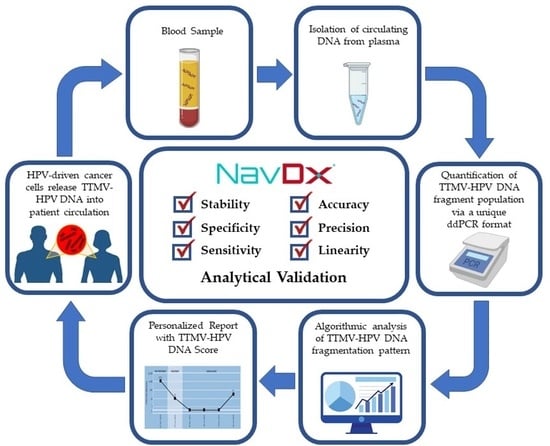
Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Characteristics
2.2. Bioinformatics
2.3. Validation Materials
2.4. Determination of Assay Performance Characteristics
2.4.1. Stability of TTMV-HPV DNA Analytes from Blood
2.4.2. Specificity (Limit of Blank)
2.4.3. Sensitivity (Detection Limit)
2.4.4. Accuracy, Precision and Linearity
3. Results
3.1. Stability of TTMV-HPV DNA Analytes in Blood
3.2. Detection Capability
3.2.1. Specificity (Limit of Blank)
3.2.2. Sensitivity (Detection Limit)
3.3. Analytical Accuracy
3.4. Precision Studies
3.5. Linearity
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| HPV16 | HPV18 | HPV31 | HPV33 | HPV35 |
|---|---|---|---|---|
| 2000 | 1900 | 2000 | 1700 | 1800 |
| 500 | 450 | 500 | 450 | 500 |
| 100 | 90 | 100 | 90 | 100 |
| 50 | 45 | 50 | 45 | 50 |
| 20 | 18 | 20 | 16 | 20 |
| 10 | 9 | 8 | 9 | 10 |
Appendix B
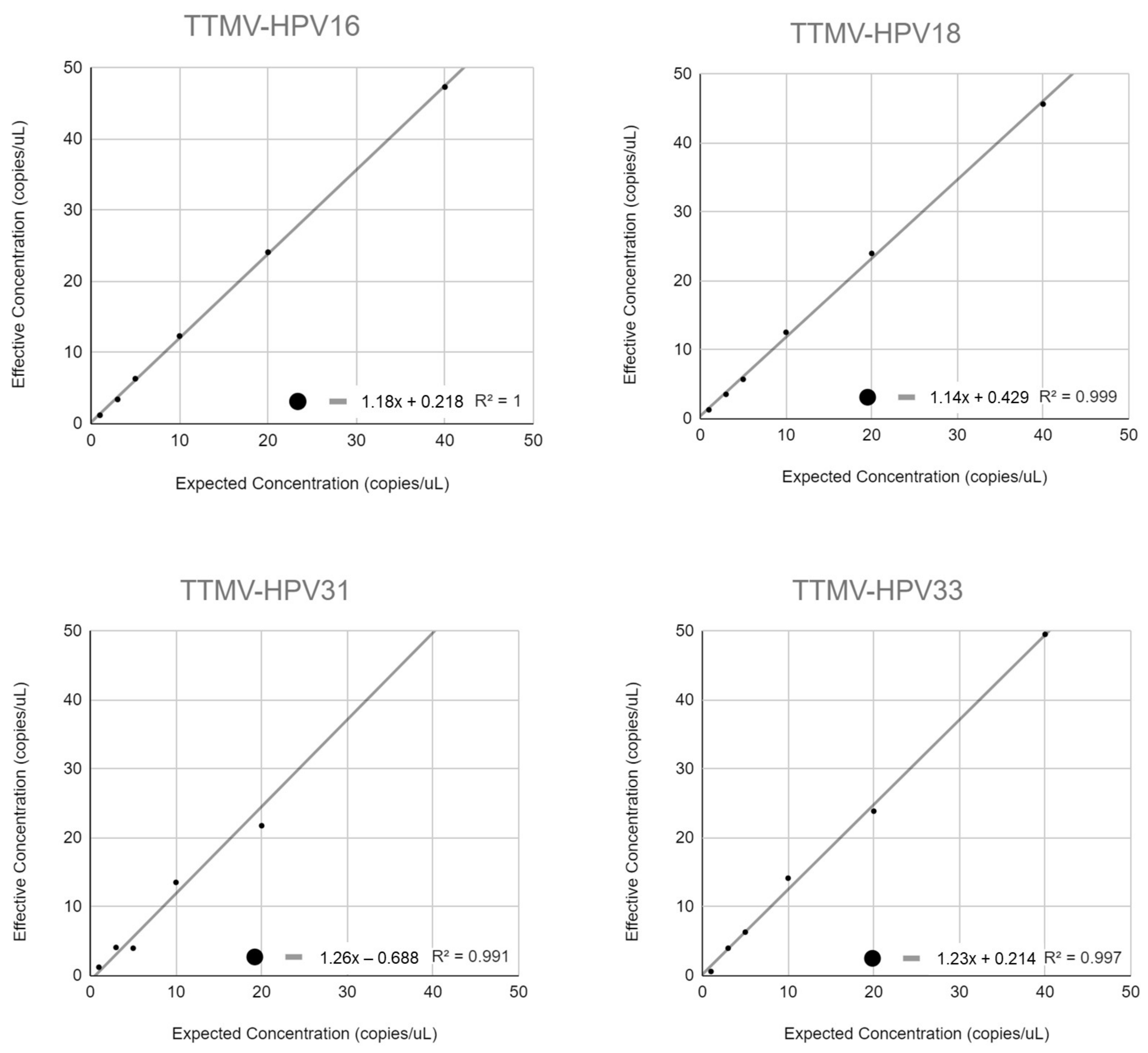
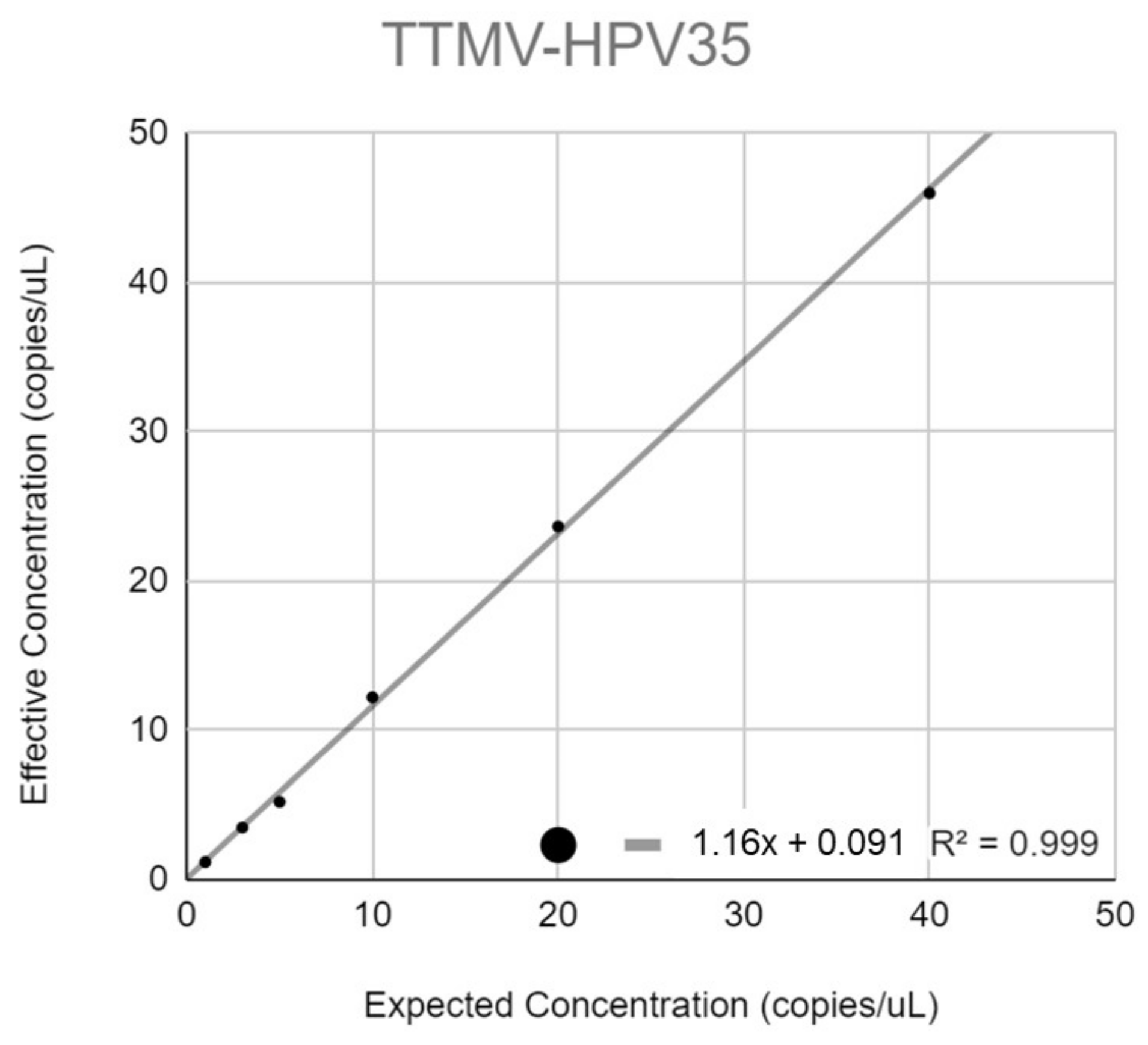
Appendix C
| Metric | Reference | Method | Result | N | Detected | 95% C.I. |
|---|---|---|---|---|---|---|
| Sensitivity | Chera, 2019 [34] | OPSCC tissue HPV+ by p16 1 | 89.3% | 103 | 92 | |
| Rettig, 2022 [38] | OPSCC tissue HPV+ by p16 and/or RNA in situ hybridization (ISH) 2 | 89.1% | 110 | 98 | ||
| Routman, 2022 [36] | OPSCC tissue HPV+ by p16 or RNA in situ hybridization (ISH) | 88.9% | 45 | 40 | ||
| Chung, 2022 [39] | OPSCC tissue HPV+ by p16 | 94.6% | 37 | 35 | ||
| Echevarria, 2022 [42] | OPSCC tissue HPV+ by p16 | 90.9% | 33 | 30 | ||
| Gerndt, 2021 [41] | OPSCC tissue HPV+ by p16 | 93.5% | 46 | 43 | ||
| Cumulative Sensitivity Data | 90.4% | 374 | 338 | 87.4–93.4 | ||
| Metric | Reference | Method | Result | N | Detected | 95% C.I. |
| Specificity | Chera, 2019 [34] | Healthy donors, (n = 55) 3 banked blood, non-HPV related malignancy patients (n = 60) | 97.4% | 115 | 3 | |
| Routman, 2022 [36] | OPSCC tissue HPV negative by p16 and/or ISH | 100% | 7 | 0 | ||
| Rettig, 2022 [37] | Academic center biobank samples, prospectively continuously curated, with no cancer or HPV related disease matched 10:1 with 10 cases of HPV-driven HNCs (n = 100) confirmed with p16 and/or ISH | 100% | 100 | 0 | ||
| Cumulative Specificity Data | 98.6% | 222 | 3 | 97.1–100 | ||
| Metric | Reference | Method | Result | N | Detected | 95% C.I. |
|---|---|---|---|---|---|---|
| PPV | Chera 2020 [35] | Biopsy proven recurrent HPV-driven OPSCC 1 | 100% | 16 | 16 | |
| Berger 2022 [40] | Biopsy and/or imaging confirmed recurrent HPV-driven OPSCC 2 | 97.5% | 80 | 78 | ||
| Cumulative PPV Data | 97.9% | 96 | 94 | 95.1–100 | ||
| Reference | Method | Result | N | Detected | 95% C.I. | |
| NPV | Chera 2020 [35] | Imaging and clinical examination negative for recurrent OPSCC | 100% | 99 | 99 | |
| Berger 2022 [40] | Clinician reported status as “no evidence of disease” 3 | 95.4% | 1256 | 1198 | ||
| Cumulative NPV Data | 95.7% | 1355 | 1297 | 94.6–96.8 | ||
References
- How Many Cancers Are Linked with HPV Each Year? | CDC. Available online: https://www.cdc.gov/cancer/hpv/statistics/cases.htm (accessed on 14 December 2022).
- Van Dyne, E.A.; Henley, S.J.; Saraiya, M.; Thomas, C.C.; Markowitz, L.E.; Benard, V.B. Trends in Human Papillomavirus–Associated Cancers—United States, 1999–2015. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human Papillomavirus and Rising Oropharyngeal Cancer Incidence in the United States. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-Associated Oropharyngeal Cancer: Epidemiology, Molecular Biology and Clinical Management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Jones, O.S.; Breeze, C.E.; Gilson, R. Gender-Neutral HPV Vaccination in the UK, Rising Male Oropharyngeal Cancer Rates, and Lack of HPV Awareness. Lancet Infect. Dis. 2019, 19, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Faraji, F.; Rettig, E.M.; Tsai, H.-L.; El Asmar, M.; Fung, N.; Eisele, D.W.; Fakhry, C. The Prevalence of Human Papillomavirus in Oropharyngeal Cancer Is Increasing Regardless of Sex or Race, and the Influence of Sex and Race on Survival Is Modified by Human Papillomavirus Tumor Status. Cancer 2019, 125, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, C.; Zhang, Q.; Nguyen-Tan, P.F.; Rosenthal, D.; El-Naggar, A.; Garden, A.S.; Soulieres, D.; Trotti, A.; Avizonis, V.; Ridge, J.A.; et al. Human Papillomavirus and Overall Survival after Progression of Oropharyngeal Squamous Cell Carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 3365–3373. [Google Scholar] [CrossRef]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved Survival of Patients with Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma in a Prospective Clinical Trial. J. Natl. Cancer Inst. 2008, 100, 261–269. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human Papillomavirus and Survival of Patients with Oropharyngeal Cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Huang, S.H.; Perez-Ordonez, B.; Weinreb, I.; Hope, A.; Massey, C.; Waldron, J.N.; Kim, J.; Bayley, A.J.; Cummings, B.; John Cho, B.C.; et al. Natural Course of Distant Metastases Following Radiotherapy or Chemoradiotherapy in HPV-Related Oropharyngeal Cancer. Oral Oncol. 2013, 49, 79–85. [Google Scholar] [CrossRef]
- Guo, T.; Kang, S.Y.; Cohen, E.E.W. Current Perspectives on Recurrent HPV-Mediated Oropharyngeal Cancer. Front. Oncol. 2022, 12, 966899. [Google Scholar] [CrossRef]
- Asheer, J.; Jensen, J.S.; Grønhøj, C.; Jakobsen, K.K.; Buchwald, C. von Rate of Locoregional Recurrence among Patients with Oropharyngeal Squamous Cell Carcinoma with Known HPV Status: A Systematic Review. Acta Oncol. Stockh. Swed. 2020, 59, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Tiedemann, D.; Jakobsen, K.K.; von Buchwald, C.; Grønhøj, C. Systematic Review on Location and Timing of Distant Progression in Human Papillomavirus-Positive and Human Papillomavirus-Negative Oropharyngeal Squamous Cell Carcinomas. Head Neck 2019, 41, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.C.J.; Kelly, J.R.; Park, H.S.; An, Y.; Judson, B.L.; Burtness, B.A.; Husain, Z.A. Patterns of Failure in High-Metastatic Node Number Human Papillomavirus-Positive Oropharyngeal Carcinoma. Oral Oncol. 2018, 85, 35–39. [Google Scholar] [CrossRef]
- Guidelines for Patients Details. Available online: https://www.nccn.org/patientresources/patient-resources/guidelines-for-patients/guidelines-for-patients-details (accessed on 14 December 2022).
- Nocon, C.C.; Kennedy, A.; Jaffe, J.; Pruitt, J.; Kuchta, K.; Bhayani, M.K. Costs Associated with Imaging Surveillance after Treatment for Head and Neck Cancer. JAMA Otolaryngol. Neck Surg. 2021, 147, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Westra, W.H. Detection of Human Papillomavirus (HPV) in Clinical Samples: Evolving Methods and Strategies for the Accurate Determination of HPV Status of Head and Neck Carcinomas. Oral Oncol. 2014, 50, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Marur, S.; D’Souza, G.; Westra, W.H.; Forastiere, A.A. HPV-Associated Head and Neck Cancer: A Virus-Related Cancer Epidemic. Lancet Oncol. 2010, 11, 781–789. [Google Scholar] [CrossRef]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. CtDNA and CTCs in Liquid Biopsy – Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating Tumor DNA and Liquid Biopsy in Oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Cao, H.; Banh, A.; Kwok, S.; Shi, X.; Wu, S.; Krakow, T.; Khong, B.; Bavan, B.; Bala, R.; Pinsky, B.A.; et al. Quantitation of Human Papillomavirus DNA in Plasma of Oropharyngeal Carcinoma Patients. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e351–e358. [Google Scholar] [CrossRef]
- Ahn, S.M.; Chan, J.Y.K.; Zhang, Z.; Wang, H.; Khan, Z.; Bishop, J.A.; Westra, W.; Koch, W.M.; Califano, J.A. Saliva and Plasma Quantitative Polymerase Chain Reaction-Based Detection and Surveillance of Human Papillomavirus-Related Head and Neck Cancer. JAMA Otolaryngol.- Head Neck Surg. 2014, 140, 846–854. [Google Scholar] [CrossRef]
- Dahlstrom, K.R.; Li, G.; Hussey, C.; Vo, J.; Wei, Q.; Zhao, C.; Sturgis, E.M. Circulating HPV DNA as a Marker for Disease Extent and Recurrence among Patients with Oropharyngeal Cancer. Cancer 2015, 121, 3455–3464. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of Somatic Mutations and HPV in the Saliva and Plasma of Patients with Head and Neck Squamous Cell Carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Garcia-Murillas, I.; Cutts, R.J.; De Castro, D.G.; Grove, L.; Hurley, T.; Wang, F.; Nutting, C.; Newbold, K.; Harrington, K.; et al. Predicting Response to Radical (Chemo)Radiotherapy with Circulating HPV DNA in Locally Advanced Head and Neck Squamous Carcinoma. Br. J. Cancer 2017, 117, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Damerla, R.R.; Lee, N.Y.; You, D.; Soni, R.; Shah, R.; Reyngold, M.; Katabi, N.; Wu, V.; McBride, S.M.; Tsai, C.J.; et al. Detection of Early Human Papillomavirus-Associated Cancers by Liquid Biopsy. JCO Precis. Oncol. 2019, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Meehan, K.; Pereira, M.R.; Mirzai, B.; Lim, S.H.; Leslie, C.; Clark, M.; Sader, C.; Friedland, P.; Lindsay, A.; et al. A Comparative Study of Extracellular Vesicle-Associated and Cell-Free DNA and RNA for HPV Detection in Oropharyngeal Squamous Cell Carcinoma. Sci. Rep. 2020, 10, 6083. [Google Scholar] [CrossRef] [PubMed]
- Reder, H.; Taferner, V.F.; Wittekindt, C.; Bräuninger, A.; Speel, E.-J.M.; Gattenlöhner, S.; Wolf, G.; Klussmann, J.P.; Wuerdemann, N.; Wagner, S. Plasma Cell-Free Human Papillomavirus Oncogene E6 and E7 DNA Predicts Outcome in Oropharyngeal Squamous Cell Carcinoma. J. Mol. Diagn. 2020, 22, 1333–1343. [Google Scholar] [CrossRef]
- Mazurek, A.M.; Rutkowski, T.; Fiszer-Kierzkowska, A.; Małusecka, E.; Składowski, K. Assessment of the Total CfDNA and HPV16/18 Detection in Plasma Samples of Head and Neck Squamous Cell Carcinoma Patients. Oral Oncol. 2016, 54, 36–41. [Google Scholar] [CrossRef]
- Jeannot, E.; Latouche, A.; Bonneau, C.; Calméjane, M.-A.; Beaufort, C.; Ruigrok-Ritstier, K.; Bataillon, G.; Larbi Chérif, L.; Dupain, C.; Lecerf, C.; et al. Circulating HPV DNA as a Marker for Early Detection of Relapse in Patients with Cervical Cancer. Clin. Cancer Res. 2021, 27, 5869–5877. [Google Scholar] [CrossRef]
- Rutkowski, T.; Mazurek, A.; Snietura, M. Post-Treatment Circulating Free HPV DNA As a Marker of Treatment Outcome in Patients with HPV-Related Propharyngeal Cancer After Radio(Chemo)Therapy. Cell. Mol. Med. Open Access 2017, 4, 12. [Google Scholar] [CrossRef]
- Hanna, G.J.; Supplee, J.G.; Kuang, Y.; Mahmood, U.; Lau, C.J.; Haddad, R.I.; Jänne, P.A.; Paweletz, C.P. Plasma HPV Cell-Free DNA Monitoring in Advanced HPV-Associated Oropharyngeal Cancer. Ann. Oncol. 2018, 29, 1980–1986. [Google Scholar] [CrossRef]
- Veyer, D.; Wack, M.; Mandavit, M.; Garrigou, S.; Hans, S.; Bonfils, P.; Tartour, E.; Bélec, L.; Wang-Renault, S.-F.; Laurent-Puig, P.; et al. HPV Circulating Tumoral DNA Quantification by Droplet-Based Digital PCR: A Promising Predictive and Prognostic Biomarker for HPV-Associated Oropharyngeal Cancers. Int. J. Cancer 2020, 147, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Chera, B.S.; Kumar, S.; Beaty, B.T.; Marron, D.; Jefferys, S.; Green, R.; Goldman, E.C.; Amdur, R.; Sheets, N.; Dagan, R.; et al. Rapid Clearance Profile of Plasma Circulating Tumor HPV Type 16 DNA during Chemoradiotherapy Correlates with Disease Control in HPV-Associated Oropharyngeal Cancer. Clin. Cancer Res. 2019, 25, 4682–4690. [Google Scholar] [CrossRef] [PubMed]
- Chera, B.S.; Kumar, S.; Shen, C.; Amdur, R.; Dagan, R.; Green, R.; Goldman, E.; Weiss, J.; Grilley-Olson, J.; Patel, S.; et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J. Clin. Oncol. 2020, 38, 1050–1058. [Google Scholar] [CrossRef]
- Routman, D.M.; Kumar, S.; Chera, B.S.; Jethwa, K.R.; Abel, K.M.V.; Frechette, K.; DeWees, T.; Golafshar, M.; Garcia, J.J.; Price, D.L.; et al. Detectable Postoperative Circulating Tumor Human Papillomavirus DNA and Association with Recurrence in Patients with HPV-Associated Oropharyngeal Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Rettig, E.M.; Faden, D.L.; Sandhu, S.; Wong, K.; Faquin, W.C.; Warinner, C.; Stephens, P.; Kumar, S.; Kuperwasser, C.; Richmon, J.D.; et al. Detection of Circulating Tumor Human Papillomavirus DNA before Diagnosis of HPV-positive Head and Neck Cancer. Int. J. Cancer 2022, 151, 1081–1085. [Google Scholar] [CrossRef]
- Rettig, E.M.; Wang, A.A.; Tran, N.-A.; Carey, E.; Dey, T.; Schoenfeld, J.D.; Sehgal, K.; Guenette, J.P.; Margalit, D.N.; Sethi, R.; et al. Association of Pretreatment Circulating Tumor Tissue–Modified Viral HPV DNA with Clinicopathologic Factors in HPV-Positive Oropharyngeal Cancer. JAMA Otolaryngol. Neck Surg. 2022, 148, 1120–1130. [Google Scholar] [CrossRef]
- Chung, C.H.; Li, J.; Steuer, C.E.; Bhateja, P.; Johnson, M.; Masannat, J.; Poole, M.I.; Song, F.; Hernandez-Prera, J.C.; Molina, H.; et al. Phase II Multi-Institutional Clinical Trial Result of Concurrent Cetuximab and Nivolumab in Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2022, 28, 2329–2338. [Google Scholar] [CrossRef]
- Berger, B.M.; Hanna, G.J.; Posner, M.R.; Genden, E.M.; Lautersztain, J.; Naber, S.P.; Del Vecchio Fitz, C.; Kuperwasser, C. Detection of Occult Recurrence Using Circulating Tumor Tissue Modified Viral HPV DNA among Patients Treated for HPV-Driven Oropharyngeal Carcinoma. Clin. Cancer Res. 2022, 28, 4292–4301. [Google Scholar] [CrossRef]
- Gerndt, S.P.; Ramirez, R.J.; Wahle, B.M.; Kuperwasser, C.; Gunning, A.; Chaudhuri, A.A.; Zevallos, J.P. Evaluating a Clinically Validated Circulating Tumor HPV DNA Assay in Saliva as a Proximal Biomarker in HPV+ Oropharyngeal Squamous Cell Carcinoma. J. Clin. Oncol. 2021, 39, 6063. [Google Scholar] [CrossRef]
- Echevarria, M.; Chung, C.H.; Giuliano, A.; Slebos, R.; Yang, G.Q.; Stevens, P.J.; Caudell, J.J. Kinetics of Circulating Human Papillomavirus (CHPC) DNA in Plasma and Oral Gargles from Patients with HPV-Positive Oropharyngeal Cancer (OPC) Treated with Definitive Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2022, 112, e2–e3. [Google Scholar] [CrossRef]
- SelectScience Cell-Free DNA BCT. Available online: https://www.selectscience.net/products/cell-free-dna-bct/?prodID=205183 (accessed on 14 December 2022).
- Norton, S.E.; Luna, K.K.; Lechner, J.M.; Qin, J.; Fernando, M.R. A New Blood Collection Device Minimizes Cellular DNA Release During Sample Storage and Shipping When Compared to a Standard Device. J. Clin. Lab. Anal. 2013, 27, 305–311. [Google Scholar] [CrossRef] [PubMed]


| Day(s) | Average (Min-Max) ESR1 Values (frg/mL) | #TTMV-HPV Positive Cases | Sample Size (N) | % Positivity |
|---|---|---|---|---|
| 1 | 3528 (504–436,345) | 981 | 3396 | 28.9 |
| 2 | 3578 500–843,085) | 1915 | 7321 | 26.2 |
| 3 | 4779 (506–920,213) | 675 | 2322 | 29.1 |
| 4 | 4224 (528–834,135) | 1126 | 4322 | 26.1 |
| 5 | 4531 (506–433,125) | 617 | 2115 | 29.2 |
| 6 | 7693 (623–683,446) | 200 | 615 | 32.5 |
| 7 | 5005 (505–70,565) | 48 | 179 | 26.8 |
| 8–14 | 7581 (675–161,538) | 38 | 128 | 29.7 |
| Totals | 5600 | 20,398 | 27.5 |
| Copies/μL | |||
|---|---|---|---|
| Type | LOB | LOQ | LOD |
| TTMV-HPV16 | 0.32 | <1.20 | 0.56 |
| TTMV-HPV18 | 0.17 | 3.56 | 1.31 |
| TTMV-HPV31 | 0 | 4.11 | 0.63 |
| TTMV-HPV33 | 0.15 | 4.00 | 1.10 |
| TTMV-HPV35 | 0.19 | 3.50 | 0.57 |
| Mean % Recoveries | |||||
|---|---|---|---|---|---|
| Dilution * | TTMV- HPV16 | TTMV- HPV18 | TTMV- HPV31 | TTMV- HPV33 | TTMV- HPV35 |
| 1:1 | 107.2 | 99.7 | 102.6 | 101.1 | 99.9 |
| 1:4 | 111.5 | 101.9 | 103.6 | 101.2 | 110.5 |
| 1:20 | 110.6 | 101.4 | 100.5 | 100.4 | 98.9 |
| 1:40 | 110.5 | 96.5 | 104.0 | 96.6 | 96.5 |
| 1:100 | 110.3 | 101.1 | 92.7 | 99.4 | 94.5 |
| 1:200 | 104.3 | 112.6 | 100.5 | 96.3 | 107.7 |
| Dilution * | %CVs of Mean Effective Concentration (Copies/μL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intra-Assay | Inter-Assay | |||||||||
| TTMV-HPV16 | TTMV-HPV18 | TTMV-HPV31 | TTMV-HPV33 | TTMV-HPV35 | TTMV-HPV16 | TTMV-HPV18 | TTMV-HPV31 | TTMV-HPV33 | TTMV-HPV35 | |
| 1:1 | 2.1 | 2.7 | 2.2 | 3.0 | 1.4 | 2.9 | 2.4 | 1.9 | 2.5 | 1.9 |
| 1:4 | 2.1 | 2.1 | 1.0 | 1.5 | 3.5 | 3.3 | 3.6 | 2.6 | 4.3 | 3.8 |
| 1:20 | 3.3 | 2.7 | 3.0 | 5.4 | 1.2 | 4.7 | 2.9 | 6.3 | 4.9 | 3.0 |
| 1:40 | 2.0 | 6.2 | 8.0 | 11.5 | 1.1 | 4.6 | 5.4 | 7.5 | 8.4 | 6.4 |
| 1:100 | 12.2 | 10.1 | 9.0 | 5.3 | 7.1 | 7.5 | 11.9 | 10.8 | 10.8 | 5.8 |
| 1:200 | 13.3 | 13.8 | 7.2 | 13.9 | 4.7 | 9.0 | 20.0 | 16.5 | 13.1 | 11.2 |
| Type | Time | Series | Equation | R2 Value |
|---|---|---|---|---|
| TTMV-HPV16 | Day 1 | ⬤ | y = 1.07x + 4.61 | 1 |
| Day 3 | ▲ | y = 1.11x + 4.65 | 1 | |
| Day 5 | ◾ | y = 1.08x + 7.96 | 1 | |
| TTMV-HPV18 | Day 1 | ⬤ | y = 0.998x + 1.91 | 1 |
| Day 3 | ▲ | y = 1.04x + 0.962 | 1 | |
| Day 5 | ◾ | y = 1.01x + 0.706 | 1 | |
| TTMV-HPV31 | Day 1 | ⬤ | y = 1.03x − 0.11 | 1 |
| Day 3 | ▲ | y = 1.04x − 2.3 | 1 | |
| Day 5 | ◾ | y = 1.05x − 1.46 | 1 | |
| TTMV-HPV33 | Day 1 | ⬤ | y = 1.01x − 0.687 | 1 |
| Day 3 | ▲ | y = 1.01x − 0.579 | 1 | |
| Day 5 | ◾ | y = 0.997x − 2.31 | 1 | |
| TTMV-HPV35 | Day 1 | ⬤ | y = 0.999x − 0.581 | 1 |
| Day 3 | ▲ | y = 1.03x − 0.681 | 1 | |
| Day 5 | ◾ | y = 1.03x − 1.62 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gunning, A.; Kumar, S.; Williams, C.K.; Berger, B.M.; Naber, S.P.; Gupta, P.B.; Del Vecchio Fitz, C.; Kuperwasser, C. Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers. Diagnostics 2023, 13, 725. https://doi.org/10.3390/diagnostics13040725
Gunning A, Kumar S, Williams CK, Berger BM, Naber SP, Gupta PB, Del Vecchio Fitz C, Kuperwasser C. Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers. Diagnostics. 2023; 13(4):725. https://doi.org/10.3390/diagnostics13040725
Chicago/Turabian StyleGunning, Alicia, Sunil Kumar, Cassin Kimmel Williams, Barry M. Berger, Stephen P. Naber, Piyush B. Gupta, Catherine Del Vecchio Fitz, and Charlotte Kuperwasser. 2023. "Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers" Diagnostics 13, no. 4: 725. https://doi.org/10.3390/diagnostics13040725
APA StyleGunning, A., Kumar, S., Williams, C. K., Berger, B. M., Naber, S. P., Gupta, P. B., Del Vecchio Fitz, C., & Kuperwasser, C. (2023). Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers. Diagnostics, 13(4), 725. https://doi.org/10.3390/diagnostics13040725