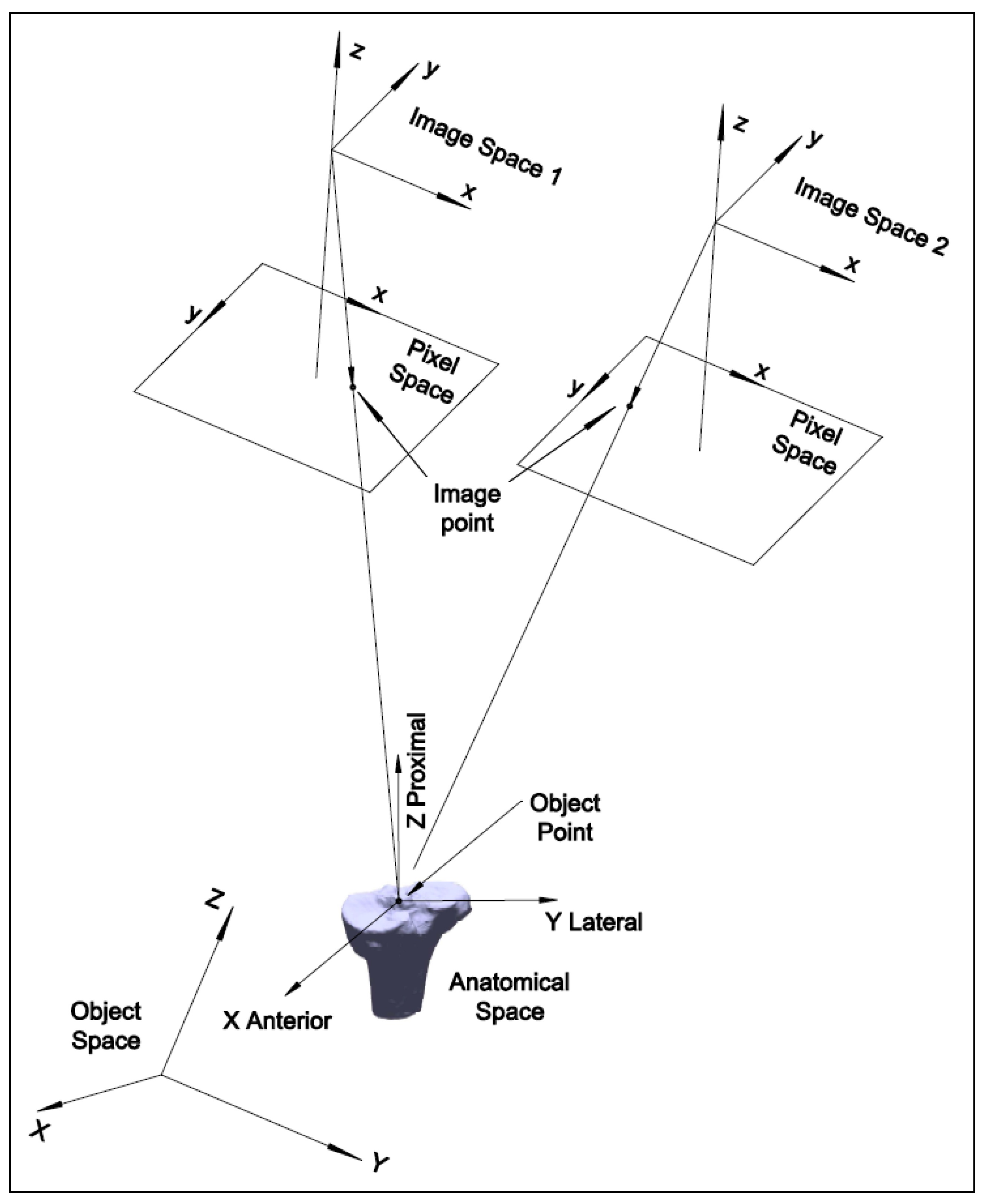
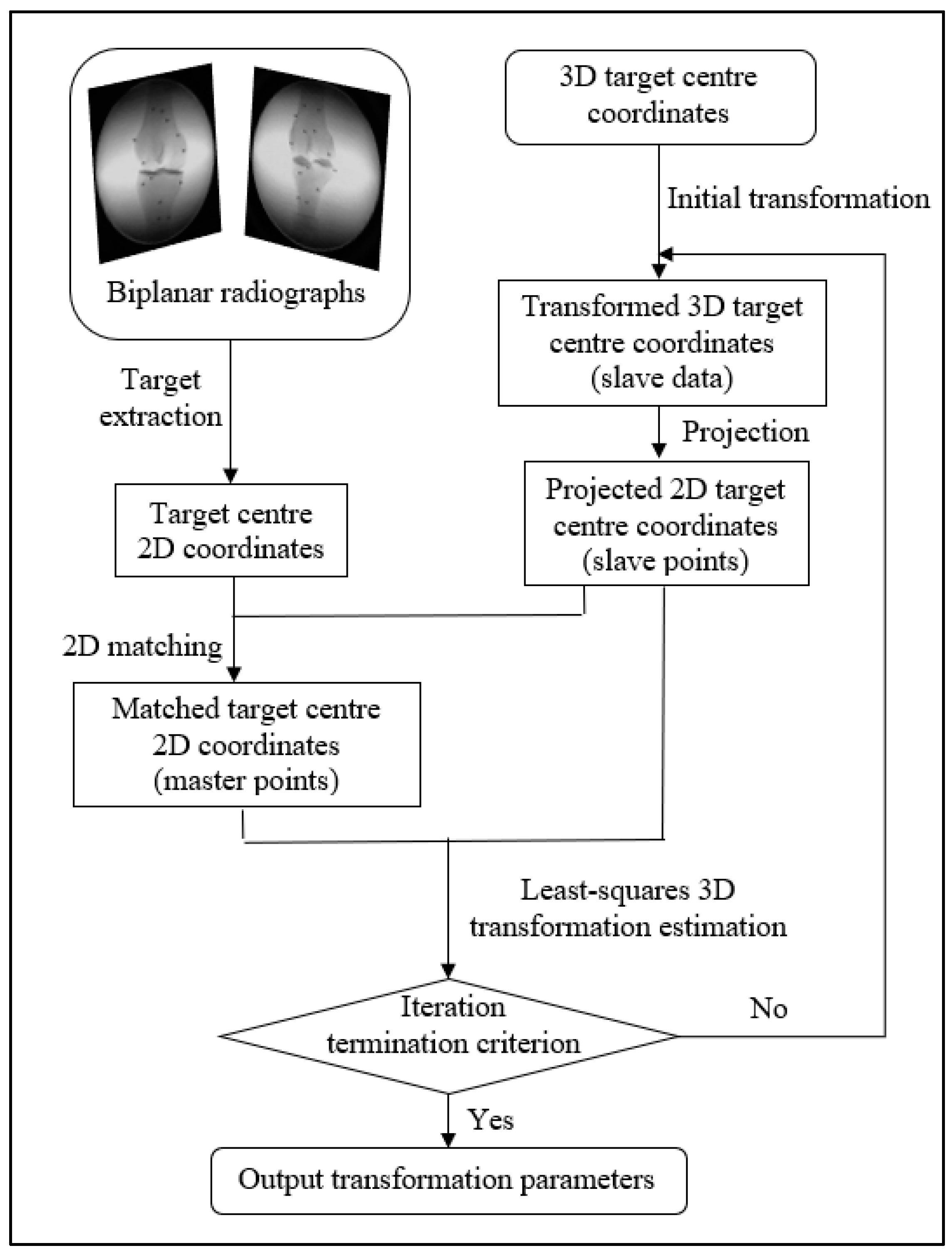
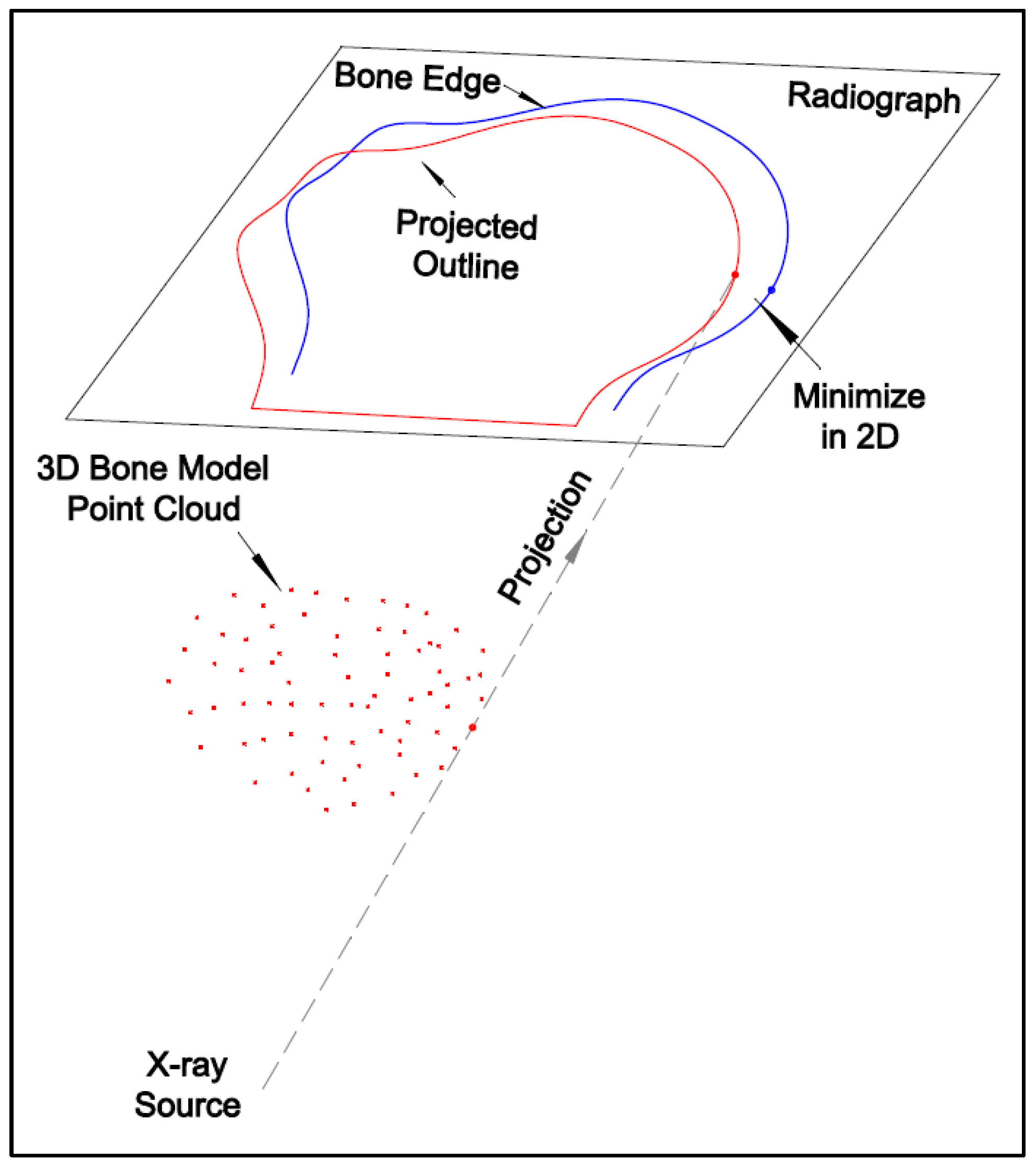
The 2D–3D registration is the process of determining the six-degree-of-freedom parameters (three translation and three rotation parameters) for the 3D bone model. This can be realized by matching the 3D bone model with the image pair at each frame, which can be conducted by either marker-based or model-based registration.
The Roentgen Stereophotogrammetry Analysis (RSA) system has been well-established for marker-based registration. It can estimate the 3D position of the markers implanted in bones and the kinematics of the skeletal segments [
8]. It first estimates the 3D object coordinates from the image pair and then the transformation parameters via Horn’s method [
9]. The error from the first estimation is propagated in the second step, so the optimization in the second step is biased. Therefore, there may be more accurate methods for marker-based registration than the RSA method.
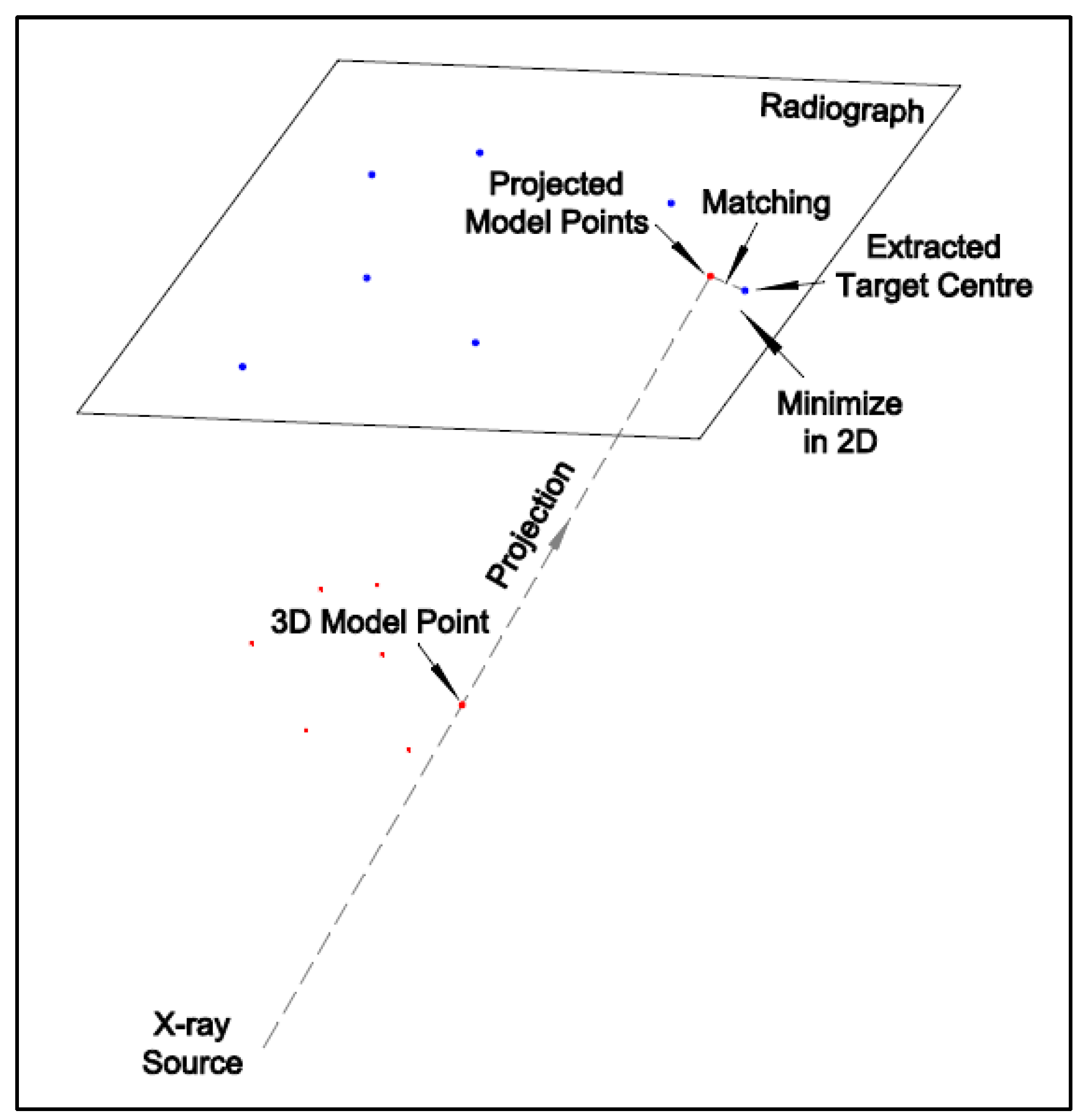
There are two commonly used model-based registration methods: intensity-based and feature-based. The intensity-based method matches the intensities of the pixels and voxels between 2D and 3D images and requires digitally reconstructed radiographs (DRRs) generated from 3D CT data. An intensity-based registration accuracy for a dynamic trial was reported [
10]. The root mean squared (RMS) errors were 0.88, 0.41, and 0.32 mm in the X-, Y-, and Z-axis for the tibia, and they were 0.60, 0.23, and 0.32 mm for the femur [
10]. This is equivalent to the distance of RMS errors of 1.02 mm for the tibia and 0.72 mm for the femur. However, this method is not suitable for MRI data as there is generally no physical correspondence between MRI-based DRRs and radiographs [
11,
12]. The feature-based method matches the feature locations in 2D and 3D images and minimizes the distances between the corresponding points. A feature-based registration for a dual fluoroscopic imaging system (DFIS), consisting of two C-arms and one treadmill, was reported [
13]. The orthogonal images acquired from C-arm are easier for registration. However, the fixed configuration limits the patient’s movement space and, thus, was unsuitable for the free-form HSBV setup in the current study. A modified iterative closest point (ICP) [
14] method was used for the 2D–3D registration with the Broyden–Fletcher–Goldfarb–Shanno (BFGS) optimization function in MATLAB. Reference [
15] tested the knee kinematics accuracy of a manually flexed-extended cycle from the DFIS system. The difference with the RSA method was 0.24 ± 0.16 mm for the posterior femoral translation and 0.16 ± 0.61° for the internal–external tibial rotation over a flexion-extension cycle [
15]. However, this comparison was conducted on the knee joint kinematics, representing a compound effect of the tibia and femur registration. The 3D reconstruction accuracy of the 2D–3D registration was not reported.
A rigid body is a solid body where the distance between any two points on the body remains constant throughout the movement. The rigid-body transformation, with only six parameters, is widely adopted due to its simplicity and stability. However, a non-rigid transformation is also adopted under the following circumstances [
13,
16]: (1) when distortion or deformation occurs between the data acquisitions; (2) registration for the deformable anatomical structure changes with time, e.g., cardiovascular or tumor; or (3) 3D reconstruction is conducted based on statistical models from a population of subjects. The use of non-rigid transformation for the unchanged bone model has not been seen since a bone is considered a rigid body. However, in a 2D–3D registration, a rigid-body transformation may not be valid if discrepancies exist between the 2D radiographs and the 3D bone model.
The purpose of this study was to develop a 2D–3D registration method that works for both marker-based and model-based registration with high accuracy using MRI-derived bone models and to assess the proposed method’s registration accuracy. The non-rigid transformation needs to be considered and tested to see if it improves registration accuracy. Whereas the model-based registration with submillimeter accuracy is required, as stated previously, marker-based registration is needed to provide the ground truth for the model-based method validation. Therefore, the accuracy requirement of the marker-based registration should be at least three times better than the model-based accuracy.