Application of Artificial Intelligence in Shoulder Pathology
Abstract
1. Introduction
2. Methods
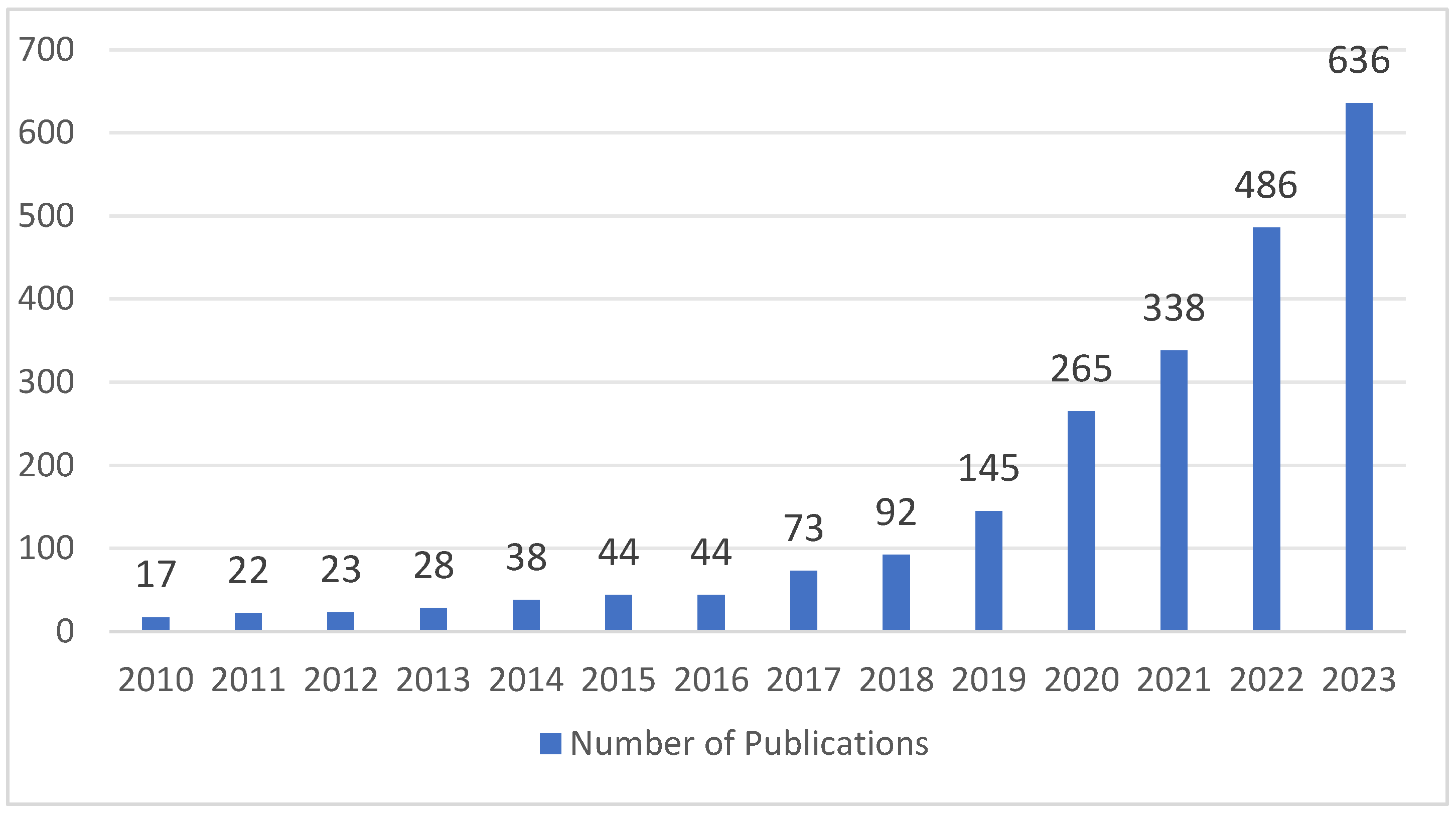
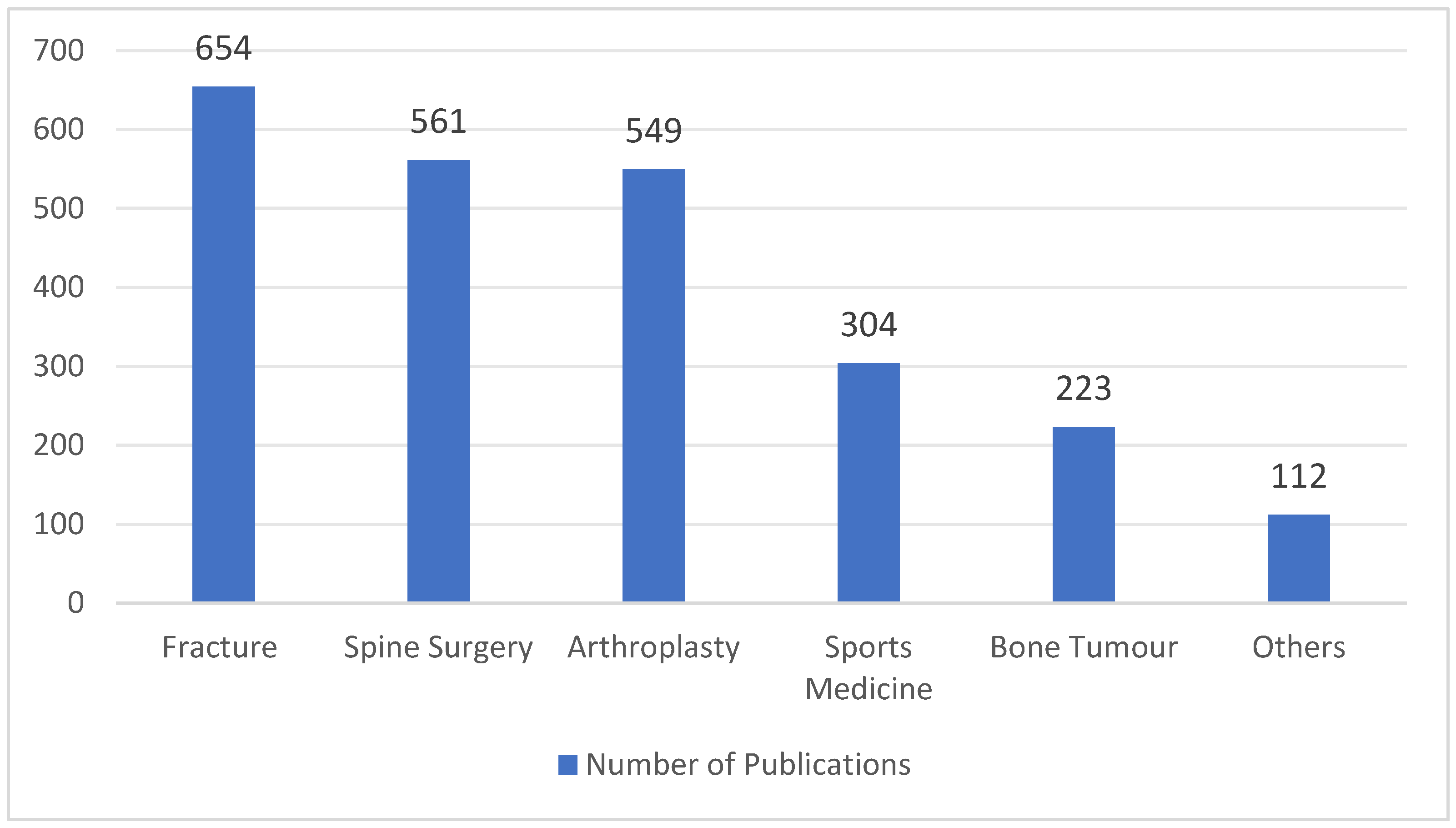
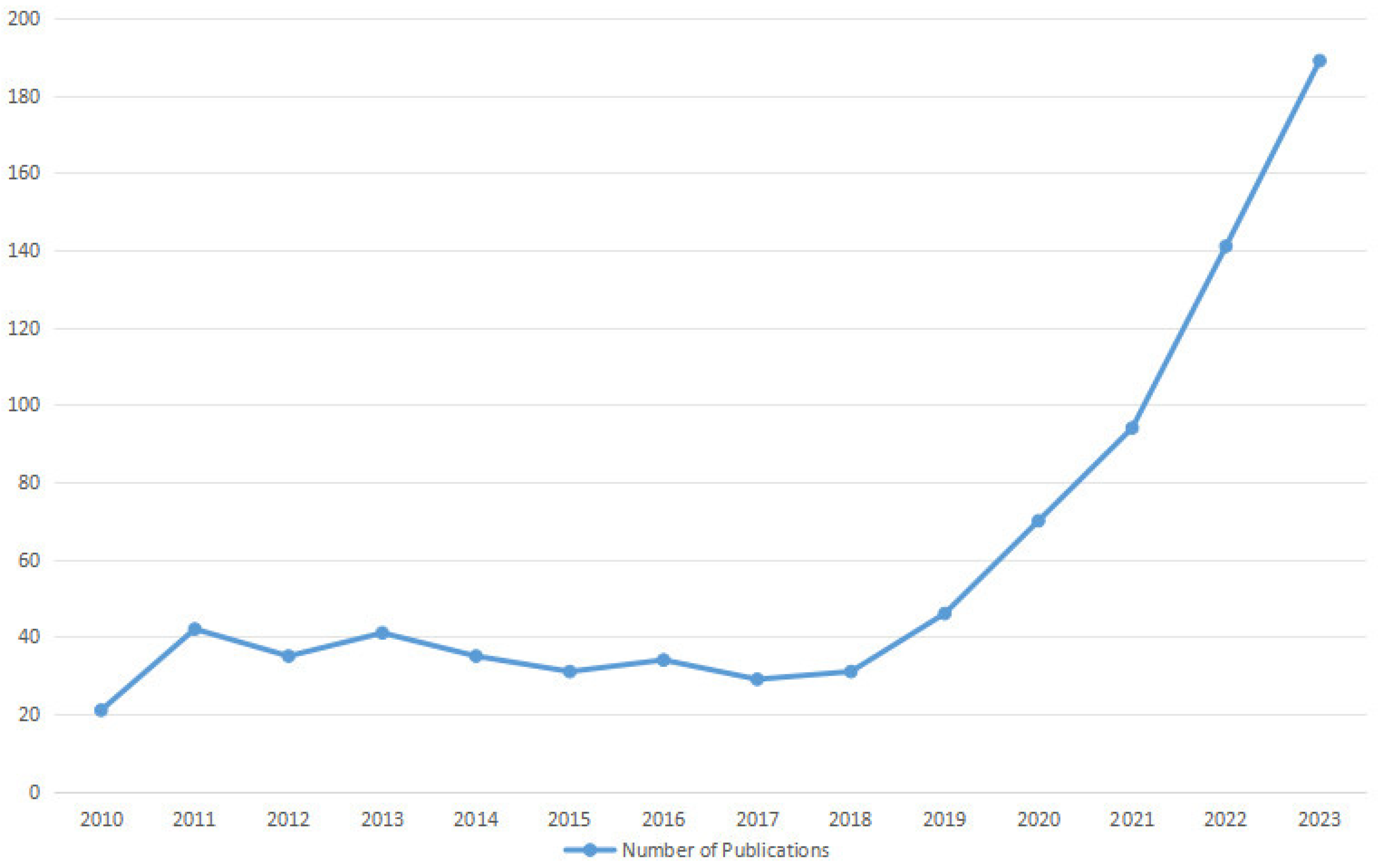
2.1. Search Strategy
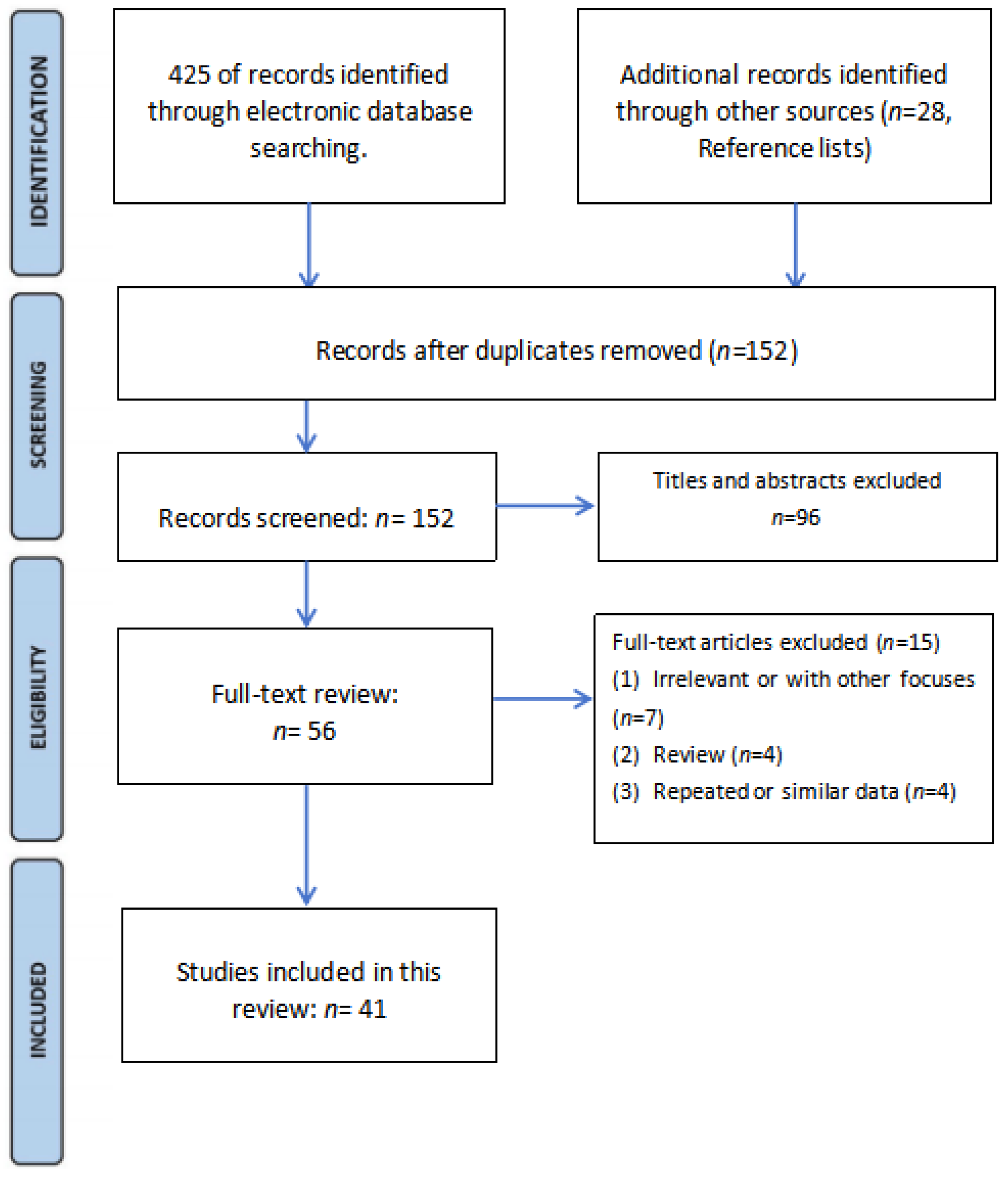
2.2. Eligibility Criteria and Article Selection
3. Rotator Cuff Tears (RCTs)
3.1. Diagnosis
3.2. Predictive Models and Prognosis
3.3. Physical Therapy
4. Shoulder Instability
4.1. Diagnosis
4.2. Predictive Models and Prognosis
5. Rotator Cuff Calcific Tendinopathy (RCCT)
6. Proximal Humeral Fractures (PHFs)
7. Other Shoulder Pathologies
8. Future of AI
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, P.R.; Lu, L.; Zhang, J.Y.; Huo, T.T.; Liu, S.X.; Ye, Z.W. Application of Artificial Intelligence in Medicine: An Overview. Curr. Med. Sci. 2021, 41, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Poduval, M.; Ghose, A.; Manchanda, S.; Bagaria, V.; Sinha, A. Artificial Intelligence and Machine Learning: A New Disruptive Force in Orthopaedics. Indian J. Orthop. 2020, 54, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Zhou, Y.B. Artificial Intelligence and Machine Learning in Clinical Medicine. N. Engl. J. Med. 2023, 388, 2397–2398. [Google Scholar] [PubMed]
- Chen, A.F.; Zoga, A.C.; Vaccaro, A.R. Point/Counterpoint: Artificial Intelligence in Healthcare. Healthc. Transform. 2017, 2, 84–92. [Google Scholar] [CrossRef]
- McCarthy, J.; Minsky, M.L.; Rochester, N.; Shannon, C.E. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence. AI Mag. 2006, 27, 4. [Google Scholar]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Mintz, Y.; Brodie, R. Introduction to Artificial Intelligence in Medicine. Minim. Invasive Ther. Allied Technol. 2019, 28, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, P.N.; Kunze, K.N.; Haeberle, H.S.; Karnuta, J.M.; Luu, B.C.; Nwachukwu, B.U.; Williams, R.J. Clinical and Research Medical Applications of Artificial Intelligence. Arthroscopy 2021, 37, 1694–1697. [Google Scholar] [CrossRef]
- Samuel, A.L. Some Studies in Machine Learning Using the Game of Checkers. IBM J. Res. Dev. 2000, 44, 206–226. [Google Scholar] [CrossRef]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine Learning in Medicine: A Practical Introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [PubMed]
- Razavian, N.; Knoll, F.; Geras, K.J. Artificial Intelligence Explained for Nonexperts. Semin. Musculoskelet. Radiol. 2020, 24, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Cabitza, F.; Locoro, A.; Banfi, G. Machine Learning in Orthopedics: A Literature Review. Front. Bioeng. Biotechnol. 2018, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.; Sandholm, T. Superhuman AI for Multiplayer Poker. Science 2019, 365, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Myers, T.G.; Ramkumar, P.N.; Ricciardi, B.F.; Urish, K.L.; Kipper, J.; Ketonis, C. Artificial Intelligence and Orthopaedics: An Introduction for Clinicians. J. Bone Jt. Surg. Am. 2020, 102, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A Review of Medical Image Data Augmentation Techniques for Deep Learning Applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Erickson, B.J. Basic Artificial Intelligence Techniques: Machine Learning and Deep Learning. Radiol. Clin. N. Am. 2021, 59, 933–940. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [PubMed]
- LeCun, Y. The Power and Limits of Deep Learning. Res. Technol. Manag. 2018, 61, 22–27. [Google Scholar] [CrossRef]
- Egger, J.; Gsaxner, C.; Pepe, A.; Pomykala, K.L.; Jonske, F.; Kurz, M.; Li, J.; Kleesiek, J. Medical Deep Learning—A Systematic Meta-Review. Comput. Methods Programs Biomed. 2022, 221, 106874. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Diao, Z.; Shi, T.; Zhou, Y.; Wang, F.; Hu, W.; Zhu, X.; Luo, S.; Tong, G.; Yao, Y.D. A Review of Deep Learning-Based Multiple-Lesion Recognition from Medical Images: Classification, Detection, and Segmentation. Comput. Biol. Med. 2023, 157, 106726. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis Using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Stotter, C.; Klestil, T.; Nehrer, S. Artificial Intelligence in Orthopedic Radiography Analysis: A Narrative Review. Diagnostics 2022, 12, 2235. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, X.C.; Wang, Y.S. Artificial Neural Network Models for Predicting 1-Year Mortality in Elderly Patients with Intertrochanteric Fractures in China. Braz. J. Med. Biol. Res. 2013, 46, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Salimi, M.; Parry, J.A.; Shahrokhi, R.; Mosalamiaghili, S. Application of Artificial Intelligence in Trauma Orthopedics: Limitation and Prospects. World J. Clin. Cases 2023, 11, 4231–4240. [Google Scholar] [CrossRef] [PubMed]
- Haug, C.J.; Drazen, J.M. Artificial Intelligence and Machine Learning in Clinical Medicine, 2023. N. Engl. J. Med. 2023, 388, 1201–1208. [Google Scholar] [CrossRef]
- Familiari, F.; Galasso, O.; Massazza, F.; Mercurio, M.; Fox, H.; Srikumaran, U.; Gasparini, G. Artificial Intelligence in the Management of Rotator Cuff Tears. Int. J. Environ. Res. Public Health 2022, 19, 16779. [Google Scholar] [CrossRef]
- Bakhsh, W.; Nicandri, G. Anatomy and Physical Examination of the Shoulder. Sports Med. Arthrosc. Rev. 2018, 26, e10–e22. [Google Scholar] [CrossRef]
- Wessel, L.E.; Eliasberg, C.D.; Bowen, E.; Sutton, K.M. Shoulder and Elbow Pathology in the Female Athlete: Sex-specific Considerations. J. Shoulder Elb. Surg. 2021, 30, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Choi, D.; Lee, K.J.; Kang, Y.; Ahn, J.M.; Lee, E.; Lee, J.W.; Kang, H.S. Ruling out Rotator Cuff Tear in Shoulder Radiograph Series Using Deep Learning: Redefining the Role of Conventional Radiograph. Eur. Radiol. 2020, 30, 2843–2852. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Choi, D.; Lee, K.J.; Oh, J.H.; Kim, B.R.; Ahn, J.M. Evaluating Subscapularis Tendon Tears on Axillary Lateral Radiographs Using Deep Learning. Eur. Radiol. 2021, 31, 9408–9417. [Google Scholar] [CrossRef] [PubMed]
- Iio, R.; Ueda, D.; Matsumoto, T.; Manaka, T.; Nakazawa, K.; Ito, Y.; Hirakawa, Y.; Yamamoto, A.; Shiba, M.; Nakamura, H. Deep Learning-based Screening Tool for Rotator Cuff Tears on Shoulder Radiography. J. Orthop. Sci. 2023; online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.J.; Schwier, M.; Geiger, B.; Raithel, E.; von Busch, H.; Fritz, J.; Kline, M.; Brooks, M.; Dunham, K.; Shukla, M. Deep Learning Diagnosis and Classification of Rotator Cuff Tears on Shoulder MRI. Investig. Radiol. 2023, 58, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Chepelev, L.; Nisha, Y.; Sathiadoss, P.; Rybicki, F.J.; Sheikh, A.M. Evaluation of a Deep Learning Method for the Automated Detection of Supraspinatus Tears on MRI. Skeletal Radiol. 2022, 51, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Liu, X.; Wang, D.; Tang, X.; Qin, Y. Development and clinical validation of deep learning for automatic diagnosis of supraspinatus tears. J. Orthop. Surg. Res. 2023, 18, 426. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.; Oh, K.S.; Yoon, J.P.; Seo, A.; Jeong, Y.; Chung, S.W. Automated 3-dimensional MRI Segmentation for the Posterosuperior Rotator Cuff Tear Lesion Using Deep Learning Algorithm. PLoS ONE 2023, 18, e0284111. [Google Scholar] [CrossRef]
- Shim, E.; Kim, J.Y.; Yoon, J.P.; Ki, S.Y.; Lho, T.; Kim, Y.; Chung, S.W. Automated Rotator Cuff Tear Classification Using 3D Convolutional Neural Network. Sci. Rep. 2020, 10, 15632. [Google Scholar] [CrossRef]
- Hahn, S.; Yi, J.; Lee, H.J.; Lee, Y.; Lim, Y.J.; Bang, J.Y.; Kim, H.; Lee, J. Image Quality and Diagnostic Performance of Accelerated Shoulder MRI With Deep Learning-Based Reconstruction. AJR Am. J. Roentgenol. 2022, 218, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, W.; Li, Z.; Yang, J.; Wang, K.; Cao, X.; Qin, N.; Xue, K.; Dai, Y.; Wu, P.; et al. Magnetic Resonance Shoulder Imaging Using Deep Learning-based Algorithm. Eur. Radiol. 2023, 33, 4864–4874. [Google Scholar] [CrossRef] [PubMed]
- Kaniewska, M.; Deininger-Czermak, E.; Getzmann, J.M.; Wang, X.; Lohezic, M.; Guggenberger, R. Application of Deep Learning-based Image Reconstruction in MR Imaging of the Shoulder Joint to Improve Image Quality and Reduce Scan Time. Eur. Radiol. 2023, 33, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Nunna, B., Jr.; Parihar, P.; Wanjari, M.; Shetty, N.; Bora, N. High-Resolution Imaging Insights into Shoulder Joint Pain: A Comprehensive Review of Ultrasound and Magnetic Resonance Imaging (MRI). Cureus 2023, 15, e48974. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, J.Y.; Lee, M.H.; Choi, C.H.; Hwang, J.Y. Imbalanced Loss-Integrated Deep-Learning-Based Ultrasound Image Analysis for Diagnosis of Rotator-Cuff Tear. Sensors 2021, 21, 2214. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.T.; Kim, G.T.; Kim, T.; Choi, S.; Park, E.K. Classification of Rotator Cuff Tears in Ultrasound Images Using Deep Learning Models. Med. Biol. Eng. Comput. 2022, 60, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Ro, K.; Kim, J.Y.; Park, H.; Cho, B.H.; Kim, I.Y.; Shim, S.B.; Choi, I.Y.; Yoo, J.C. Deep-learning Framework and Computer Assisted Fatty Infiltration Analysis for the Supraspinatus Muscle in MRI. Sci. Rep. 2021, 11, 15065. [Google Scholar] [CrossRef] [PubMed]
- Goutallier, D.; Postel, J.M.; Bernageau, J.; Lavau, L.; Voisin, M.C. Fatty Infiltration of Disrupted Rotator Cuff Muscles. Rev. Rhum. Engl. Ed. 1995, 62, 415–422. [Google Scholar] [PubMed]
- Kim, J.Y.; Ro, K.; You, S.; Nam, B.R.; Yook, S.; Park, H.S.; Yoo, J.C.; Park, E.; Cho, K.; Cho, B.H.; et al. Development of an Automatic Muscle Atrophy Measuring Algorithm to Calculate the Ratio of Supraspinatus in Supraspinous Fossa Using Deep Learning. Comput. Methods Programs Biomed. 2019, 182, 105063. [Google Scholar] [CrossRef]
- Taghizadeh, E.; Truffer, O.; Becce, F.; Eminian, S.; Gidoin, S.; Terrier, A.; Farron, A.; Büchler, P. Deep Learning for the Rapid Automatic Quantification and Characterization of Rotator Cuff Muscle Degeneration from Shoulder CT Datasets. Eur. Radiol. 2021, 31, 181–190. [Google Scholar] [CrossRef]
- Medina, G.; Buckless, C.G.; Thomasson, E.; Oh, L.S.; Torriani, M. Deep Learning Method for Segmentation of Rotator Cuff Muscles on MR Images. Skeletal Radiol. 2021, 50, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Alike, Y.; Hou, J.; Long, Y.; Zheng, Z.; Meng, K.; Yang, R. Machine Learning Model Successfully Identifies Important Clinical Features for Predicting Outpatients with Rotator Cuff Tears. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 2615–2623. [Google Scholar] [CrossRef]
- Potty, A.G.; Potty, A.S.R.; Maffulli, N.; Blumenschein, L.A.; Ganta, D.; Mistovich, R.J.; Fuentes, M.; Denard, P.J.; Sethi, P.M.; Shah, A.A.; et al. Approaching Artificial Intelligence in Orthopaedics: Predictive Analytics and Machine Learning to Prognosticate Arthroscopic Rotator Cuff Surgical Outcomes. J. Clin. Med. 2023, 12, 2369. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.M.; Leung, N.; Hardisty, M.; Whyne, C.M.; Henry, P.; McLachlin, S. Shoulder Physiotherapy Exercise Recognition: Machine Learning the Inertial Signals from a Smartwatch. Physiol. Meas. 2018, 39, 075007. [Google Scholar] [CrossRef]
- Croci, E.; Hess, H.; Warmuth, F.; Künzler, M.; Börlin, S.; Baumgartner, D.; Müller, A.M.; Gerber, K.; Mündermann, A. Fully Automatic Algorithm for Detecting and Tracking Anatomical Shoulder Landmarks on Fluoroscopy Images with Artificial Intelligence. Eur. Radiol. 2024, 34, 270–278. [Google Scholar] [CrossRef]
- Kompel, A.J.; Li, X.; Guermazi, A.; Murakami, A.M. Radiographic Evaluation of Patients with Anterior Shoulder Instability. Curr. Rev. Musculoskelet. Med. 2017, 10, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Stillwater, L.; Koenig, J.; Maycher, B.; Davidson, M. 3D-MR vs. 3D-CT of the Shoulder in Patients with Glenohumeral Instability. Skelet. Radiol. 2017, 46, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Cantarelli Rodrigues, T.; Deniz, C.M.; Alaia, E.F.; Gorelik, N.; Babb, J.S.; Dublin, J.; Gyftopoulos, S. Three-dimensional MRI Bone Models of the Glenohumeral Joint Using Deep Learning: Evaluation of Normal Anatomy and Glenoid Bone Loss. Radiol. Artif. Intell. 2020, 2, e190116. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, D.; Sing, D.C.; Beeram, I.; Puvanesarajah, V.; Tornetta, P., 3rd; Fritz, J.; Yi, P.H. Detecting Upper Extremity Native Joint Dislocations Using Deep Learning: A Multicenter Study. Clin. Imaging 2022, 92, 38–43. [Google Scholar] [CrossRef]
- Till, S.E.; Lu, Y.; Reinholz, A.K.; Boos, A.M.; Krych, A.J.; Okoroha, K.R.; Camp, C.L. Artificial Intelligence Can Define and Predict the “Optimal Observed Outcome” After Anterior Shoulder Instability Surgery: An Analysis of 200 Patients With 11-Year Mean Follow-Up. Arthrosc. Sports Med. Rehabil. 2023, 5, e100773. [Google Scholar] [CrossRef]
- Chianca, V.; Albano, D.; Messina, C.; Midiri, F.; Mauri, G.; Aliprandi, A.; Catapano, M.; Pescatori, L.C.; Monaco, C.G.; Gitto, S.; et al. Rotator Cuff Calcific Tendinopathy: From Diagnosis to Treatment. Acta Biomed. 2018, 89 (Suppl. S1), 186–196. [Google Scholar] [PubMed]
- Bechay, J.; Lawrence, C.; Namdari, S. Calcific Tendinopathy of the Rotator Cuff: A Review of Operative Versus Nonoperative Management. Physician Sportsmed. 2020, 48, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Vassalou, E.E.; Klontzas, M.E.; Marias, K.; Karantanas, A.H. Predicting Long-term Outcomes of Ultrasound-guided Percutaneous Irrigation of Calcific Tendinopathy with the Use of Machine Learning. Skelet. Radiol. 2022, 51, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Jawa, A.; Burnikel, D. Treatment of Proximal Humeral Fractures: A Critical Analysis Review. JBJS Rev. 2016, 4, e2. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.W.; Han, S.S.; Lee, J.W.; Oh, K.S.; Kim, N.R.; Yoon, J.P.; Kim, J.Y.; Moon, S.H.; Kwon, J.; Lee, H.J.; et al. Automated Detection and Classification of the Proximal Humerus Fracture by Using Deep Learning Algorithm. Acta Orthop. 2018, 89, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Magnéli, M.; Ling, P.; Gislén, J.; Fagrell, J.; Demir, Y.; Arverud, E.D.; Hallberg, K.; Salomonsson, B.; Gordon, M. Deep Learning Classification of Shoulder Fractures on Plain Radiographs of the Humerus, Scapula and Clavicle. PLoS ONE 2023, 18, e0289808. [Google Scholar] [CrossRef] [PubMed]
- Dipnall, J.F.; Lu, J.; Gabbe, B.J.; Cosic, F.; Edwards, E.; Page, R.; Du, L. Comparison of State-of-the-Art Machine and Deep Learning Algorithms to Classify Proximal Humeral Fractures Using Radiology Text. Eur. J. Radiol. 2022, 153, 110366. [Google Scholar] [CrossRef]
- Guan, H.; Wu, Q.; Zhou, Y.; Fan, X.; Zheng, K.; Si, T.; Zhao, J. A Retrospective Study of Ultrasound-Guided Intervention for Frozen Shoulder in the Frozen Stage. Front. Surg. 2022, 9, 998590. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, Y.; Wang, X.F.; Zhang, Z.Q. Analysis of the Value of Artificial Intelligence Combined with Musculoskeletal Ultrasound in the Differential Diagnosis of Pain Rehabilitation of Scapulohumeral Periarthritis. Medicine 2023, 102, e33125. [Google Scholar] [CrossRef]
- Shu, Y.C.; Lo, Y.C.; Chiu, H.C.; Chen, L.R.; Lin, C.Y.; Wu, W.T.; Özçakar, L.; Chang, K.V. Deep Learning Algorithm for Predicting Subacromial Motion Trajectory: Dynamic Shoulder Ultrasound Analysis. Ultrasonics 2023, 134, 107057. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, L.; Zhao, Y.J.; Lin, Z.Y.; Yang, H. Machine Learning-Based Ultrasomics for Predicting Subacromial Impingement Syndrome Stages. J. Ultrasound Med. 2022, 41, 2279–2285. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.V.; Wu, W.T.; Özçakar, L. Association of bicipital peritendinous effusion with subacromial impingement: A dynamic ultrasonographic study of 337 shoulders. Sci. Rep. 2016, 6, 38943. [Google Scholar] [CrossRef]
- Chang, K.V.; Chen, W.S.; Wang, T.G.; Hung, C.Y.; Chien, K.L. Associations of sonographic abnormalities of the shoulder with various grades of biceps peritendinous effusion (BPE). Ultrasound Med. Biol. 2014, 40, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.S.; Chen, J.L.; Tu, Y.H.; Shih, Y.X.; Lin, Y.C.; Chi, W.L.; Wu, Y.C. Using Deep Learning in Ultrasound Imaging of Bicipital Peritendinous Effusion to Grade Inflammation Severity. IEEE J. Biomed. Health Inform. 2020, 24, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Grauhan, N.F.; Niehues, S.M.; Gaudin, R.A.; Keller, S.; Vahldiek, J.L.; Adams, L.C.; Bressem, K.K. Deep Learning for Accurately Recognizing Common Causes of Shoulder Pain on Radiographs. Skelet. Radiol. 2022, 51, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Canseco, J.A.; Nicholson, K.J.; Patel, N.; Vaccaro, A.R. The Role of Machine Learning in Spine Surgery: The Future Is Now. Front. Surg. 2020, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Loftus, T.J.; Tighe, P.J.; Filiberto, A.C.; Efron, P.A.; Brakenridge, S.C.; Mohr, A.M.; Rashidi, P.; Upchurch, G.R., Jr.; Bihorac, A. Artificial Intelligence and Surgical Decision-making. JAMA Surg. 2020, 155, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Makhni, E.C.; Makhni, S.; Ramkumar, P.N. Artificial Intelligence for the Orthopaedic Surgeon: An Overview of Potential Benefits, Limitations, and Clinical Applications. J. Am. Acad. Orthop. Surg. 2021, 29, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Char, D.S.; Shah, N.H.; Magnus, D. Implementing machine learning in health care—Addressing ethical challenges. N. Engl. J. Med. 2018, 378, 981–983. [Google Scholar] [CrossRef]
- Kumar, V.; Patel, S.; Baburaj, V.; Vardhan, A.; Singh, P.K.; Vaishya, R. Current Understanding on Artificial Intelligence and Machine Learning in Orthopaedics—A Scoping Review. J. Orthop. 2022, 34, 201–206. [Google Scholar] [CrossRef]

| Term | Definition |
|---|---|
| Area under the curve (AUC) | A valuable metric for evaluating the performance of binary classification models, which provides a concise measure of the model’s ability to discriminate between positive and negative classes and is widely used for comparing and assessing the overall performance of predictive models. |
| Class activation map (CAM) | A technique that generates a heatmap to visualize the important regions of an input image for predicting a specific class in a deep convolutional neural network, and helps in interpreting model decisions and understanding the features learned by the network during the classification process. |
| DenseNet121 | DenseNet is a deep learning architecture characterized by dense connectivity patterns, where each layer receives direct input from all preceding layers, leading to improved feature reuse, parameter efficiency, and gradient flow during training. DenseNet121 has 121 layers in total and is commonly used for image classification tasks on the ImageNet dataset. |
| Dice similarity coefficient (DSC) | A statistical measure used to quantify the similarity between two sets, often employed in the context of image segmentation to evaluate the overlap between predicted and ground truth masks, ranging from 0 to 1, with 1 indicating perfect overlap between the two sets and 0 indicating no overlap. |
| F1-score | A performance metric used to evaluate the accuracy of binary classification models, which is to predict one of two possible outcomes based on input data, with values ranging from 0 to 1, where 1 indicates perfect precision and recall and 0 indicates poor performance. |
| Gradient-weighted class activation mapping (Grad-CAM) | A technique that extends the class activation map (CAM) approach to provide better visual explanations for the predictions made by deep convolutional neural networks. |
| nnU-Net | An extension of the original U-Net architecture and a framework for 3D biomedical image segmentation that aims to provide a standardized and automated way to train and evaluate deep learning models on various datasets. |
| Otsu thresholding technique | An image processing technique used for automatic image thresholding, and the goal of thresholding is to separate objects or regions of interest from the background in an image by converting it into a binary image (black and white). |
| Segmentation Model Adopting a pre-trained Classification Architecture (SMART-CA) | A deep learning algorithm that improves the efficiency and accuracy of CNNs by adaptively refining the network architecture during training based on the complexity of the input data, which uses a self-modulating mechanism and a measure of network capacity called the channel attention score to achieve this. |
| Shapley plot | A valuable tool for explaining and interpreting machine learning models by attributing the model’s predictions to individual features, help data scientists and stakeholders gain insights into the model’s decision-making process, and understand the significance of each feature in driving the model’s output. |
| U-Net | A convolutional neural network architecture that was designed for biomedical image segmentation tasks. The U-Net architecture consists of a contracting path to capture context and a symmetric expanding path to enable precise localization. |
| Voxception-ResNet (VRN) | A hybrid neural network architecture that merges the strengths of Voxception and ResNet to tackle tasks that require processing 3D image data. |
| XGBoost model | A versatile and efficient algorithm that excels in handling structured/tabular data and is widely used for tasks such as regression, classification, ranking, and more. |
| Youden index | A single statistic that captures the performance of a binary classification test, which takes into account both the sensitivity and specificity of the test to provide an overall measure of its accuracy, with 1 indicating perfect performance and 0 indicating no discriminatory power. |
| Author (Year) | Input Feature | Model/Algorithm | Dataset | Type of Outcome | Results |
|---|---|---|---|---|---|
| Kim et al., 2020 [33] | X-ray | ResNet-based CNN (1) | 6793 radiograph series | Rule out significant RCTs (2) | The sensitivity, NPV (3), and LR- (4) were 97.3%, 96.6%, and 0.06, respectively. |
| Kang et al., 2021 [34] | X-ray | ResNet-based CNN | 2779 radiograph series | Rule out subscapularis tendon tears | The AUC (4), sensitivity, NPV, and LR- were 0.83 91.4%, 90.4%, and 0.21 in Test Set 1, and 0.82 90.2%, 89.5%, and 0.21 in Test Set 2, respectively. |
| Iio et al., 2023 [35] | X-ray | EfficientNet-based CNN | 2803 radiograph series | Rule out significant RCTs | The sensitivity, NPV, and LR- were 94.5%, 96.2%, and 0.10, respectively. |
| Lin et al., 2023 [36] | MRI | ResNet-based CNN | 11,925 MRI scans | Detection and classification of RCTs | The AUCs for supraspinatus, infraspinatus, and subscapularis tendon tears were 0.93, 0.89, and 0.90, respectively. The model performed best for full-thickness supraspinatus, infraspinatus, and subscapularis tears with AUCs of 0.98, 0.99, and 0.95, respectively. |
| Yao et al., 2022 [37] | MRI | ResNet-based CNN | 200 MRI scans | Detection and segmentation of supraspinatus tears | The sensitivity and specificity were 85.0% and 85.0%, respectively. The AUC for classification was 0.943; DSC (5) for segmentation was 0.814. |
| Guo et al., 2023 [38] | MRI | Xception-based CNN | 701 MRI scans for training and 69 MRI scans for clinical validation | Detection of supraspinatus tears | The model showed high F1-scores and sensitivity on both surgery and internal test sets. Subgroup analyses confirmed its robustness across tear degrees and MRI field strengths. |
| Lee et al., 2020 [39] | MRI | U-Net-based CNN | 303 MRI scans | Segmentation of RCTs | The model reached 94.3% DSC, 97.1% sensitivity, 95.0% specificity, 84.9% precision, 90.5% F1-score, and a Youden index of 91.8%. |
| Shim et al., 2020 [40] | MRI | VRN (6)-based CNN | 2124 MRI scans | Detect the presence or absence of RCTs, classify the tear size, and provide 3D visualization of the tear location. | The model outperformed orthopedists in binary accuracy (92.5% vs. 76.4% and 68.2%), top-1 accuracy (69.0% vs. 45.8% and 30.5%), top-1 ± 1 accuracy (87.5% vs. 79.8% and 71.0%), sensitivity (0.94 vs. 0.86 and 0.90), and specificity (0.90 vs. 0.58 and 0.29). The generated 3D CAM (7) provided effective information regarding the 3D location and size of the tear. |
| Lee et al., 2021 [45] | Ultrasound imaging | VGG19-basedCNN, denoted as SMART-CA (8) | 1400 ultrasound images | Segmentation of RCTs | The precision, recall, and DSC were 0.604% (+38.4%), 0.942% (+14.0%), and 0.736% (+38.6%), respectively. |
| Ho et al., 2022 [46] | Ultrasound imaging | CNN (based on VGG19, ResNet50, InceptionV3, DenseNet121, or Xception) | 194 ultrasound images | Segmentation of RCTs | DenseNet121 demonstrated the best performance, with 88.2% accuracy, 93.8% sensitivity, 83.6% specificity, and an AUC score of 0.832. |
| Ro et al., 2021 [47] | MRI | VGG19-based CNN | 240 MRI scans | Segmentation of the supraspinatus muscle and fossa, and calculation of the amount of fatty infiltration of the supraspinatus muscle | The mean DSC, accuracy, sensitivity, specificity, and relative area difference for the segmented lesion were 0.97, 99.84, 96.89, 99.92, and 0.07, respectively, for the supraspinatus fossa and 0.94, 99.89, 93.34, 99.95, and 2.03, respectively, for the supraspinatus muscle. |
| Kim et al., 2019 [49] | MRI | CNN (fully convolutional network) | 240 MRI scans | Segmentation of the supraspinatus muscle and fossa | The DSC is 0.9718 ± 0.012 in the fossa region and 0.9463 ± 0.047 in the muscle region. |
| Taghizadeh et al., 2020 [50] | CT | U-Net-based CNN | 103 CT scans | Segmentation of RC (9) muscle and calculation of muscle atrophy and degeneration. | Average DSC for muscle segmentations (88 ± 9%) and manually by human raters (89 ± 6%) were comparable. The model provided good–very good estimates of muscle atrophy (R2 = 0.87), fatty infiltration (R2 = 0.91), and overall muscle degeneration (R2 = 0.91) |
| Medina et al., 2020 [51] | MRI | U-Net-based CNN | 258 cases of model A (Y-view selection) and 1048 sagittal T1 Y-views for model (muscle segmentation) | Segmentation of RC muscles on a Y-view | Model A showed top-3 accuracy >98% to select an appropriate Y-view. Model B produced accurate RC muscle segmentations with mean DSC > 0.93. |
| Li et al., 2023 [52] | Questionnaires and physical examinations | ML (stacking, gradient boosting machine, bagging, random forest, XGBoost, and adaptive boosting) | 1684 patients | Identify best model and important clinical variables for predicting patients with RCTs in outpatient settings. | The XGBoost model showed superior performance, with accuracy, AUC, and Brier scores of 0.85, 0.92, and 0.15, respectively. The most important variables were Jobe test, bear hug test, and age for prediction, with mean SHAP (10) values of 1.458, 0.950, and 0.790, respectively. |
| Potty et al., 2023 [53] | Patient-related and surgical-related factors | ML (linear regression, ridge regression, lasso, support vector regression, K-nearest neighbor, random forest, and XGBoost) | 631 patients | Identify important clinical variables for predicting patients with repairing RCTs. | The XGBoost model predicted post-operative outcomes accurately. The most essential variables were pre-operative ASES (11) score, pre-operative pain score, BMI (12), age, and tendon quality. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.; Liang, X.; Guo, D.; Xie, D. Application of Artificial Intelligence in Shoulder Pathology. Diagnostics 2024, 14, 1091. https://doi.org/10.3390/diagnostics14111091
Cheng C, Liang X, Guo D, Xie D. Application of Artificial Intelligence in Shoulder Pathology. Diagnostics. 2024; 14(11):1091. https://doi.org/10.3390/diagnostics14111091
Chicago/Turabian StyleCheng, Cong, Xinzhi Liang, Dong Guo, and Denghui Xie. 2024. "Application of Artificial Intelligence in Shoulder Pathology" Diagnostics 14, no. 11: 1091. https://doi.org/10.3390/diagnostics14111091
APA StyleCheng, C., Liang, X., Guo, D., & Xie, D. (2024). Application of Artificial Intelligence in Shoulder Pathology. Diagnostics, 14(11), 1091. https://doi.org/10.3390/diagnostics14111091







