Homologous Recombination Deficiency Score Determined by Genomic Instability in a Romanian Cohort
Abstract
1. Introduction
- A.
- A direct, enzymatic, rapidly exhaustible repair that acts specifically to correct the modifications brought on by various alkylating agents.
- B.
- Multiple subtypes of excision repair:
- -
- Repair by excision of a single nitrogenous base (base excision repair, BER) for chain break or small change changes (non-bulky, small);
- -
- Repair by excision of a nucleotide fragment (20–30 nucleotides) for larger DNA lesions that cause double helix distortion;
- -
- Reconstruction of lesions caused by mismatch repair and small insertions/deletions [23].
- A.
- Abnormalities in the repair of strand breaks and replication errors—the most frequently involved are mutations in the BRCA1 and BRCA2 genes that act through the homologous repair mechanism, forming together with other proteins (encoded by the PALB2, BRIP1, RAD51C, RAD51D genes) the complex BASC (BRCA-associated protein) involved in the recognition and repair of abnormal DNA structures. Pathogenic variants in the BRCA1 and BRCA2 genes appear in different types of cancer, among which they are more frequently found in breast, ovarian, pancreatic, or prostate cancer [27].
- B.
- Defects in the genes that control the cell cycle and identify lesions in the genetic material (ATM, ATR, CHEK1, and CHEK2).
- C.
HRD Testing
- Predispositional insights: HRD testing can identify a person’s family members’ risk of getting ovarian cancer by detecting germline mutations of the BRCA1/2 genes. It can also help identify ovarian cancer patients at risk of other cancers;
- Prognostic insights: The test provides insight into the course of the disease and identifies whether the tumor has variants that may cause HRD;
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| ATM | ataxia telangiectasia mutated |
| BASC | BRCA1-Associated Genome Surveillance Complex |
| BARD1 | BRCA1 Associated RING Domain 1 |
| BER | base excision repair |
| BRCA1/2 | breast cancer gene 1/2 |
| BRIP1 | BRCA1 interacting protein 1 |
| FANC | Fanconi anemia complementation |
| FFPE | formalin-fixed paraffin-embedded |
| HRD | homologous recombination deficiency |
| HRR | homologous recombination repair |
| LOH | loss of heterozygosity |
| LST | large scale state transitions |
| MMR | mismatch repair |
| NHEJ | non-homologous end joining |
| PALB2 | partner and localizer of BRCA2 |
| SNP | single nucleotide polymorphism |
| TAI | telomere allelic imbalance |
References
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Wilczyński, J.R.; Wilczyński, M.; Paradowska, E. Cancer Stem Cells in Ovarian Cancer-A Source of Tumor Success and a Challenging Target for Novel Therapies. Int. J. Mol. Sci. 2022, 23, 2496. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.M.; Robson, M.E.; Ventz, S.; Santa-Maria, C.A.; Nanda, R.; Marcom, P.K.; Shah, P.D.; Ballinger, T.J.; Yang, E.S.; Vinayak, S.; et al. TBCRC 048: Phase II Study of Olaparib for Metastatic Breast Cancer and Mutations in Homologous Recombination-Related Genes. J. Clin. Oncol. 2020, 38, 4274–4282. [Google Scholar] [CrossRef]
- Koshiyama, M.; Matsumura, N.; Konishi, I. Recent concepts of ovarian carcinogenesis: Type I and type II. Biomed. Res. Int. 2014, 2014, 934261. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Lord, C.J.; Serra, V.; Tutt, A.; Balmaña, J.; Castroviejo-Bermejo, M.; Cruz, C.; Oaknin, A.; Kaye, S.B.; de Bono, J.S. A Decade of Clinical Development of PARP Inhibitors in Perspective. Ann. Oncol. 2019, 30, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous recombination deficiency: Exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- Janysek, D.C.; Kim, J.; Duijf, P.H.G.; Dray, E. Clinical use and mechanisms of resistance for PARP inhibitors in homologous recombination-deficient cancers. Transl. Oncol. 2021, 14, 101012. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.; Karakasis, K.; Madariaga, A.; Oza, A.M. Overcoming Platinum and PARP-Inhibitor Resistance in Ovarian Cancer. Cancers 2020, 12, 1607. [Google Scholar] [CrossRef]
- Vaidyanathan, A.; Sawers, L.; Gannon, A.L.; Chakravarty, P.; Scott, A.L.; Bray, S.E.; Ferguson, M.J.; Smith, G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 2016, 115, 431–441. [Google Scholar] [CrossRef]
- Yap, T.A.; Plummer, R.; Azad, N.S.; Helleday, T. The DNA Damaging Revolution: PARP Inhibitors and Beyond. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Perol, D.; Gonzalez-Martin, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Maenpaa, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.-Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 3970. [Google Scholar] [CrossRef] [PubMed]
- Wagener-Ryczek, S.; Merkelbach-Bruse, S.; Siemanowski, J. Biomarkers for Homologous Recombination Deficiency in Cancer. J. Pers. Med. 2021, 11, 612. [Google Scholar] [CrossRef]
- Reid, B.M.; Fridley, B.L. DNA Methylation in Ovarian Cancer Susceptibility. Cancers 2021, 13, 108. [Google Scholar] [CrossRef]
- Paulet, L.; Trecourt, A.; Leary, A.; Peron, J.; Descotes, F.; Devouassoux-Shisheboran, M.; Leroy, K.; You, B.; Lopez, J. Cracking the homologous recombination deficiency code: How to identify responders to PARP inhibitors. Eur. J. Cancer 2022, 166, 87–99. [Google Scholar] [CrossRef]
- Rondinelli, B.; Gogola, E.; Yücel, H.; Duarte, A.A.; van de Ven, M.; van der Sluijs, R.; Konstantinopoulos, P.A.; Jonkers, J.; Ceccaldi, R.; Rottenberg, S.; et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017, 19, 1371–1378. [Google Scholar] [CrossRef]
- Miller, R.E.; Leary, A.; Scott, C.L.; Serra, V.; Lord, C.J.; Bowtell, D.; DChang, K.; Garsed, D.W.; Jonkers, J.; Ledermann, J.A.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef]
- Plon, S.E.; Eccles, D.M.; Easton, D.; Foulkes, W.D.; Genuardi, M.; Greenblatt, M.S.; Hogervorst, F.B.; Hoogerbrugge, N.; Spurdle, A.B.; Tavtigian, S.V. Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008, 29, 1282–1291. [Google Scholar] [CrossRef]
- Paik, J. Olaparib: A Review as First-Line Maintenance Therapy in Advanced Ovarian Cancer. Target Oncol. 2021, 16, 847–856. [Google Scholar] [CrossRef]
- AstraZeneca Pharmaceuticals LP. LYNPARZA® (Olaparib)—Prescribing Information; AstraZeneca Pharmaceuticals LP: Wilmington, DE, USA, 2021. [Google Scholar]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M. DNA Repair. In The Cell: A Molecular Approach, 2nd ed.; Chapter 5; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Gee, M.E.; Faraahi, Z.; McCormick, A.; Edmondson, R.J. DNA damage repair in ovarian cancer: Unlocking the heterogeneity. J. Ovarian Res. 2018, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, N.; Miyagawa, K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014, 105, 370–388. [Google Scholar] [CrossRef]
- Angeli, D.; Salvi, S.; Tedaldi, G. Genetic Predisposition to Breast and Ovarian Cancers: How Many and Which Genes to Test? Int. J. Mol. Sci. 2020, 21, 1128. [Google Scholar] [CrossRef] [PubMed]
- Tomasova, K.; Cumova, A.; Seborova, K.; Horák, J.; Koucka, K.; Vodickova, L.; Vaclavikova, R.; Vodicka, P. DNA Repair and Ovarian Carcinogenesis: Impact on Risk, Prognosis and Therapy Outcome. Cancers 2020, 12, 1713. [Google Scholar] [CrossRef]
- Pacheco-Barcia, V.; Muñoz, A.; Castro, E.; Ballesteros, A.I.; Marquina, G.; González-Díaz, I.; Colomer, R.; Romero-Laorden, N. The Homologous Recombination Deficiency Scar in Advanced Cancer: Agnostic Targeting of Damaged DNA Repair. Cancers 2022, 14, 2950. [Google Scholar] [CrossRef] [PubMed]
- Mangogna, A.; Munari, G.; Pepe, F.; Maffii, E.; Giampaolino, P.; Ricci, G.; Fassan, M.; Malapelle, U.; Biffi, S. Homologous Recombination Deficiency in Ovarian Cancer: From the Biological Rationale to Current Diagnostic Approaches. J. Pers. Med. 2023, 13, 284. [Google Scholar] [CrossRef]
- Hoppe, M.M.; Sundar, R.; Tan, D.S.P.; Jeyasekharan, A.D. Biomarkers for Homologous Recombination Deficiency in Cancer. J. Natl. Cancer. Inst. 2018, 110, 704–713. [Google Scholar] [CrossRef]
- Vergote, I.; Gonzalez-Martin, A.; Ray-Coquard, I.; Harter, P.; Colombo, N.; Pujol, P.; Lorusso, D.; Mirza, M.R.; Brasiuniene, B.; Madry, R.; et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann. Oncol. 2022, 33, 276–287. [Google Scholar] [CrossRef]
- Hauke, J.; Horvath, J.; Groß, E.; Gehrig, A.; Honisch, E.; Hackmann, K.; Schmidt, G.; Arnold, N.; Faust, U.; Sutter, C.; et al. Gene panel Testing of 5589 BRCA1/2-Negative Index Patients with Breast Cancer in a Routine Diagnostic Setting: Results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. 2018, 7, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; Wu, X. The role of patient-derived ovarian cancer organoids in the study of PARP inhibitors sensitivity and resistance: From genomic analysis to functional testing. J. Exp. Clin. Cancer Res. 2021, 40, 338. [Google Scholar] [CrossRef] [PubMed]
- Kondrashova, O.; Nguyen, M.; Shield-Artin, K.; Tinker, A.V.; Teng, N.N.H.; Harrell, M.I.; Kuiper, M.J.; Ho, G.Y.; Barker, H.; Jasin, M.; et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017, 7, 984–998. [Google Scholar] [CrossRef] [PubMed]
- Herzog, T.J.; Vergote, I.; Gomella, L.G.; Milenkova, T.; French, T.; Tonikian, R.; Poehlein, C.; Hussain, M. Testing for homologous recombination repair or homologous recombination deficiency for poly (ADP-ribose) polymerase inhibitors: A current perspective. Eur. J. Cancer 2023, 179, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Creeden, J.F.; Nanavaty, N.S.; Einloth, K.R.; Gillman, C.E.; Stanbery, L.; Hamouda, D.M.; Dworkin, L.; Nemunaitis, J. Homologous recombination proficiency in ovarian and breast cancer patients. BMC Cancer 2021, 21, 1154. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhu, J.; Zhao, W.; Huang, Y.; An, R.; Zheng, H.; Qu, P.; Wang, L.; Zhou, Q.; Wang, D.; et al. Olaparib Maintenance Monotherapy in Asian Patients with Platinum-Sensitive Relapsed Ovarian Cancer: Phase III Trial (L-MOCA). Clin. Cancer Res. 2022, 28, 2278–2285. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Brown, J.; Barnicle, A.; Rowe, P.; Lao-Sirieix, P.; Criscione, S.; du Bois, A.; Lorusso, D.; Romero, I.; Petru, E.; et al. Homologous recombination repair mutation gene panels (excluding BRCA) are not predictive of maintenance olaparib plus bevacizumab efficacy in the first-line PAOLA1/ENGOT-ov25 trial. Gynecol. Oncol. 2021, 162, S26–S27. [Google Scholar] [CrossRef]
- Goel, N.; Foxall, M.E.; Scalise, C.B.; Wall, J.A.; Arend, R.C. Strategies in Overcoming Homologous Recombination Proficiency and PARP Inhibitor Resistance. Mol. Cancer Ther. 2021, 20, 1542–1549. [Google Scholar] [CrossRef]
- Onstad, M.; Coleman, R.L.; Westin, S.N. Movement of Poly-ADP Ribose (PARP) Inhibition into Frontline Treatment of Ovarian Cancer. Drugs 2020, 80, 1525–1535. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef]
- Myriad MyChoice® CDx—Technical Information, Myriad Genetic Laboratories, Inc. Available online: https://bit.ly/myChoiceCDxSpecs (accessed on 12 April 2020).
- Patel, J.N.; Braicu, I.; Timms, K.M.; Solimeno, C.; Tshiaba, P.; Reid, J.; Lanchbury, J.S.; Darb-Esfahani, S.; Ganapathi, M.K.; Sehouli, J.; et al. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br. J. Cancer 2018, 119, 1060–1066. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.; de la Hoya, M.; Bellosillo, B.; de Juan, I.; Matías-Guiu, X.; Lázaro, C.; Palanca, S.; Osorio, A.; Rojo, F.; Rosa-Rosa, J.; et al. Mutational screening of BRCA1/2 genes as a predictive factor for therapeutic response in epithelial ovarian cancer: A consensus guide from the Spanish Society of Pathology (SEAP-IAP) and the Spanish Society of Human Genetics (AEGH). Virchows Arch. 2020, 476, 195–207. [Google Scholar] [CrossRef]
- Cline, M.S.; Liao, R.G.; Parsons, M.T.; Paten, B.; Alquaddoomi, F.; Antoniou, A.; Baxter, S.; Brody, L.; Cook-Deegan, R.; Coffin, A.; et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018, 14, e1007752. [Google Scholar] [CrossRef] [PubMed]
- Heitz, F.; Ataseven, B.; Staniczok, C.; Denkert, C.; Rhiem, K.; Hahnen, E.; Heikaus, S.; Moubarak, M.; Welz, J.; Dagres, T.; et al. Implementing HRD Testing in Routine Clinical Practice on Patients with Primary High-Grade Advanced Ovarian Cancer. Cancers 2023, 15, 818. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Gonzalez-Martin, A.; Harter, P.; Lorusso, D.; Moore, K.N.; Oaknin, A.; Ray-Coquard, I. First-line PARP inhibitors in ovarian cancer: Summary of an ESMO Open-Cancer Horizons round-table discussion. ESMO Open 2020, 5, e001110. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, B.A.; Lai, Z.; Hodgson, D.R.; Orr, M.C.; Hawryluk, M.; Sun, J.; Yelensky, R.; Spencer, S.K.; Robertson, J.D.; Ho, T.W.; et al. Biological and clinical evidence for somatic mutations in BRCA1 and BRCA2 as predictive markers for olaparib response in high-grade serous ovarian cancers in the maintenance setting. Oncotarget 2017, 8, 43653–43661. [Google Scholar] [CrossRef]
- Lheureux, S.; Lai, Z.; Dougherty, B.A.; Runswick, S.; Hodgson, D.R.; Timms, K.M.; Lanchbury, J.S.; Kaye, S.; Gourley, C.; Bowtell, D.; et al. Long-Term Responders on Olaparib Maintenance in High-Grade Serous Ovarian Cancer: Clinical and Molecular Characterization. Clin. Cancer Res. 2017, 23, 4086–4094. [Google Scholar] [CrossRef]
- Chiang, Y.C.; Lin, P.H.; Cheng, W.F. Homologous Recombination Deficiency Assays in Epithelial Ovarian Cancer: Current Status and Future Direction. Front. Oncol. 2021, 11, 675972. [Google Scholar] [CrossRef]
- Ruscito, I.; Dimitrova, D.; Vasconcelos, I.; Gellhaus, K.; Schwachula, T.; Bellati, F.; Zeillinger, R.; Benedetti-Panici, P.; Vergote, I.; Mahner, S.; et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients—A study of the tumour Bank ovarian cancer (T.O.C.) and ovarian cancer diagnosis consortium (OVCAD). Eur. J. Cancer 2014, 50, 2090–2098. [Google Scholar] [CrossRef]
- Swisher, E.M.; Kwan, T.T.; Oza, A.M.; Tinker, A.V.; Ray-Coquard, I.; Oaknin, A.; Coleman, R.L.; Aghajanian, C.; Konecny, G.E.; O’malley, D.M.; et al. Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2). Nat. Commun. 2021, 12, 2487. [Google Scholar] [CrossRef]
- Callens, C.; Vaur, D.; Soubeyran, I.; Rouleau, E.; Just, P.A.; Guillerm, E.; Golmard, L.; Goardon, N.; Sevenet, N.; Cabaret, O.; et al. Concordance between Tumor and Germline BRCA Status in High-Grade Ovarian Carcinoma Patients in the Phase III PAOLA-1/ENGOT-ov25 Trial. J. Natl. Cancer Inst. 2021, 113, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, F.; Gieldon, L.; Rump, A.; Seifert, M.; Grützmann, K.; Krüger, A.; Loos, S.; Zeugner, S.; Hackmann, K.; Porrmann, J.; et al. Targeted capture-based NGS is superior to multiplex PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE tumor samples. BMC Cancer 2019, 19, 396. [Google Scholar] [CrossRef]
- McDonough, S.J.; Bhagwate, A.; Sun, Z.; Wang, C.; Zschunke, M.; Gorman, J.A.; Kopp, K.J.; Cunningham, J.M. Use of FFPE-derived DNA in next generation sequencing: DNA extraction methods. PLoS ONE 2019, 14, e0211400. [Google Scholar] [CrossRef] [PubMed]
- Ngoi, N.Y.L.; Tan, D.S.P. The role of homologous recombination deficiency testing in ovarian cancer and its clinical implications: Do we need it? ESMO Open 2021, 6, 100144. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.M.; Hinchcliff, E.; Avila, M.; Westin, S.N. The Clinical Challenges, Trials, and Errors of Combatting Poly(ADP-Ribose) Polymerase Inhibitors Resistance. Cancer J. 2021, 27, 491–500. [Google Scholar] [CrossRef]
- Damia, G.; Broggini, M. Platinum Resistance in Ovarian Cancer: Role of DNA Repair. Cancers 2019, 11, 119. [Google Scholar] [CrossRef]
- Pettitt, S.J.; Krastev, D.B.; Brandsma, I.; Dréan, A.; Song, F.; Aleksandrov, R.; Harrell, M.I.; Menon, M.; Brough, R.; Campbell, J.; et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 2018, 9, 1849. [Google Scholar] [CrossRef]

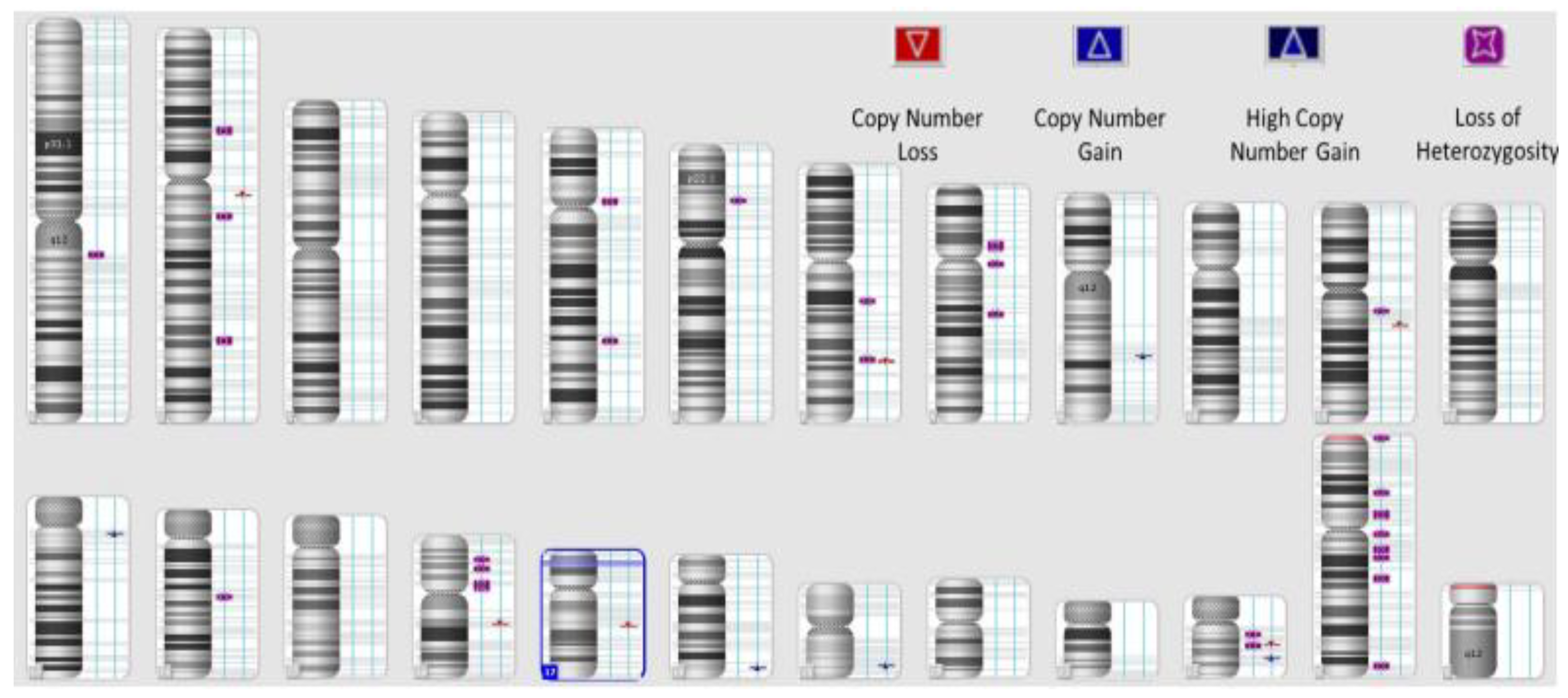
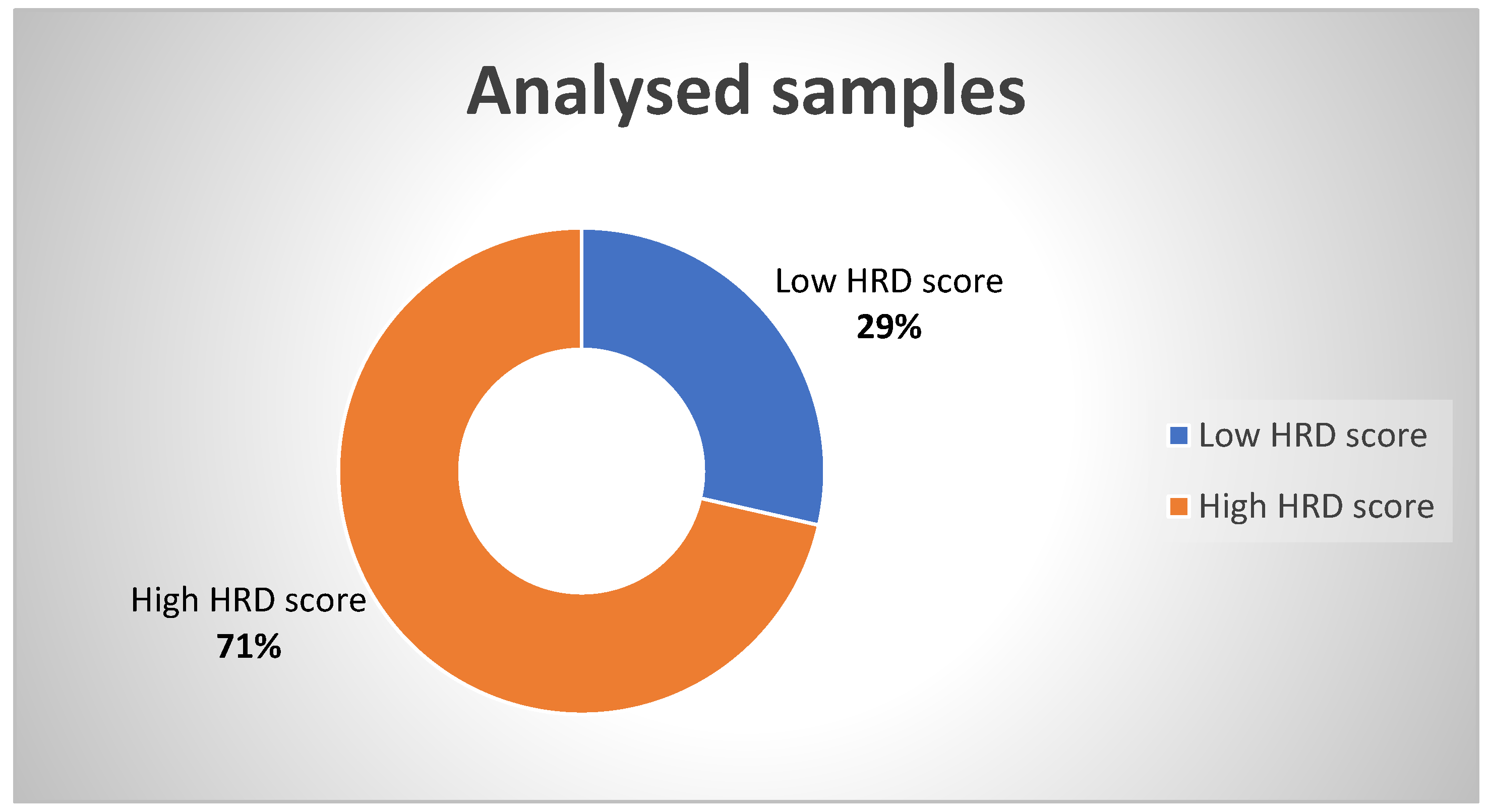
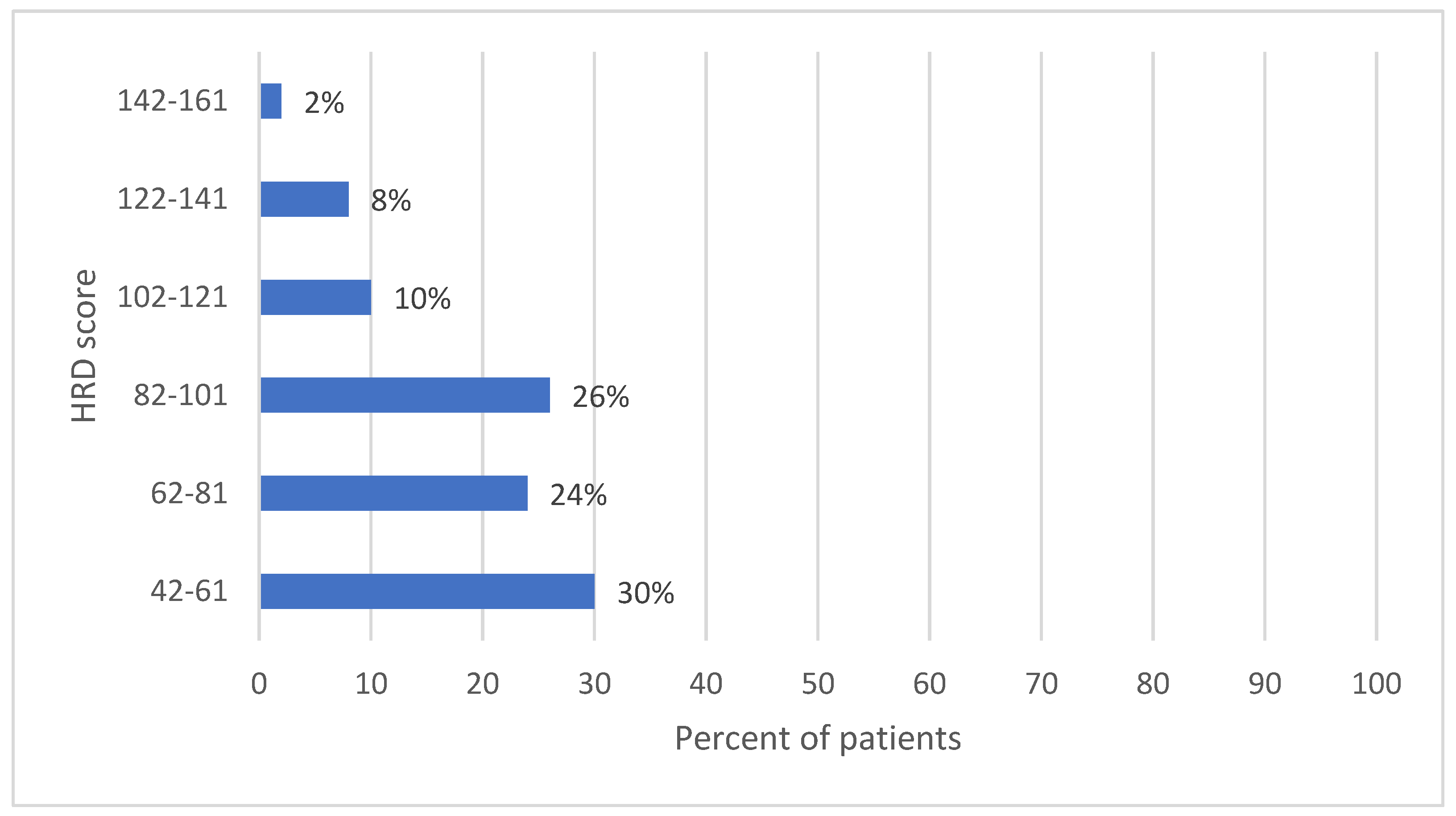
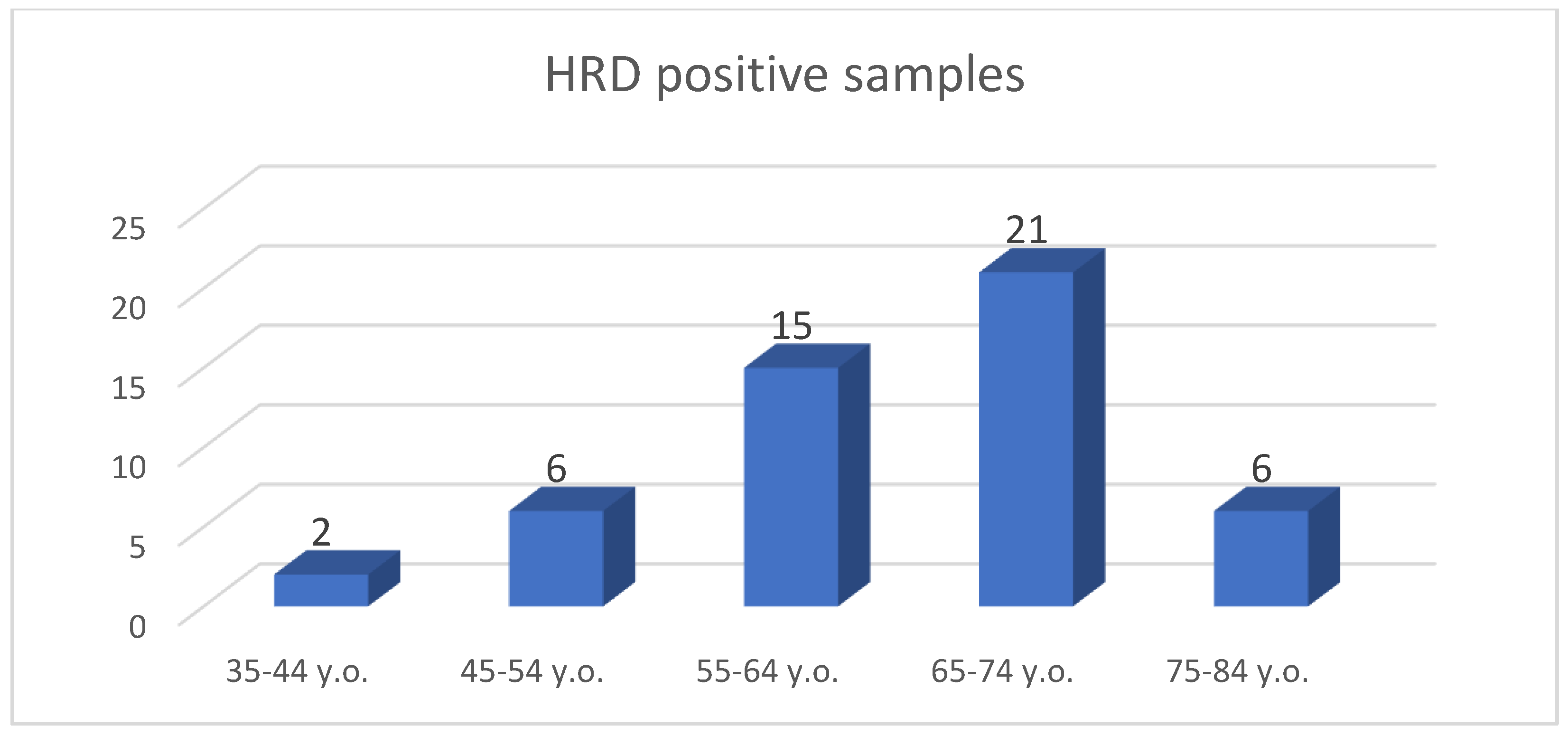
| FIGO Stage of Ovarian Cancer | Frequency | Percent | Exact 95% LCL | Exact 95% UCL | Median HRD Score | Std. Dev. |
|---|---|---|---|---|---|---|
| IIIA | 6 | 12.00% | 4.53% | 24.31% | 59 | 18.5876 |
| IIIB | 6 | 12.00% | 4.53% | 24.31% | 64.5 | 38.6506 |
| IIIC | 29 | 58.00% | 43.21% | 71.81% | 85 | 25.4200 |
| IV | 9 | 18.00% | 8.58% | 31.44% | 73 | 27.6667 |
| TOTAL | 50 | 100.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rădoi, V.-E.; Țurcan, M.; Maioru, O.V.; Dan, A.; Bohîlțea, L.C.; Dumitrescu, E.A.; Gheorghe, A.S.; Stănculeanu, D.L.; Thodi, G.; Loukas, Y.L.; et al. Homologous Recombination Deficiency Score Determined by Genomic Instability in a Romanian Cohort. Diagnostics 2023, 13, 1896. https://doi.org/10.3390/diagnostics13111896
Rădoi V-E, Țurcan M, Maioru OV, Dan A, Bohîlțea LC, Dumitrescu EA, Gheorghe AS, Stănculeanu DL, Thodi G, Loukas YL, et al. Homologous Recombination Deficiency Score Determined by Genomic Instability in a Romanian Cohort. Diagnostics. 2023; 13(11):1896. https://doi.org/10.3390/diagnostics13111896
Chicago/Turabian StyleRădoi, Viorica-Elena, Mihaela Țurcan, Ovidiu Virgil Maioru, Andra Dan, Laurentiu Camil Bohîlțea, Elena Adriana Dumitrescu, Adelina Silvana Gheorghe, Dana Lucia Stănculeanu, Georgia Thodi, Yannis L. Loukas, and et al. 2023. "Homologous Recombination Deficiency Score Determined by Genomic Instability in a Romanian Cohort" Diagnostics 13, no. 11: 1896. https://doi.org/10.3390/diagnostics13111896
APA StyleRădoi, V.-E., Țurcan, M., Maioru, O. V., Dan, A., Bohîlțea, L. C., Dumitrescu, E. A., Gheorghe, A. S., Stănculeanu, D. L., Thodi, G., Loukas, Y. L., & Săbău, I.-D. (2023). Homologous Recombination Deficiency Score Determined by Genomic Instability in a Romanian Cohort. Diagnostics, 13(11), 1896. https://doi.org/10.3390/diagnostics13111896






