Persistently Elevated HBV Viral-Host Junction DNA in Urine as a Biomarker for Hepatocellular Carcinoma Minimum Residual Disease and Recurrence: A Pilot Study
Abstract
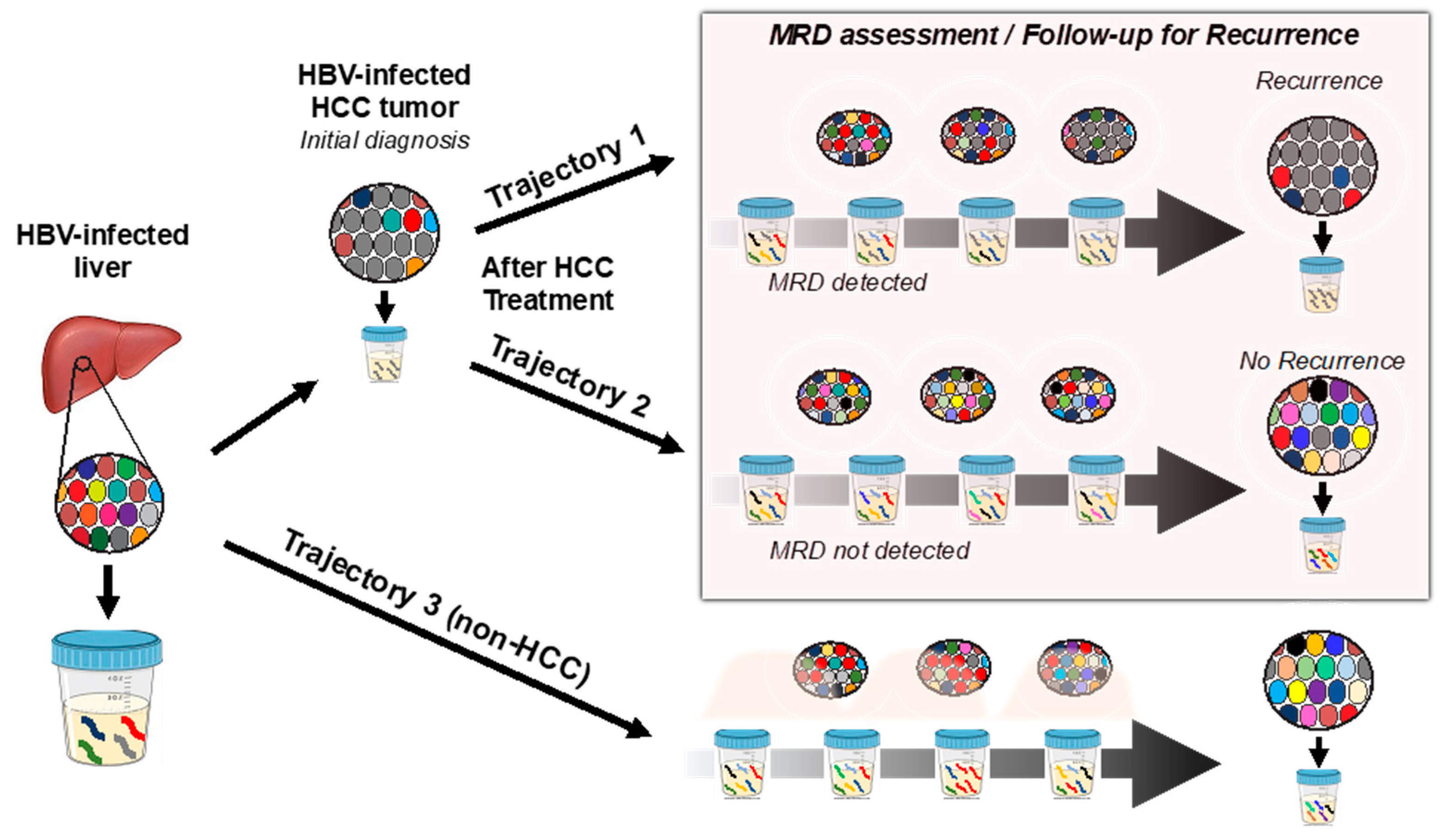
1. Introduction
2. Case Presentations
2.1. Case Selection
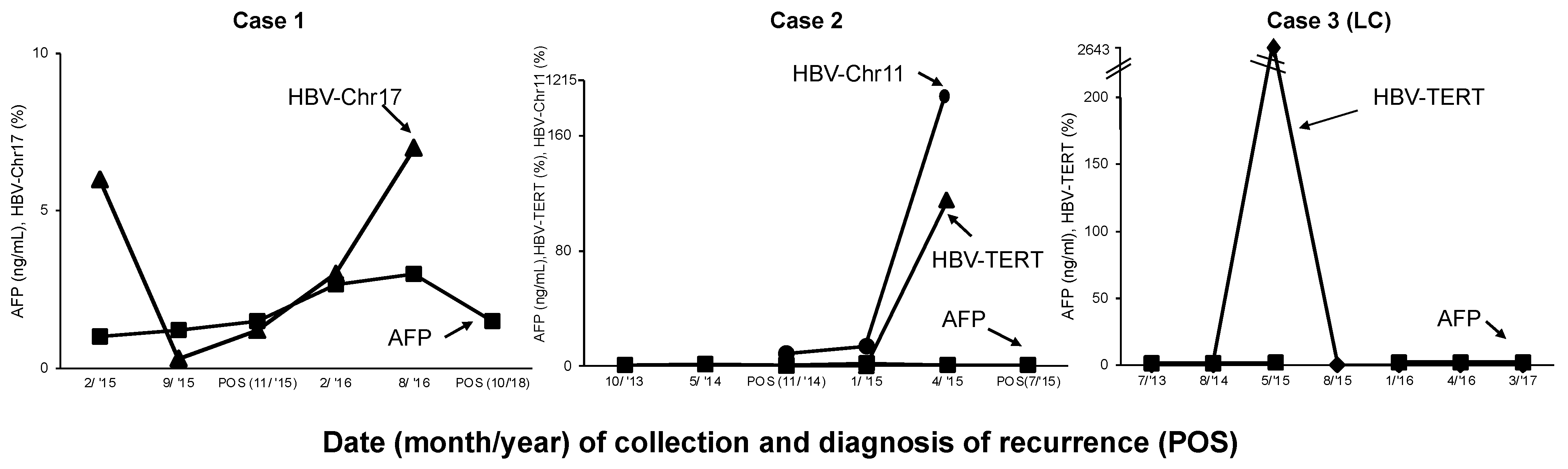
2.2. Case 1
2.3. Case 2
2.4. Case 3
2.5. Case 4–6
3. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fund, W.C.R. Liver Cancer Statistics. Available online: https://www.wcrf.org/dietandcancer/cancer-trends/liver-cancer-statistics (accessed on 5 June 2022).
- Hung, I.F.-N.; Wong, D.K.-H.; Poon, R.T.-P.; Fong, D.Y.-T.; Chui, A.H.-W.; Seto, W.-K.; Fung, J.Y.-Y.; Chan, A.C.-Y.; Yuen, J.C.-H.; Tiu, R.; et al. Risk Factors and Post-Resection Independent Predictive Score for the Recurrence of Hepatitis B-Related Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0148493. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.C.-N.; Lee, K.-f.; Ip, P.C.-T.; Wong, J.S.-W.; Cheung, S.Y.-S.; Wong, J.; Ho, S.C.; Lai, P.B.-S. Pre-operative predictors of post-hepatectomy recurrence of hepatocellular carcinoma: Can we predict earlier? Surgeon 2012, 10, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Pelletier, S. Hepatocellular carcinoma. Clin. Liver Dis. 2006, 10, 339–351. [Google Scholar] [CrossRef]
- Lok, A.; McMahon, B. Chronic hepatitis B. Hepatology 2001, 34, 1225–1241. [Google Scholar] [CrossRef]
- Sherman, M. Recurrence of hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 2045–2047. [Google Scholar] [CrossRef]
- Portolani, N.; Coniglio, A.; Ghidoni, S.; Giovanelli, M.; Benetti, A.; Tiberio, G.A.M.; Giulini, S.M. Early and Late Recurrence After Liver Resection for Hepatocellular Carcinoma: Prognostic and Therapeutic Implications. Ann. Surg. 2006, 243, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, T.; Nakanishi, K.; Yokoo, H.; Kamachi, H.; Tahara, M.; Suzuki, T.; Shimamura, T.; Furukawa, H.; Matsushita, M.; Todo, S. Recurrence Patterns After Hepatectomy of Hepatocellular Carcinoma: Implication of Milan Criteria Utilization. Ann. Surg. Oncol. 2009, 16, 1560–1571. [Google Scholar] [CrossRef]
- Minami, Y.; Nishida, N.; Kudo, M. Therapeutic response assessment of RFA for HCC: Contrast-enhanced US, CT and MRI. World J. Gastroenterol. 2014, 20, 4160–4166. [Google Scholar] [CrossRef]
- Minami, Y.; Kudo, M. Imaging Modalities for Assessment of Treatment Response to Nonsurgical Hepatocellular Carcinoma Therapy: Contrast-Enhanced US, CT, and MRI. Liver Cancer 2015, 4, 106–114. [Google Scholar] [CrossRef]
- Piao, C.Y. Lamivudine treatment in patients with HBV-related hepatocellular carcinoma--using an untreated, matched control cohort. Acta Med. Okayama 2005, 59, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, J.M. Prediction models of hepatocellular carcinoma recurrence after liver transplantation: A comprehensive review. Clin. Mol. Hepatol. 2022, 28, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jin, Y.J.; Shin, S.K.; Kwon, J.H.; Kim, S.G.; Suh, Y.J.; Jeong, Y.; Yu, J.H.; Lee, J.W.; Kwon, O.S.; et al. Surgery versus radiofrequency ablation in patients with Child- Pugh class-A/single small (≤3 cm) hepatocellular carcinoma. Clin. Mol. Hepatol. 2022, 28, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Kuzuya, T.; Katano, Y.; Kumada, T.; Toyoda, H.; Nakano, I.; Hirooka, Y.; Itoh, A.; Ishigami, M.; Hayashi, K.; Honda, T.; et al. Efficacy of antiviral therapy with lamivudine after initial treatment for hepatitis B virus-related hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2007, 22, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Park, J.Y. Clinical significance of AFP and PIVKA-II responses for monitoring treatment outcomes and predicting prognosis in patients with hepatocellular carcinoma. BioMed. Res. Int. 2013, 2013, 310427. [Google Scholar] [CrossRef] [PubMed]
- Willatt, J.M.; Hussain, H.K.; Adusumilli, S.; Marrero, J.A. MR Imaging of Hepatocellular Carcinoma in the Cirrhotic Liver: Challenges and Controversies. Radiology 2008, 247, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-S. Hepatocellular Carcinoma after Transcatheter Arterial Chemoembolization: Difficulties on Imaging Follow-up. Korean J. Radiol. 2005, 6, 134–135. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-J.; Ju, Q.; Li, G.-C. Tumor markers for hepatocellular carcinoma. Mol. Clin. Oncol. 2013, 1, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Korean Liver Cancer Association. 2022 KLCA-NCC Korea practice guidelines for the management of hepatocellular carcinoma. Clin. Mol. Hepatol. 2022, 28, 583–705. [Google Scholar] [CrossRef]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.-S.; Mortensen, F.V.; Stribolt, K. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-H.; Song, J.; Wang, Z.; Wang, X.; Wang, M.; Brenner, D.E.; Block, T.M. Removal of high molecular weight DNA by carboxylated magnetic beads enhances the detection of mutated K-ras DNA in urine. Ann. N. Y. Acad. Sci. 2008, 1137, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-H.; Wang, M.; Aiamkitsumrit, B.; Brenner, D.E.; Block, T.M. Detection of K-ras mutation in urine of patients with colorectal cancer. Cancer Biomark. 2005, 1, 177–182. [Google Scholar] [CrossRef]
- Su, Y.-H.; Wang, M.; Brenner, D.E.; Norton, P.A.; Block, T.M. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann. N. Y. Acad. Sci. 2008, 1137, 197–206. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chang, T.-T.; Steffen, J.D.; Chen, S.; Jain, S.; Song, W.; Lin, Y.-J.; Su, Y.-H. Detection of CTNNB1 Hotspot Mutations in Cell-Free DNA from the Urine of Hepatocellular Carcinoma Patients. Diagnostics 2021, 11, 1475. [Google Scholar] [CrossRef]
- Lin, S.Y.; Dhillon, V.; Jain, S.; Chang, T.-T.; Hu, C.-T.; Lin, Y.-J.; Chen, S.-H.; Yu, L.; Block, T.M.; Su, Y.-H. A locked nucleic acid clamp-mediated PCR assay for detection of a p53 codon 249 hotspot mutation in urine. J. Mol. Diagn. 2011, 13, 474–484. [Google Scholar] [CrossRef]
- Lin, S.Y.; Su, Y.-P.; Trauger, E.R.; Song, B.P.; Thompson, E.G.C.; Hoffman, M.C.; Chang, T.-T.; Lin, Y.-J.; Kao, Y.-L.; Cui, Y.; et al. Detection of Hepatitis B Virus–Host Junction Sequences in Urine of Infected Patients. Hepatol. Commun. 2021, 5, 1649–1659. [Google Scholar] [CrossRef]
- Hann, H.W.; Jain, S.; Park, G.; Steffen, J.D.; Song, W.; Su, Y.-H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res. 2017, 3, 105–111. [Google Scholar] [CrossRef]
- Kim, A.K.; Hamilton, J.P.; Lin, S.Y.; Chang, T.-T.; Hann, H.-W.; Hu, C.-T.; Lou, Y.; Lin, Y.-J.; Gade, T.P.; Park, G. Urine DNA biomarkers for hepatocellular carcinoma screening. Br. J. Cancer 2022, 126, 1432–1438. [Google Scholar] [CrossRef]
- Sung, W.K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Kan, Z.; Zheng, H.; Liu, X.; Li, S.; Barber, T.D.; Gong, Z.; Gao, H.; Hao, K.; Willard, M.D.; Xu, J.; et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013, 23, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Esumi, M.; Aritaka, T.; Arii, M.; Suzuki, K.; Tanikawa, K.; Mizuo, H.; Mima, T.; Shikata, T. Clonal origin of human hepatoma determined by integration of hepatitis B virus DNA. Cancer Res. 1986, 46, 5767–5771. [Google Scholar] [PubMed]
- Esumi, M.; Tanaka, Y.; Tozuka, S.; Shikata, T. Clonal state of human hepatocellular carcinoma and non-tumorous hepatocytes. Cancer Chemother. Pharmacol. 1989, 23, S1–S3. [Google Scholar] [CrossRef] [PubMed]
- (Korean Association for the Study of the Liver. KASL clinical practice guidelines for management of chronic hepatitis B. Clin. Mol. Hepatol. 2022, 28, 276–331. [Google Scholar] [CrossRef]
- Li, C.L.; Ho, M.C.; Lin, Y.Y.; Tzeng, S.T.; Chen, Y.J.; Pai, H.Y.; Wang, Y.C.; Chen, C.L.; Lee, Y.H.; Chen, D.S.; et al. Cell-Free Virus-Host Chimera DNA From Hepatitis B Virus Integration Sites as a Circulating Biomarker of Hepatocellular Cancer. Hepatology 2020, 72, 2063–2076. [Google Scholar] [CrossRef]
- Mohme, M.; Riethdorf, S.; Pantel, K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017, 14, 155–167. [Google Scholar] [CrossRef]
- Lin, S.Y.; Zhang, A.; Lian, J.; Wang, J.; Chang, T.-T.; Lin, Y.-J.; Song, W.; Su, Y.-H. Recurrent HBV Integration Targets as Potential Drivers in Hepatocellular Carcinoma. Cells 2021, 10, 1294. [Google Scholar] [CrossRef]
- Garrido, D.; Block, P.; Lin, S.; Halegoua-DeMarzio, D.; Hann, H.-W. Survival Disparity between Antiviral-Treated and Antiviral-Naïve Patients Who Develop Their First HBV-Associated Hepatocellular Carcinoma. Arch. Gastroenterol. Res. 2021, 2, 86–94. [Google Scholar] [CrossRef]
- Rizzo, A.; Cusmai, A.; Gadaleta-Caldarola, G.; Palmiotti, G. Which role for predictors of response to immune checkpoint inhibitors in hepatocellular carcinoma? Expert Rev. Gastroenterol. Hepatol. 2022, 16, 333–339. [Google Scholar] [CrossRef]
- Viscardi, G.; Tralongo, A.C.; Massari, F.; Lambertini, M.; Mollica, V.; Rizzo, A.; Comito, F.; Di Liello, R.; Alfieri, S.; Imbimbo, M.; et al. Comparative assessment of early mortality risk upon immune checkpoint inhibitors alone or in combination with other agents across solid malignancies: A systematic review and meta-analysis. Eur. J. Cancer 2022, 177, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Ricci, A.D.; Di Federico, A.; Frega, G.; Palloni, A.; Tavolari, S.; Brandi, G. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy in Hepatocellular Carcinoma: Where Do We Stand? Front. Oncol. 2021, 11, 803133. [Google Scholar] [CrossRef] [PubMed]
| Case | Diagnosis | Age | Gender | Antiviral Therapy | ALT (IU/L) | Serum AFP (ng/mL) | HBV Serum (IU/mL) | HBe Ag (−/+) | Hbe Ab | HCC Stage | Tumor Size (cm) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HCC | 78 | F | LAM, TDF | NA | 1 | ND | - | + | I | 0.74 |
| 2 | HCC | 72 | M | LAM, TDF | 30 | 1.4 | <20 | - | - | NA | 3.4 |
| 3 | LC | 53 | M | LAM, TAF, TDF | 27 | 1.5 | <20 | ND | ND | NA | |
| 4 | LC | 68 | F | - | NA | 9.1 | <10 | ND | ND | ||
| 5 | LC | 60 | M | LAM, ENT | 18 | 3 | <20 | - | - | ||
| 6 | LC | 57 | M | IFN, LAM, TDF | 83 | 1.8 | <20 | + | - | ||
| Case | Sample Used (Month/Year) | Disease Status | HBV-JS * Gene (Supporting Read) |
|---|---|---|---|
| 1 | 11/2015 | 3 years pre-recur (HCC) | Chr17_NC (2) |
| 2 | 4/2015 | 3 months pre-recur | TERT_1 (22), chr11_NC (63), GCSHP2 (3) |
| 3 | 5/2015 | LC | TERT_1 (9), Chr7_NC (2) |
| 4 | 7/2019 | LC | NA |
| 5 | 9/2013 | LC | Simple repeat (9), LTR (6) |
| 6 | 7/2015 | LC | SINE (17), Simple repeat (6) |
| Case | Data Collection Timepoint (Month/Year) | Follow-Up (Months) | Disease Status (HCC Treatment) | Serum AFP (ng/mL) | LI-RADS | HCC Stage | Tumor Size (cm) |
|---|---|---|---|---|---|---|---|
| 1 | 2/2015 | 0 | Pre-recur | 1.0 | NA | NA | NA |
| 9/2015 | 7 | Pre-recur | 1.21 | NA | NA | NA | |
| 11/2015 | 9 | POS (MW) | 1.21 | NA | I | 0.74 | |
| 2/2016 | 12 | Pre-recur | 2.65 | 3 | NA | NA | |
| 8/2016 | 18 | Pre-recur | 1.08 | 4 | NA | NA | |
| 10/2018 | 44 | POS | 1.5 | 5 | I | 1.2 | |
| 2 | 10/2013 | 0 | Pre-recur | 1.4 | NA | NA | NA |
| 5/2014 | 7 | Pre-recur | 1.2 | NA | NA | NA | |
| 11/2014 | 13 | Pre-recur | 1.2 | NA | NA | NA | |
| 1/2015 | 15 | POS (MW) | 1.5 | 5 | NA | 3.4 | |
| 4/2015 | 18 | Pre-recur | 1 | NA | NA | NA | |
| 7/2015 | 21 | POS (TACE) | 1 | NA | NA | 0.6 | |
| 3 | 7/2013 | 0 | Cirrhosis | 1.5 | NA | NA | NA |
| 8/2014 | 14 | Cirrhosis | 1.5 | NA | NA | NA | |
| 5/2015 | 22 | Cirrhosis | 1.8 | NA | NA | NA | |
| 8/2015 | 25 | Cirrhosis | 1.8 | NA | NA | NA | |
| 1/2016 | 31 | Cirrhosis | 2.1 | NA | NA | NA | |
| 4/2016 | 33 | Cirrhosis | 1.9 | NA | NA | NA | |
| 3/2017 | 45 | Cirrhosis | 1.9 | NA | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.Y.; Halegoua-DeMarzio, D.; Block, P.; Kao, Y.-L.; Civan, J.; Shieh, F.-S.; Song, W.; Hann, H.-W.; Su, Y.-H. Persistently Elevated HBV Viral-Host Junction DNA in Urine as a Biomarker for Hepatocellular Carcinoma Minimum Residual Disease and Recurrence: A Pilot Study. Diagnostics 2023, 13, 1537. https://doi.org/10.3390/diagnostics13091537
Lin SY, Halegoua-DeMarzio D, Block P, Kao Y-L, Civan J, Shieh F-S, Song W, Hann H-W, Su Y-H. Persistently Elevated HBV Viral-Host Junction DNA in Urine as a Biomarker for Hepatocellular Carcinoma Minimum Residual Disease and Recurrence: A Pilot Study. Diagnostics. 2023; 13(9):1537. https://doi.org/10.3390/diagnostics13091537
Chicago/Turabian StyleLin, Selena Y., Dina Halegoua-DeMarzio, Peter Block, Yu-Lan Kao, Jesse Civan, Fwu-Shan Shieh, Wei Song, Hie-Won Hann, and Ying-Hsiu Su. 2023. "Persistently Elevated HBV Viral-Host Junction DNA in Urine as a Biomarker for Hepatocellular Carcinoma Minimum Residual Disease and Recurrence: A Pilot Study" Diagnostics 13, no. 9: 1537. https://doi.org/10.3390/diagnostics13091537
APA StyleLin, S. Y., Halegoua-DeMarzio, D., Block, P., Kao, Y.-L., Civan, J., Shieh, F.-S., Song, W., Hann, H.-W., & Su, Y.-H. (2023). Persistently Elevated HBV Viral-Host Junction DNA in Urine as a Biomarker for Hepatocellular Carcinoma Minimum Residual Disease and Recurrence: A Pilot Study. Diagnostics, 13(9), 1537. https://doi.org/10.3390/diagnostics13091537







