Identification of New Prognostic Genes and Construction of a Prognostic Model for Lung Adenocarcinoma
Abstract
1. Introduction
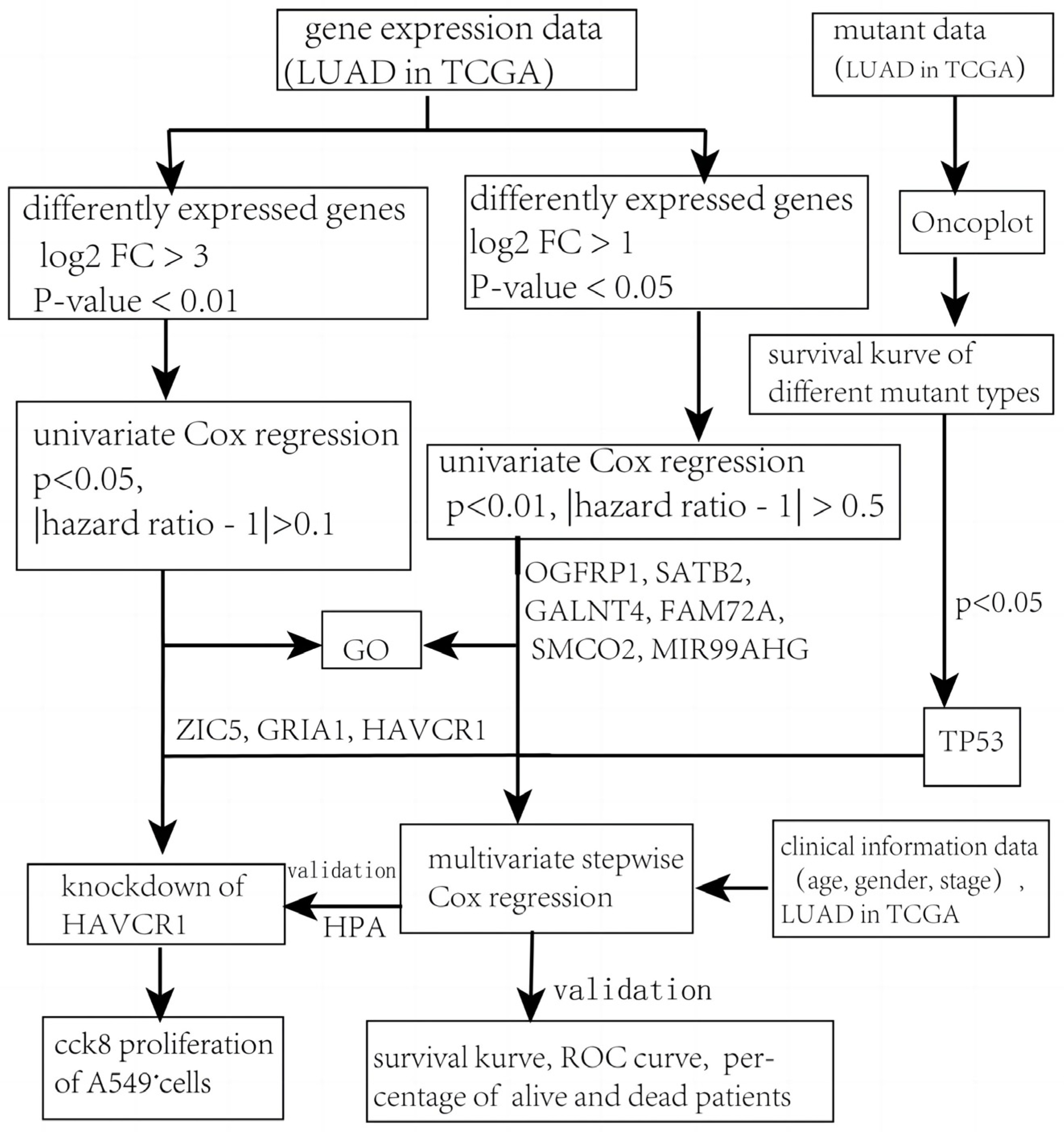
2. Materials and Methods
2.1. The Source of Materials
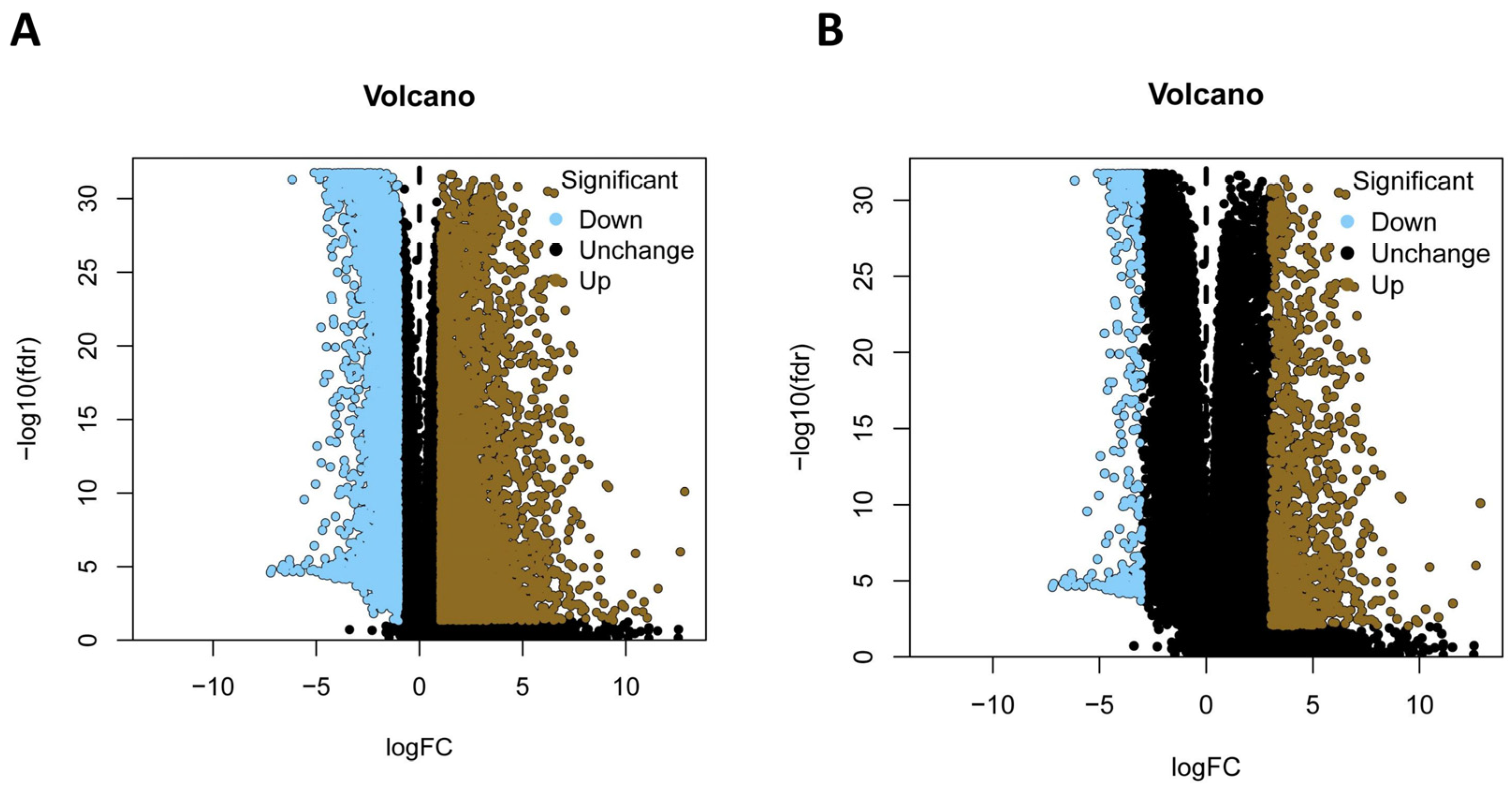
2.2. Screening Differentially Expressed Genes (DEGs)
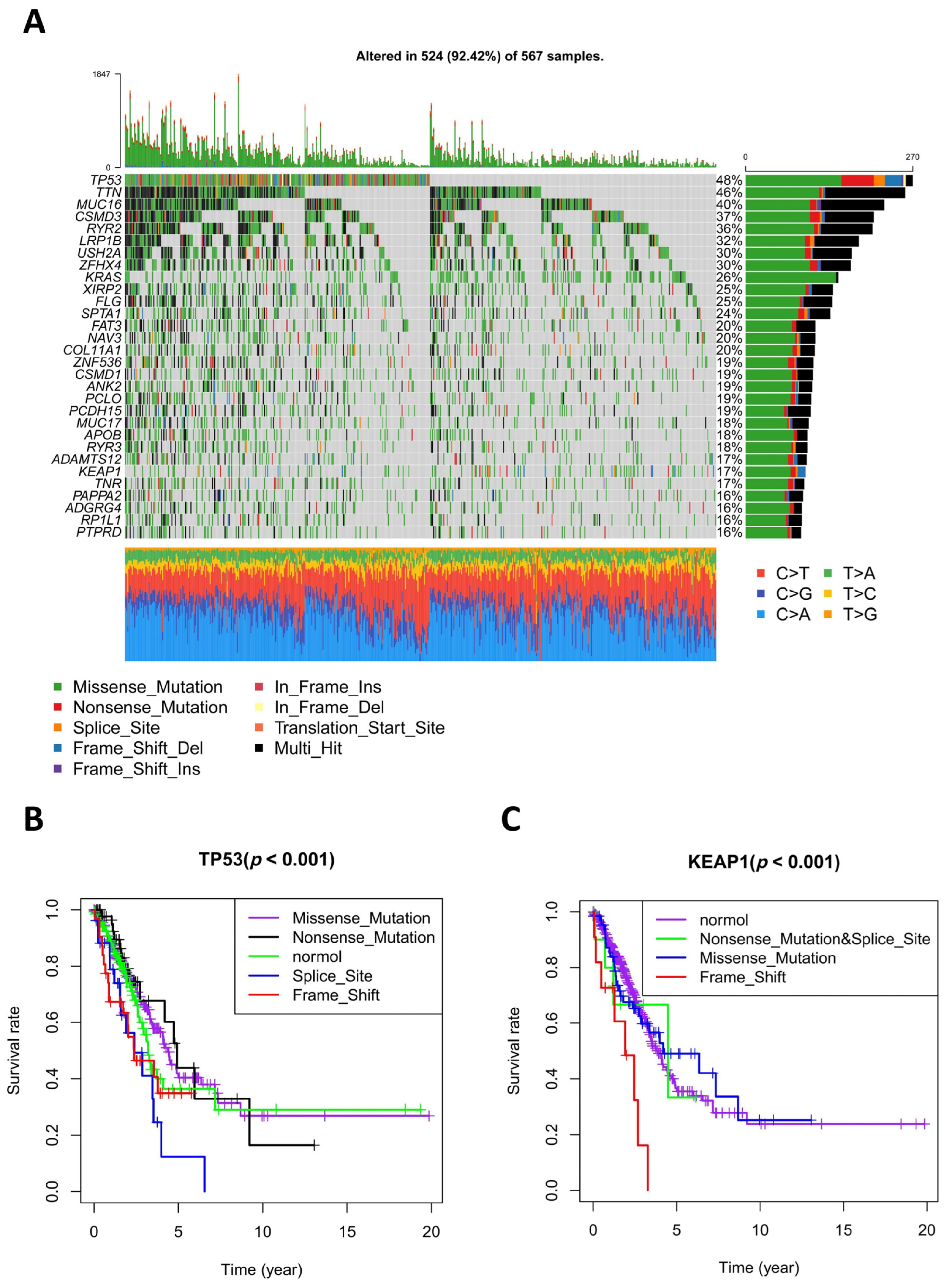
2.3. Screening Genes Whose Mutant Subtypes Were Meaningful to Overall Survival
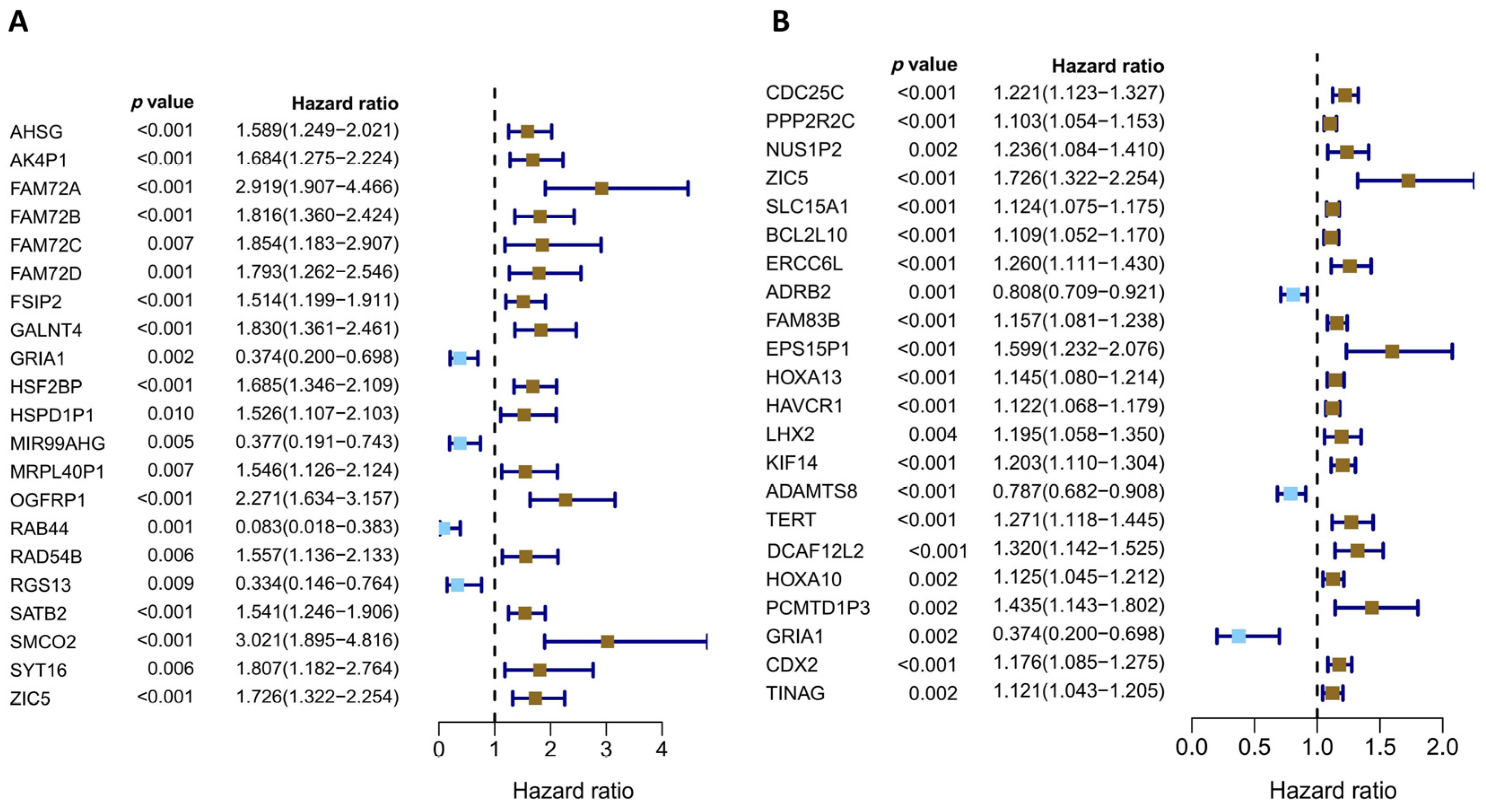
2.4. Construction of the Prognostic Model for LUAD Patients
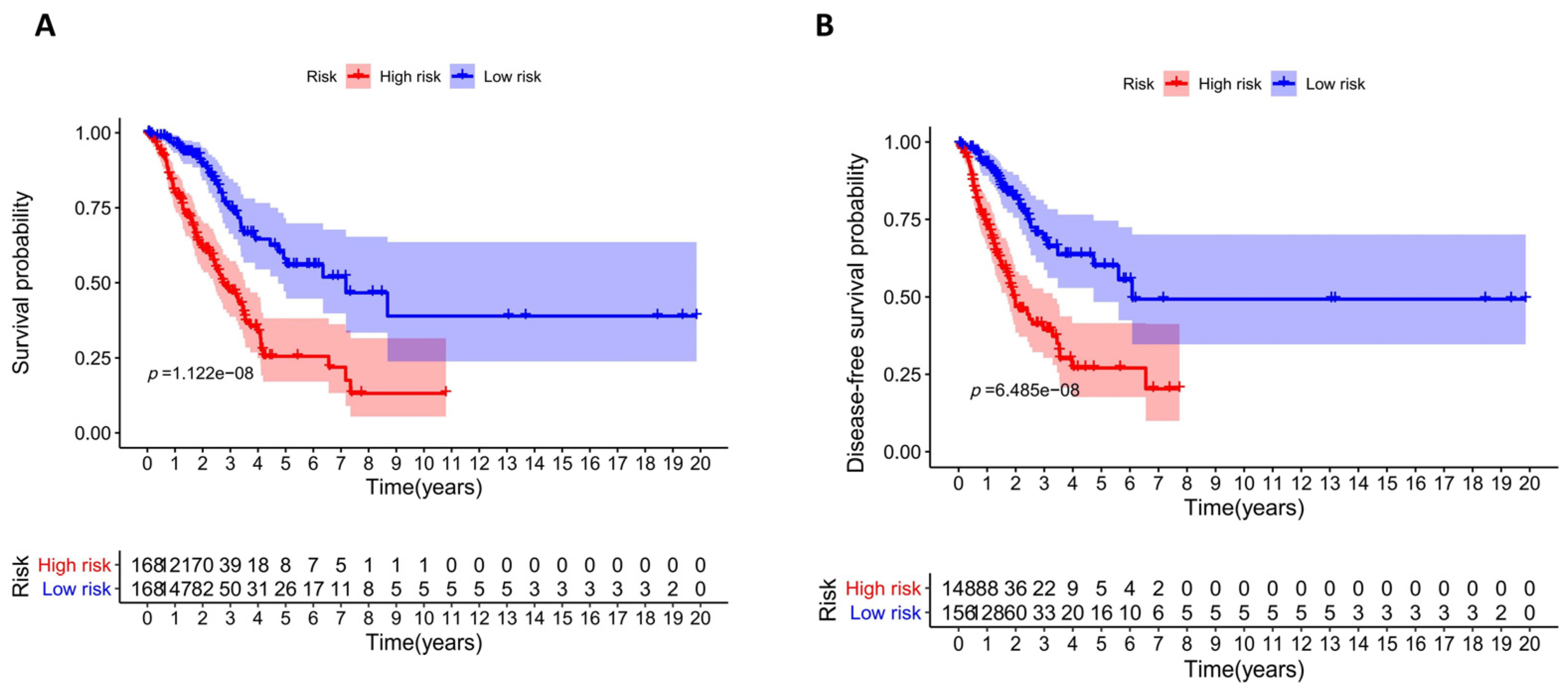
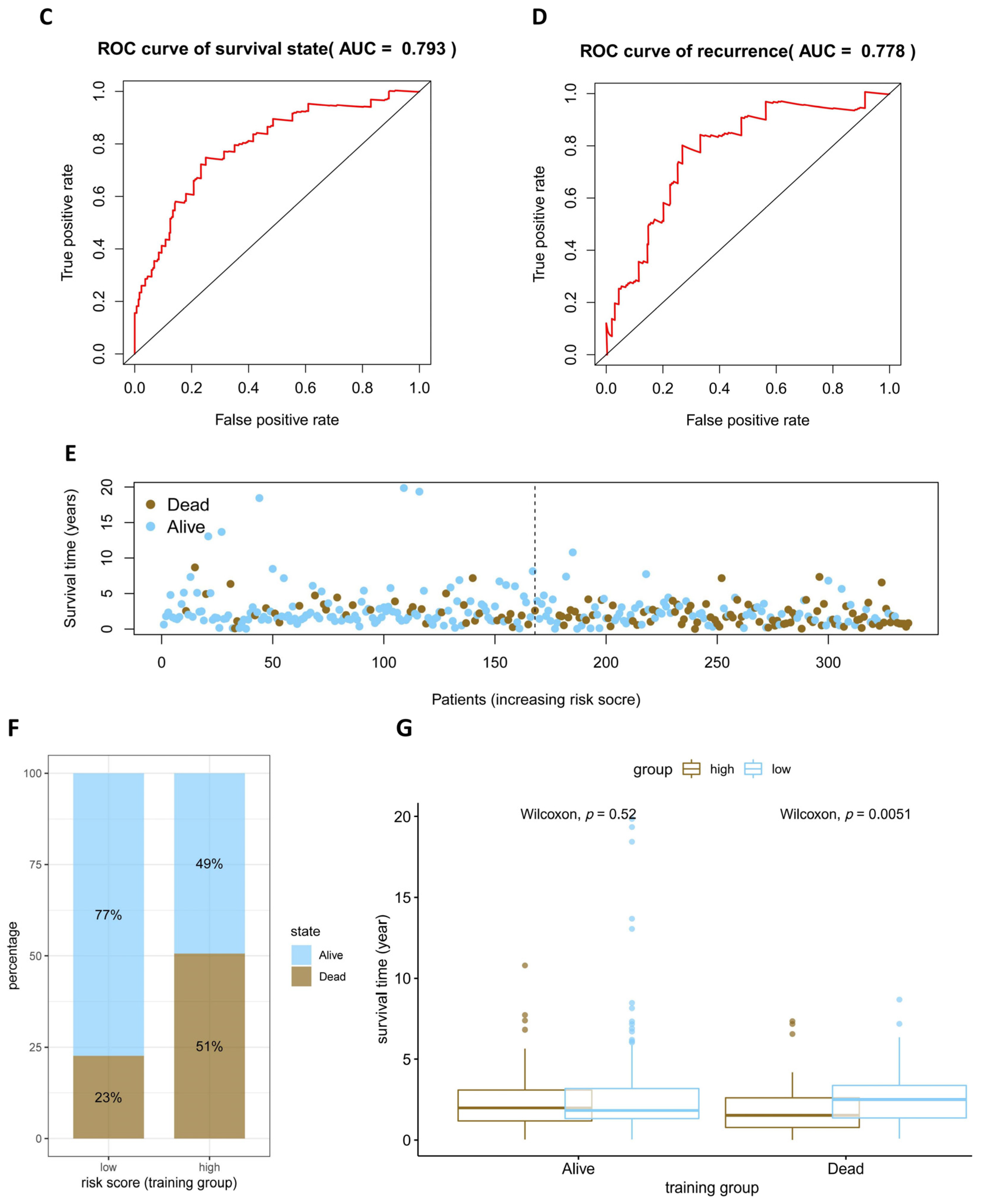
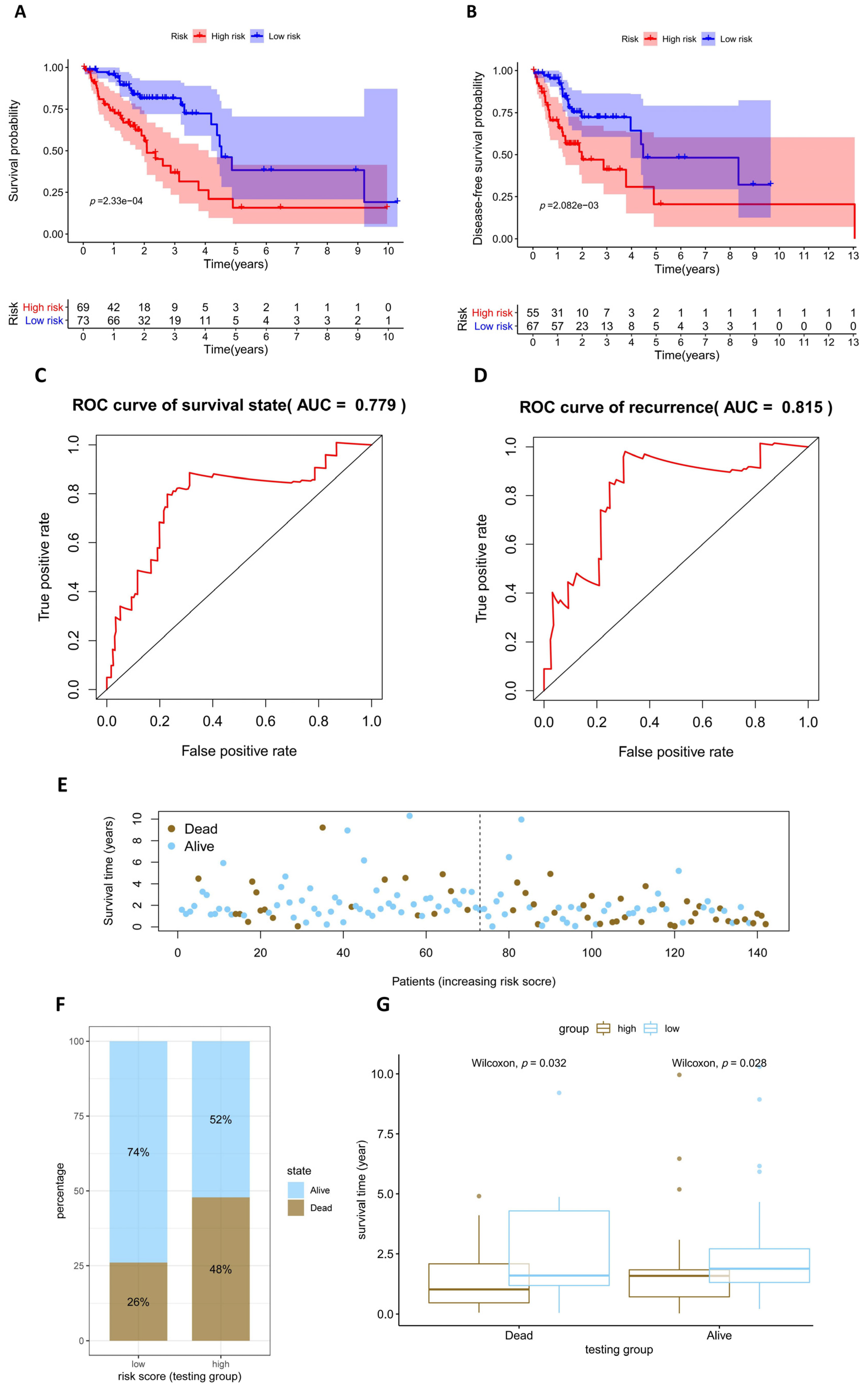
2.5. Assessment and Validation of the Prognostic Model
2.6. Gene Ontology (GO) Term Analysis
2.7. Cell Experiments
2.7.1. Design and Synthesis of siRNA
2.7.2. Cell Culture and Transfection
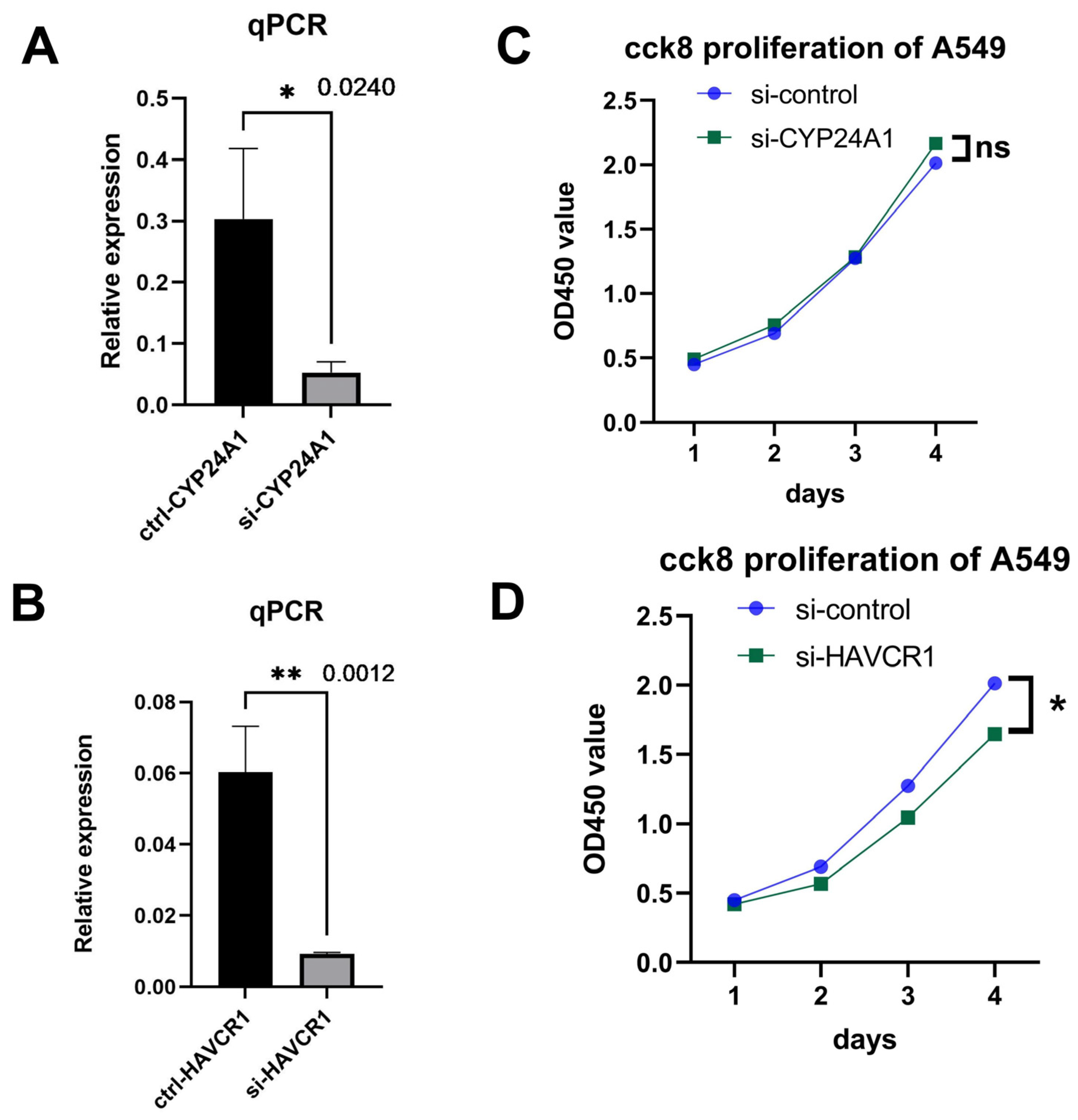
2.7.3. Quantitative RT-PCR after Knockdown of CYP24A1 or HAVCR1 in A549 Cells
2.7.4. CCK8 Proliferation Assay after Knockdown of CYP24A1 or HAVCR1 in A549 Cells
3. Results
3.1. Identification of Differentially-Expressed Genes
3.2. Different Mutant Subtypes of TP53 and KEAP1 were Significantly Related to Prognosis
3.3. Construction of Prognostic Model for LUAD Patients
3.4. Assessment and Validation of the Prognostic Model
3.5. Knockdown of HAVCR1 Suppressed A549 Cell Proliferation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Chen, J.; Yao, L.; Hao, Z.; Chang, Q. A Deep Learning Approach for Prognostic Evaluation of Lung Adenocarcinoma Based on Cuproptosis-Related Genes. Biomedicines 2023, 11, 1479. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Che, H.; Zhang, M.; Lv, J.; Pu, C.; Wu, J.; Zhang, Y.; Gu, Y. Developing CuS for Predicting Aggressiveness and Prognosis in Lung Adenocarcinoma. Genes 2023, 14, 1055. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Liu, L.; Guo, Y.; Shui, Y.; Wei, Q. A Comprehensive Analysis of the Effects of Key Mitophagy Genes on the Progression and Prognosis of Lung Adenocarcinoma. Cancers 2023, 15, 57. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, L.; Zhang, L.; Zheng, X.; Xu, H.; Wang, K.; Weng, X. Identification of a Four-Gene Signature Associated with the Prognosis Prediction of Lung Adenocarcinoma Based on Integrated Bioinformatics Analysis. Genes 2022, 13, 238. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Hughes, M. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 2.2021. J. Natl. Compr. Cancer Netw. 2021, 19, 254–266. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Mardis, E.R.; Steinberg, K.M.; Larson, D.E.; Wilson, R.K. The next-generation sequencing revolution and its impact on genomics. Cell 2013, 155, 27–38. [Google Scholar] [CrossRef]
- Wadowska, K.; Bil-Lula, I.; Trembecki, Ł.; Śliwińska-Mossoń, M. Genetic Markers in Lung Cancer Diagnosis: A Review. Int. J. Mol. Sci. 2020, 21, 4569. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, L.; Wang, Q.; Zhang, M.; Wang, B.; Jiang, K.; Ye, Y.; Wang, S.; Shen, Z. Molecular Characterization and Clinical Relevance of RNA Binding Proteins in Colorectal Cancer. Front. Genet. 2020, 11, 580149. [Google Scholar] [CrossRef]
- Aca, C.A.; Shareef, O.H. Knockdown of tigar induces apoptosis and autophagy with modulates nf-κb and ho-1 expression in a549 lung cancer cells. J. Inst. Sci. Technol. 2019, 9, 310–320. [Google Scholar] [CrossRef]
- Luo, X.; Weng, X.; Bao, X.; Bai, X.; Lv, Y.; Zhang, S.; Chen, Y.; Zhao, C.; Zeng, M.; Xu, B.; et al. Corrigendum to “A novel anti-atherosclerotic mechanism of quercetin: Competitive binding to KEAP1 via Arg483 to inhibit macrophage pyroptosis”. Redox Biol. 2022, 58, 102511–102548. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Na Niu, N.; Li, P.; Zhai, L.; Xiao, K.; Chen, W.; Zhuang, X. LncRNA OGFRP1 acts as an oncogene in NSCLC via miR-4640-5p/eIF5A axis. Cancer Cell Int. 2021, 21, 425. [Google Scholar] [CrossRef]
- Yu, F.; Liang, M.; Huang, Y.; Wu, W.; Zheng, B.; Chen, C. Hypoxic tumor-derived exosomal miR-31-5p promotes lung adenocarcinoma metastasis by negatively regulating SATB2-reversed EMT and activating MEK/ERK signaling. J. Exp. Clin. Cancer Res. 2021, 40, 179. [Google Scholar] [CrossRef]
- Xing, L.; Hong, X.; Chang, L.; Ren, P.; Zhang, H. miR-365b regulates the development of non-small cell lung cancer via GALNT4. Exp. Ther. Med. 2020, 20, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Li, H.; Ma, Z.; Dong, G.; Wang, Q.; Wang, S.; Fang, P.; Li, X.; Chen, H.; Liu, T.; et al. MIR99AHG is a noncoding tumor suppressor gene in lung adenocarcinoma. Cell Death Dis. 2021, 12, 424. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luo, X.; Cheng, C.; Amos, C.I.; Cai, G.; Xiao, F. A gene expression-based immune signature for lung adenocarcinoma prognosis. Cancer Immunol. Immunother. 2020, 69, 1881–1890. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Di Marco, M.; Mahale, S.; Jachimowicz, D.; Kosalai, S.T.; Reischl, S.; Statello, L.; Mishra, K.; Darnfors, C.; Kanduri, M.; et al. LY6K-AS lncRNA is a lung adenocarcinoma prognostic biomarker and regulator of mitotic progression. Oncogene 2021, 40, 2463–2478. [Google Scholar] [CrossRef]
- Zhu, D.; Wu, S.; Li, Y.; Zhang, Y.; Chen, J.; Ma, J.; Cao, L.; Lyu, Z.; Hou, T. Ferroptosis-related gene slc1a5 is a novel prognostic biomarker and correlates with immune infiltrates in stomach adenocarcinoma. Cancer Cell Int. 2022, 22, 124. [Google Scholar] [CrossRef]
- Wang, T.Y.; Jiang, X.Y.; Lu, Y.; Ruan, Y.M.; Wang, J.M. Identification and integration analysis of a novel prognostic signature associated with cuproptosis-related ferroptosis genes and relevant lncRNA regulatory axis in lung adenocarcinoma. Aging 2023, 15, 1543–1563. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, Z.; Zeng, Z.; Li, J.; Xie, H.; Xie, C. An integrative analysis of DNA methylation and gene expression to predict lung adenocarcinoma prognosis. Front. Genet. 2022, 13, 970507. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gao, L.N.; Song, P.P.; You, C.G. Development and validation of a RNA binding protein-associated prognostic model for lung adenocarcinoma. Aging 2020, 12, 3558–3573. [Google Scholar] [CrossRef]
- Kaplan, G.; Totsuka, A.; Thompson, P.; Akatsuka, T.; Moritsugu, Y.; Feinstone, S.M. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 1996, 15, 4282–4296. [Google Scholar] [CrossRef] [PubMed Central]
- Manangeeswaran, M.; Jacques, J.; Tami, C.; Konduru, K.; Amharref, N.; Perrella, O.; Casasnovas, J.M.; Umetsu, D.T.; DeKruyff, R.H.; Freeman, G.J.; et al. Binding of hepatitis A virus to its cellular receptor 1 inhibits T-regulatory cell functions in humans. Gastroenterology 2012, 142, 1516–1525.e3. [Google Scholar] [CrossRef]
- Meyers, J.H.; Sabatos, C.A.; Chakravarti, S.; Kuchroo, V.K. The TIM gene family regulates autoimmune and allergic diseases. Trends Mol. Med. 2005, 11, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tang, W.; Cao, J.; Shang, M.; Sun, H.; Gong, J.; Hu, B. A Comprehensive Analysis of HAVCR1 as a Prognostic and Diagnostic Marker for Pan-Cancer. Front. Genet. 2022, 13, 904114. [Google Scholar] [CrossRef]
- Li, Y.; Ge, D.; Gu, J.; Xu, F.; Zhu, Q.; Lu, C. A large cohort study identifying a novel prognosis prediction model for lung adenocarcinoma through machine learning strategies. BMC Cancer 2019, 19, 886. [Google Scholar] [CrossRef]
- Qian, C.; Jiang, Z.; Zhou, T.; Wu, T.; Zhang, Y.; Huang, J.; Ouyang, J.; Dong, Z.; Wu, G.; Cao, J. Vesicle-mediated transport-related genes are prognostic predictors and are associated with tumor immunity in lung adenocarcinoma. Front. Immunol. 2022, 13, 1034992. [Google Scholar] [CrossRef]
- Ang, G.; Brown, L.A.; Tam, S.K.E.; Davies, K.E.; Foster, R.G.; Harrison, P.J.; Sprengel, R.; Vyazovskiy, V.V.; Oliver, P.L.; Bannerman, D.M.; et al. Deletion of AMPA receptor GluA1 subunit gene (Gria1) causes circadian rhythm disruption and aberrant responses to environmental cues. Transl. Psychiatry 2021, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-B.; Gao, L.; Yang, Q.; Liu, Y.; Yu, X.-Y.; Shen, J.-H.; Zhang, W.-C.; Han, Z.-Y.; Chen, S.-Z.; Guo, S. Role of GALNT4 in protecting against cardiac hypertrophy through ASK1 signaling pathway. Cell Death Dis. 2021, 12, 980. [Google Scholar] [CrossRef] [PubMed]
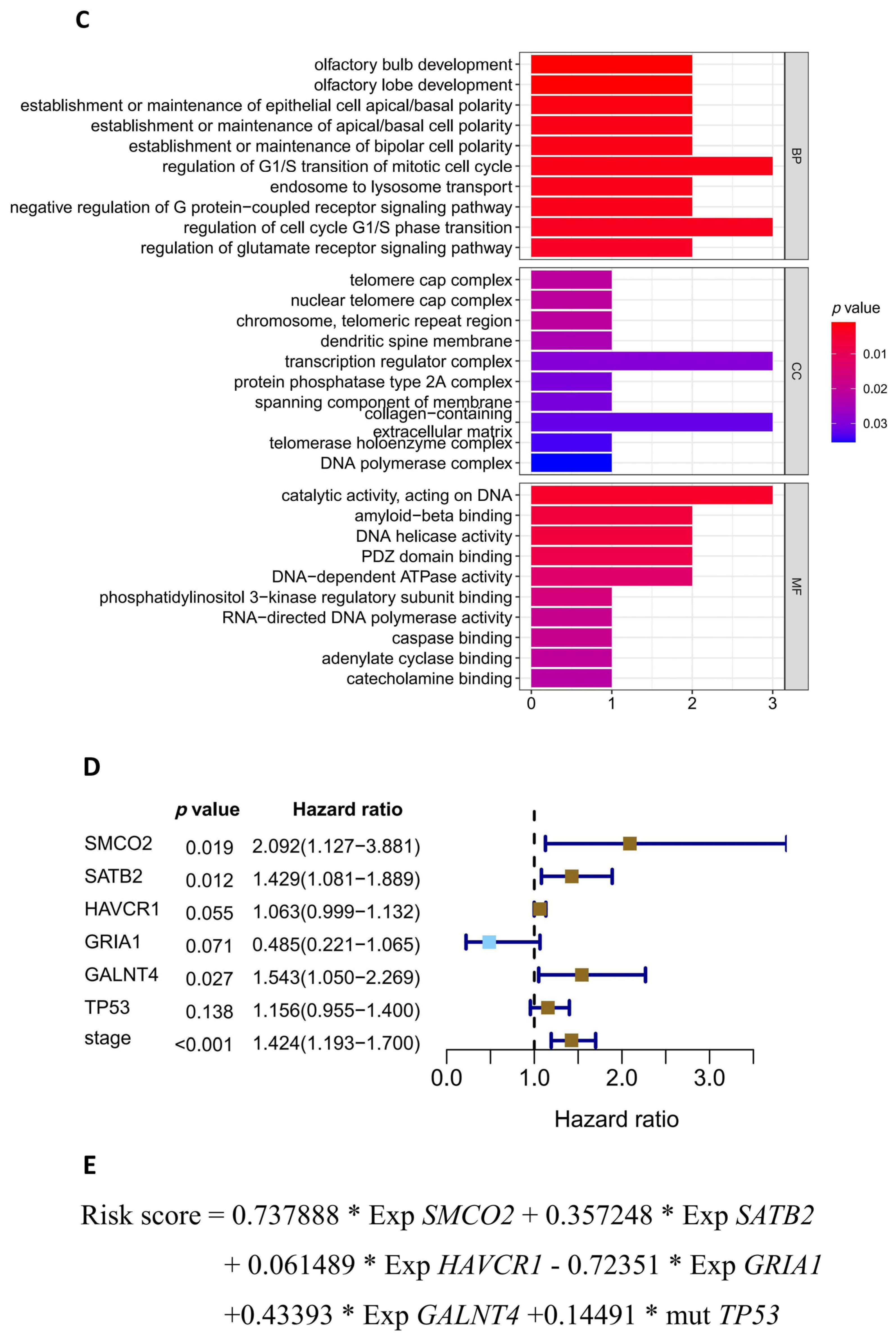
| ID | Coefficient | HR (Hazard Ratio) | HR.95L | HR.95H | p-Value |
|---|---|---|---|---|---|
| SMCO2 | 0.737888 | 2.091513 | 1.127194 | 3.88081 | 0.019305 |
| SATB2 | 0.357248 | 1.429391 | 1.081459 | 1.88926 | 0.012065 |
| HAVCR1 | 0.061489 | 1.063419 | 0.998669 | 1.132368 | 0.05506 |
| GRIA1 | −0.72351 | 0.485047 | 0.220938 | 1.064871 | 0.07134 |
| GALNT4 | 0.43393 | 1.543311 | 1.049862 | 2.268687 | 0.027279 |
| TP53 | 0.14491 | 1.155935 | 0.954653 | 1.399657 | 0.137666 |
| stage | 0.353671 | 1.424287 | 1.193488 | 1.699717 | 8.82 × 10−5 |
| Gene | Protein Domains in Pfam | Biological Functions |
|---|---|---|
| SMCO2 |  | The function is unknown. A single-pass membrane protein with two coiled-coil domains (NCBI: https://www.ncbi.nlm.nih.gov/gene/341346/ortholog/?scope=32525, accessed on 27 March 2023). |
| SATB2 |  | Controls the transcription factor expression of nuclear genes by binding to the matrix attachment regions of DNA. A docking site for several chromatin remodeling enzymes. Recruits corepressors (HDACs) or coactivators (HATs) to promoters and enhancers. (HPA: https://www.proteinatlas.org/ENSG00000119042-SATB2, accessed on 27 March 2023) |
| HAVCR1 |  | Cell recognition, immune activation, tight junction, and cancer biology. Promotes the activation and proliferation of immune cells and cytokine secretion and regulates the function of NK cells and CD8+ T for an efficient antitumor immune response [28]. |
| GRIA1 |  | An ionotropic glutamate receptor that has been proven to be related to neurotransmitter systems, sleep, and circadian rhythm [31]. |
| GALNT4 |  | Initiates the mucin-type O-glycosylation in cells. Participates in the post-translational modification, invasion, and proliferation of cancer cells [17,32]. |
| TP53 |  | A tumor suppressor protein with oligomeric domains that participates in the regulation of target gene expression, cell cycle arrest, apoptosis, senescence, changes in metabolism, and DNA repair. Mutations of this gene are related to various cancers (NCBI: https://www.ncbi.nlm.nih.gov/gene?Cmd=DetailsSearch&Term=7157, accessed on 27 March 2023). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Yu, L.; Zhang, H.; Jin, H. Identification of New Prognostic Genes and Construction of a Prognostic Model for Lung Adenocarcinoma. Diagnostics 2023, 13, 1914. https://doi.org/10.3390/diagnostics13111914
Chen X, Yu L, Zhang H, Jin H. Identification of New Prognostic Genes and Construction of a Prognostic Model for Lung Adenocarcinoma. Diagnostics. 2023; 13(11):1914. https://doi.org/10.3390/diagnostics13111914
Chicago/Turabian StyleChen, Xueping, Liqun Yu, Honglei Zhang, and Hua Jin. 2023. "Identification of New Prognostic Genes and Construction of a Prognostic Model for Lung Adenocarcinoma" Diagnostics 13, no. 11: 1914. https://doi.org/10.3390/diagnostics13111914
APA StyleChen, X., Yu, L., Zhang, H., & Jin, H. (2023). Identification of New Prognostic Genes and Construction of a Prognostic Model for Lung Adenocarcinoma. Diagnostics, 13(11), 1914. https://doi.org/10.3390/diagnostics13111914







