Abstract
An automatic pathological diagnosis is a challenging task because histopathological images with different cellular heterogeneity representations are sometimes limited. To overcome this, we investigated how the holistic and local appearance features with limited information can be fused to enhance the analysis performance. We propose an unsupervised deep learning model for whole-slide image diagnosis, which uses stacked autoencoders simultaneously feeding multiple-image descriptors such as the histogram of oriented gradients and local binary patterns along with the original image to fuse the heterogeneous features. The pre-trained latent vectors are extracted from each autoencoder, and these fused feature representations are utilized for classification. We observed that training with additional descriptors helps the model to overcome the limitations of multiple variants and the intricate cellular structure of histopathology data by various experiments. Our model outperforms existing state-of-the-art approaches by achieving the highest accuracies of 87.2 for ICIAR2018, 94.6 for Dartmouth, and other significant metrics for public benchmark datasets. Our model does not rely on a specific set of pre-trained features based on classifiers to achieve high performance. Unsupervised spaces are learned from the number of independent multiple descriptors and can be used with different variants of classifiers to classify cancer diseases from whole-slide images. Furthermore, we found that the proposed model classifies the types of breast and lung cancer similar to the viewpoint of pathologists by visualization. We also designed our whole-slide image processing toolbox to extract and process the patches from whole-slide images.
1. Introduction
Currently, pathologists use whole-slide images (WSIs) to make an accurate primary pathological diagnosis. A manual histopathological examination of such images is time-consuming, and it is difficult to reproduce them; thus, an efficient and robust automated system is needed that can achieve quick scanning and diagnosis of potential disease regions. With the advancement of digital imaging techniques and the advent of WSIs, significant attention has been devoted to the automatic detection and classification of cancers. Current research in this field focuses on machine-learning- and deep-neural-network-based methods, directly impacting clinical and related studies, as well as the progression and development of targeted therapy approaches [1]. Computational tools based on the digital slide workflow concept can help increase the efficiency and accuracy of detection while enabling a more detailed and precise disease analysis.
The challenges in deep learning for cancer or disease diagnosis on histopathological images include the use of large amounts of accurately annotated data and the generalization limitations of WSIs due to multiple variants and the intricate cellular structure of histopathology data. Publicly available WSIs with accurate annotation or label information are fewer as compared to natural image datasets because acquiring WSIs is a difficult and costly task owing to the specific nature of medical imaging modalities [2,3,4,5]. To address this issue, previous researchers used pre-trained models based on previously acquired knowledge from different domain image datasets and then applied them to the medical imaging domain.
However, for rare types of cancers, it is difficult to obtain WSI data. We thought it would be a very effective idea to use multiple descriptors such as HOG and LBPs for shape and texture feature extraction, respectively, which not only showed improved performance together [6], but can help obtain sufficientinformation when the number of data is very small. We propose an unsupervised learning model for heterogeneous feature learning for WSI classification, which showed relatively good performance even on small balanced datasets and has not yet been exploited. Our simple structured model is superior in the sense that we do not assume a specific set of pretrained features based on different classifiers to achieve high performance. Instead, we rely only on unsupervised latent spaces learned from stacked autoencoders based on any number of independent multiple descriptors, which enables the model to extract and fuse these rich spaces and can be used with different types of classifiers. Our model can classify the rare types of cancer diseases from WSIs more accurately than previous literature methods.
The contributions of our paper are as follows:
- (1)
- We propose an unsupervised deep learning model that uses stacked autoencoders to learn the multiple low-dimensional latent spaces from an input, including the histogram of oriented gradients (HOG) [7] and local binary patterns (LBPs) [8]), along with the original image to fuse the heterogeneous features. Two additional inputs as descriptors help the model learn and classify multiple variants’ cellular information robustly from less or partially available annotated data. Although only three image descriptors, which are raw images and their HOG and LBP, representations are assumed as the inputs, we can use as many as required for future research.
- (2)
- Our model outperforms the existing state-of-the-art models, yielding the best score for binary-class and multi-class classification using four metrics, and it achieved the highest overall multi-class accuracies and other metrics for two public benchmark WSI cancer datasets.
- (3)
- We also discuss the performance of the model from various perspectives using multiple-input descriptors, visualization experiments such as confusion matrices, and the t-SNE technique, which confirmed that our model classifies the WSIs similarly to the way pathologists diagnose. The AUC (ROC) curves of the model also showed our model can be used clinically for cancer diagnosis.
The remainder of this paper is organized as follows. In Section 2, we discuss related research. The architecture and technical details of the proposed model are presented in Section 3. Section 4 describes the datasets used in this study. The experimental setup, model training, and implementation details are described in Section 5. Section 6 and Section 7 provide a detailed comparison of the classification results of WSIs and examine these results from the perspective of pathological diagnosis by visualization the last layer of the model. Finally, Section 8 provides concluding remarks and potential areas for future research.
2. Related Work
With the continuous technological advances in digital scanners and image visualization methods, the diagnosis of disease through glass slides manually is rapidly replaced by WSI inspection based on AI algorithms. These changes are expected to further accelerate the development of automatic diagnosis systems through WSIs; as the development of AI techniques progresses, such methods assist pathologists in making early decisions on treatments.
Hand-crafted features and clustering techniques have been extensively researched, such as [9], which extracts the handcrafted features from annotated tumor regions of WSIs and then applies a machine learning model for the top feature selection and cancer classification, which solely relies on careful annotations of cancer regions of WSIs. Balazsi attempted to overcome the effect of fixed tiling of WSIs by using color, texture, and gradient features extracted with RF classifiers for invasive ductal carcinoma versus normal classification [10]. Barker applied the unsupervised K-means technique on phenotypes of patches, followed by feature selection via CNN and feature aggregation for the final prediction of features that represent the morphological character, texture, and statistical property of malignant tumors [11]. Spanhol designed a method to extract the image patches from the BreaKHis dataset for training the CNN and, later, the combination of these patches for final classification. The performance metrics showed improved results when compared to previous models trained with handcrafted textural descriptors [12]. In contrast, Zhu obtained the survival prediction by utilizing only the WSI-level labels [13]. Whitney developed an algorithm to provide quantitative measurements of nuclear shape and size, which could be applied across different tumor subtypes [14]. Bahlmann used color gradients of square patches with linear SVMs for the classification of diagnostically relevant versus irrelevant regions in WSIs, but used only a small set of patches [15]. Bejnordi used a 2D long short-term memory to aggregate the context information of WSIs from a grid of neighboring patches and classify spatial information [16]. Litjens designed a deep network with 128 128 pixel patches at 5 magnification for the delineation of prostate cancer [17]. Bejnordi proposed a context-aware stacked CNN to take advantage of spatial information within WSIs [18]. Lin built a framework by leveraging a fully convolutional network for efficient inference while reconstructing dense predictions to ensure detection accuracy [19]. Cruz-Roa proposed a three-layer CNN that operated on 100 100 pixel patches at 2.5 magnification, which was more successful than color-, texture-, and graph-based features with an RF classifier in the detection of invasive breast cancer regions [20]. Attallah proposed a model for the diagnosis of pediatric medulloblastomas and their subtypes from WSIs. The textural analysis features and deep learning extraction methods were used in a cascaded manner, and the best accuracy results were reported for binary and multi-class classification [21]. Attallah and Zaghlool applied a combination of textural and deep learning features to improve subtype identification of WSIs. This technique yields an increase in the accuracy of the classification of pediatric medulloblastomas with a relatively small number of features [22]. Anwar designed a CAD method, which includes a fusion of features extracted from the ResNet CNN model combined with features of wavelet packet decomposition (WPD) and HOG. Later, these features were reduced by the PCA technique and achieved the highest accuracy of 97.1% [23]. Attallah proposed a Histo-CADx model, which applied the fusion of different deep learning techniques with handcrafted feature methods to learn the set of fused features. Then, a classifier system with the fusion of outputs from different classifiers was used, which showed improved accuracy results with BreakHis and the ICIAR 2018 datasets [24].
A few weakly supervised methods have also been presented. For example, Dundar used multiple instances of learning for discrimination of benign cases from actionable (atypical ductal hyperplasia and ductal carcinoma in situ) by using WSIs with manually identified ROIs [25]. Sudharshan proposed a framework for the diagnosis of breast cancer patients, to investigate the relevance of multiple instances’ learning with sample images into bags (patients) without the need to label all the instances. The experiments were conducted on the BreaKHis dataset and achieved the highest results with 40 magnification for both images or patientswise cases [26]. Some additional studies have used multiple instance learning [27,28] to address the classification problem by automatically extracting valuable information from coarsely labeled patches. Some studies also used multiple resolutions [29], multi-field-of-view sliding windows [30], a range of magnification levels of WSIs [31], and high-level WSI classification tasks [32,33] to analyze WSIs and reported accurate survival predictions [34].
Literature studies have used supervised or weakly supervised learning methods or proposed fused models for a more accurate diagnosis of WSIs. The fused models [35,36] have demonstrated better computational performance than the specific set of optimal feature extractors, but these models are more dependent on the selected set of features or classifiers, which might not work with relatively rare types of cancer cases, extracted from WSI data. In contrast, our study helps to obtain sufficient information from WSIs, by using any number of independent multiple descriptors for unsupervised learning of features, and later, different classifiers (supervised) can be used to achieve high performance for any type of WSI classification. The fusion of latent spaces learned with multiple descriptors extracts more distinct and salient information while retaining coarse-scale features of WSIs at specific magnification levels. Our model also consists of separable modules that can be used for various medical image analysis tasks.
3. Proposed Model and Architecture
The block diagram of the proposed model is schematically shown in Figure 1. The model consists of a WSI pre-processing stage, data representation stage, and two main processing sub-blocks of the training and classification stages. The latter includes the unsupervised learning of multiple features using stacked autoencoders [37], the fusion of hidden latent spaces, and the final classification layers.
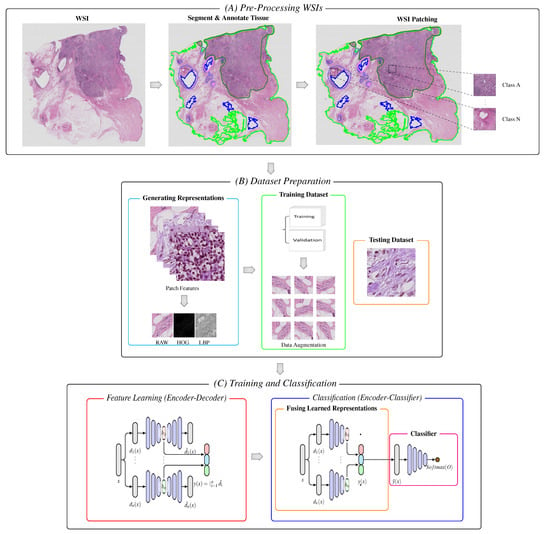
Figure 1.
Block diagram of the proposed model. (A) Following segmentation and annotation, image patches are extracted from the tissue regions of the WSIs. (B) Patches are encoded into a set of descriptive feature representations (i.e., RAW, HOG, and LBPs). The representations are further split into two portions, and data augmentation is applied to the set of descriptive representations of the training portion. (C,Left) For unsupervised learning, the set of generated descriptive features is feed into the stacked autoencoders without labels, which embeds the input vectors into a lower-dimensional space. (C,Right) The learned representations are fused together and passed with labels to the classifier with respective labels, which are used to make the final diagnostic prediction and classification.
3.1. Pre-Processing WSIs
3.1.1. Segment and Annotate Tissue
For each slide, our pipeline begins with automated segmentation of the tissue regions, and we developed our own pre-processing WSI system to obtain patches from WSIs. The WSIs are read into memory at a down-sampled resolution, for example, 64, converted from RGB to the HSV color space. A binary mask for the tissue regions is computed based on the threshold of the saturation channel of the image after median blurring to smooth the edges; subsequently, we applied an additional morphological closing operator to fill small gaps and holes. The approximate contours of the detected foreground objects are then filtered based on an area threshold and stored for downstream processing, while the segmentation mask for each slide is made available for other visual analysis in the next work. A human-readable file is also generated, which includes the list of files processed with important fields containing the set of segmentation parameters used. The segmentation parameter list includes a segment level that represents the down-sampled level of WSIs, which can be easily loaded into memory for segmenting and annotating classwise regions, thresholds, and so on.
3.1.2. WSI Patching
After segmentation, our algorithm crops 224 224 patches from within the segmented foreground contours at the user-specified magnification and stores stacks of image patches along with their coordinates, class labels, and slide metadata using the hierarchical data format (hdf5). Depending on the size of the WSIs and the user-specified magnification, the number of patches extracted from each slide range can vary from hundreds to hundreds of thousands. The number of extracted patches for each class was also saved in separate folders with the respective classwise label information.
3.2. Dataset Preparation
3.2.1. Generating Representations
The next important phase after WSI patching is extracting a set of n feature descriptors from the set of extracted patches. The main reason for using these feature descriptors is to learn the overall variant structure of the cells, such as the color, shape, and texture. We used two feature descriptors, HOG and LBPs, to extract the shape and texture features, respectively. This controls the significant loss of visual information that disappears during learning, which results in poor classification performance [38]. We re-scaled the patch raw images to a downscaled ratio (40 40) to enhance the finer visually important details and kept the size of the images in each category intact. In the next step, we further divided the input data (x) into two sub-modules: training and testing.
3.2.2. Training Dataset
The training data without augmentation were subdivided into training and validation sets. We generated an augmented dataset from a subset of three representations: RAW, HOG, and LBP. In general, pathologists study histological images from various orientations and magnifications, and there are many variations in the staining and acquisition conditions. We applied different data augmentation techniques to mimic the pathologist examination process and realistic variations, such as flipping (horizontal and vertical), rotation, shifting (width and height), brightness, zooming, and blurring. Such data augmentation can increase the size of the dataset without deteriorating its quality. These images were used to train the model from scratch without any pre-trained weights.
3.2.3. Testing Dataset
The remaining portion of the data was used to validate the performance of the proposed model.
The data were flattened into vectors from images before feeding the data to the training and classification modules because we used multi-layer autoencoders that accept a vector as the input.
3.3. Training and Classification
This is the main stage of our model, which includes a hierarchical feature contraction and fusion scheme based on autoencoders for classification.
3.3.1. Feature Learning
To learn multiple representations from input patches x, we used a set of different image descriptors that included three different characteristics (color, shape, and texture), represented as , where represents the feature vector of each representation. We wanted to convert data x into a more suitable representation that has particular distributions or characteristics, making it easier to handle. Several methods can be used to achieve a compact space, including clustering or dimensionality reduction, followed by a mapping function. Nevertheless, autoencoders demonstrated all of these capabilities using a single method [39]. They learn an identity function by embedding input vectors into a lower-dimensional space that learns the relevant features. Owing to this characteristic, they are the backbone of our proposed architecture.
Our model consists of a set of n autoencoders, , parameterized by , where Θ is the set of parameters for the complete model. Hence, for the raw image patch along with its corresponding HOG and LBPs, there should be 3n features learned for feature fusion. In the following text, we suppress the parameters from the notation of each AE for a simplified and concise representation. The goal of each AE is to compress input into a more expressive space, that is , where the ith reconstructed feature is the output of the ith AE that operates on the input feature . During the training of each AE, the objective was to minimize the error between the reconstructed and original features. Hence, we define our feature reconstruction loss as
3.3.2. Classification
We need a compact representation learned within the autoencoders, because they define a space that separates and clusters the input learned data [40]. In general, the ith AE, , is the composition of encoder , and decoder functions such that . Let the hidden compact representation of the ith AE (the output of the encoder) be represented as . We are then interested in the learned feature representation , where error is the concatenation operator. To find the optimal set of parameters Θ for our model, the loss of n autoencoders is minimized by the function defined in Equation (2), where the loss function in Equation (1) is parameterized by Θ, which corresponds to the parameters of each AE, , which were omitted in the previous subsection. These training parameters are learned through back-propagation of the training phase.
At the final stage of the proposed model, we replaced the decoder stage with a classifier stage. Our classifier stage consists of fully connected layers, which are connected to the concatenated feature space representations of the encoders. Given any learned latent representations, , we can extract the multiple feature representations to retrain our classification model with fused latent representations connected to the fully connected layers and with the help of labels of the training set to classify the samples after passing through the encoder and classifier layers. We present more details in Section 5 with regard to our experimental setup. Our model has a loosely coupled architecture that allows us to switch between various classifiers without the need to relearn the unsupervised feature representation learning process.
3.4. Model Architecture
With the following design considerations, we summarize the details of the architecture in Table 1:

Table 1.
Proposed model architecture. H, H, H∈ (Feature-X, Feature-X/K, K*Feature-X) with activation map. Abbreviations: Feature-RAW, Feature-HOG, Feature-LBP (Feature-X ∈), number of neurons (K), Number of hidden Layers (NL).
- (1)
- Normalize (N):This layer accepts the raw image to normalize the color distribution of an over-/under-stained image to a well-stained target image by using a simple H&E color normalization technique [41].
- (2)
- Input (I): This layer accepts a normalized image, which is pre-processed further to extract features using two image descriptors, HOG and LBPs, corresponding to the image from the raw images H and L, respectively.
- (3)
- Feature Representation Set (FRS): This operation is applied to flatten the output of each input image into feature vectors prior to feeding it to the network layers, which significantly reduces the layer operations and prepares the model input for learning.
- (4)
- Autoencoder Layers (AE-x): Autoencoders are mainly dimensionality reduction algorithms with a couple of important properties: autoencoders are a specific type of feedforward neural network where the output is the mirror image of the input. Each AE consists of three components: an encoder, hidden layers (latent space), and a decoder. The encoder compresses the input into a lower-dimensional representation, which is known as a latent space, and then reconstructs the output from this representation. The tissues vary in the target images; thus, to explore different features, our model architecture has hidden layers with a sufficient number of operations to represent each of these features, as shown in Table 1.
- (5)
- Fused Feature (FF): This operation, applied to concatenate the hidden learned representation of each AE hidden representation in the previous layer, significantly reduces the data handling and prepares the model for the final classification layers.
- (6)
- Output (O): The number of output neurons corresponding to each class, which are normalized using the softmax function, depends on the type of classification. In the present study, we conducted a multi-class classification (eight or nine classes).
4. Datasets
We used the following two public WSI datasets as reference standards for training and evaluating our unsupervised deep learning model. These datasets contained different variants of cancer diseases collected from different patients.
4.1. ICIAR2018
This dataset (publicly available at https://iciar2018-challenge.grand-challenge.org/Dataset; accessed on 10 May 2019) is a version of the Bioimaging 2015 breast histology classification challenge dataset [42]. The dataset consists of 30 WSIs, where 10 are pixelwise labeled and 20 are non-labeled slides. These slides were digitized under the same acquisition conditions as Leica SCN400; the magnification was 20, and the pixel dimensions were μ per pixel. We used our own WSI pre-processing toolbox to generate patchwise images from each WSI based on the pixelwise annotations of WSIs and categorized them into one of four classes: (1) benign, (2) in situ, (3) invasive carcinoma, and (4) normal; for each case, the assigned class corresponds to a predominant cancer type in the respective WSI, as shown in Figure 2.
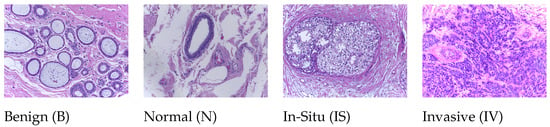
Figure 2.
Microscopic H&E patched images of four types in the ICIAR2018 dataset.
The goal of this challenge was to provide an automatic classification of WSIs into four classes. For our experiments, we used 10 labeled WSIs from which we generated a total of 32,215 patch samples, as shown in Table 2. Since the number of patches in the invasive class was more than approximately 10-times larger than the number of patches in other classes, there was a serious data imbalance. It is known that the unbalances in datasets can affect performance. Therefore, we created another balanced dataset that randomly selected only 3800 samples of the invasive class. We performed experiments on both datasets and looked at how much impact balancing the datasets had on performance. We ensured patient-based separation between the training and testing data to evaluate the performance of our model for clinical situations.

Table 2.
Structure of the ICIAR2018 with 20 magnification factor.
4.2. Dartmouth Lung Cancer
This dataset (publicly available at https://bmirds.github.io/LungCancer, accessed on 4 March 2019) is composed of 143 WSIs of lung adenocarcinoma from the Department of Pathology and Laboratory Medicine at Dartmouth-Hitchcock Medical Center (DHMC) [43]. All WSIs were labeled according to the consensus opinions of the three pathologists. These histopathology slides were scanned using a Leica Aperio whole-slide scanner at 20 magnification. A total of 31 WSIs were used to generate image patches for each WSI and categorized into five classes. Each class contains different random samples and types: (1) acinar, (2) lepidic, (3) micropapillary, (4) papillary, and (5) solid. Some examples of histological images are shown in Figure 3. We used 31 random WSIs owing to our limited hardware resources. The structural details of this dataset are listed in Table 3.
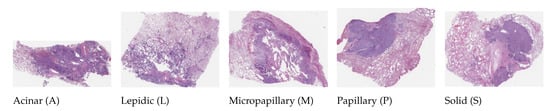
Figure 3.
Histological images of five types of lung adenocarcinoma in the Dartmouth Lung Cancer dataset.

Table 3.
Structure of the Dartmouth Lung Cancer dataset (31 WSIs only) with 20 magnification factor.
5. Experimental Setup
5.1. Aspects of Performance Evaluation
We evaluated the performance of the proposed model with respect to five different aspects: classification of binary-class and multi-class samples, comparison with literature encoders, effect of multi-input descriptors, confusion matrices with feature visualizations, and ROC curve analysis, as described in Section 6.1, Section 6.2 and Section 7.1–Section 7.3. To assess the performance of our model in clinical situations, we ensured that the same patient data were not used simultaneously during the training and testing phases. We trained the model using 5-fold cross-validation on a subset of training samples to determine the best hyperparameters for the model. For our experiments, we used a random 80:20 partitioning of the each dataset into training and testing subsets.
We used multiple solid-state drives and hard drive storages to store the raw files of digital WSIs. To perform segmentation and patching of WSIs, we used the Intel Xeon CPUs and feature extraction and learning using an NVIDIA Quadro RTX 5000 on local workstations with 128 GB RAM. The proposed model pipeline was implemented in Python and takes advantage of imaging processing libraries, such as OpenSlide, OpenCV, and NumPy. We used the Keras deep learning library to load data and train our models. Our source code is publicly available at https://github.com/AIMILab/Diagnostics, accessed on 12 June 2022).
5.2. Best Hyperparameters
Our model comprises two modules. For feature learning, four hyperparameters were applied for each autoencoder: the number of hidden layers, number of neurons per layer, type of activation function, and type of reconstruction loss. The following seven hyperparameters were tuned for the classifier: the number of layers, number of epochs, learning rate, batch size, optimizer, and batch normalization. The best selected parameters of our model with the cross-validation technique are listed in Table 4, according to the abbreviations in Table 1.

Table 4.
The best hyperparameters of our model.
For feature extraction from descriptors, we used the following hyperparameters, which demonstrated the best results. HOG parameters: cells per block (1,1), orientations (16), and pixels per cell (14,14); LBPs: method (uniform), number of patterns (16), and radius of circle (2). These both descriptors help our model learn and classify the input samples more accurately with substantial information.
5.3. Performance Evaluation Metrics
The performance of the proposed pipeline was evaluated using several evaluation metrics including accuracy, sensitivity, precision, -score, and the area under the curve of the receiving operating characteristic AUC (ROC). Equations (3)–(6) were used to compute these metrics, where the true positives (TPs) are the number of correctly predicts samples as the positive class. False positives (FPs) represent the number of incorrectly predicted samples as the positive class. True negatives (TNs) are the number of correctly predicted samples as the negative class. False negatives (FNs) represent the number of incorrectly predicted samples as the negative class.
6. Performance Results
6.1. Comparison with Deep Learning Models
Table 5 and Table 6 show the classification results of binary-class and multi-class classifications with different well-known classification models for the two datasets, ours and the state-of-the-art models for histopathology diagnosis. The accuracy of the balanced dataset increased to 90.4% when only two classes were considered, and our model performed better than the other models by a large margin for the ICIAR2018 dataset, as shown in Table 5. Our method also demonstrated the highest results for all the other metrics when compared with the other models. For multiple classifications, the accuracy of our model outperformed the state-of-the-art models on both datasets, as shown in Table 6. The accuracy increased to 87.2% for the ICIAR2018 dataset and 94.6% for the Dartmouth dataset when four and five classes were considered.

Table 5.
Comparison of four metrics on the ICIAR2018 for the binary-classification. The best results are shown in bold.

Table 6.
Comparison of four metrics on the ICIAR2018 and Dartmouth datasets for the multi-classification. The best results are shown in bold.
We also noted that the variants introduced in these databases pose a significant challenge for the descriptors we use because HOG and LBPs are not invariant to the perspective and intensity transforms of these two datasets. Nevertheless, the sensitivity, specificity, and -scores of our model outperformed those of the other CNN models. Clinically, because multi-class classification is able to determine cancer subtypes, it is more important for pathologists. For both datasets, the results of our model were higher, which indicates that our model can be developed for clinical use. It should be noted that there is a difference in performance between the unbalanced and balanced datasets. The balanced dataset improved the performance by more than 4% for all the metrics. These results demonstrate the importance of using a balanced dataset for deep learning models.
6.2. Comparison with Different Deep Encoders
We compared the performance behavior of deep feature encoders with the proposed encoder. We replaced the unsupervised stacked autoencoders of our model with different deep encoders that were trained and fine-tuned on both datasets using imagenet weights and later used these learned features in the classification stage. Table 7 shows the results for both WSI datasets. For the experiments, we used the following five deep encoders of well-known CNN models: ResNet-50 [44], Inception-V3 [45], DenseNet-121 [46], MobileNet [47], RuneCNN [48], BreastNet [49], LiverNet [50], and HCNN [51]. We report the accuracy of all the experiments on five-fold cross-validation. The maximum accuracies of 77.9% and 86.5% were achieved by the fine-tuned encoder of the HCNN model. Similarly, other encoders also obtained lower accuracy results compared to the HCNN encoder. Thus, the empirical results show that the idea of using fine-tuned deep encoders of complex pathological data is not efficient enough, and neither improve the performance results when used with the imagenet weights or deeply layer structured encoders. The use of image descriptors with stacked autoencoders obtained higher accuracy as compared to features learned from deep encoders for both datasets. It should be noted that the proposed stacked autoencoders with descriptors are more effective than the deep and complex CNN-based encoders.

Table 7.
Accuracy comparison of our encoder against literature encoders. The parenthesis values represent standard deviation.
7. Discussion
7.1. Effect of Multiple Descriptors
The behavior of our model for multi-class classification was thoroughly studied with special emphasis on factors that can affect performance. The number of inputs to autoencoders significantly affects the performance. In Figure 4, we observe that the performance improves as we feed more inputs to autoencoders in the form of image descriptors for both datasets. The input set of triplet representation shows the highest metrics in comparison to pairs or individual descriptors, which indicates that later fusion of learned representations for classification can increase the invariants of the given input descriptors. Our observation from the results is that removing the input combinations of descriptors will significantly degrade the results, as shown with the input set of single and pair representation. The performance can be further improved by employing more descriptors as inputs, which significantly boosts the metrics and classifies samples more efficiently. Based on our experiments, we concluded that our results align with the design of using multiple input descriptors as inputs to the stacked autoencoders and the later fusion of these learned latent spaces for classifying the sample of the WSIs more effectively. We believe that these results can also be applied to general medical images.
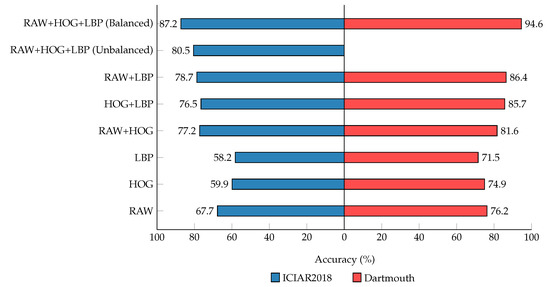
Figure 4.
Accuracy for multi-class classification on the ICIAR2018 and Dartmouth datasets. Using the different combination of generated representations.
7.2. Pathologist’s Analysis of the Results
We analyzed the results of our model from a pathological point of view and confirmed that the results of our model are very similar to the diagnosis of pathologist’s. The performance of the ICIAR2018 dataset was slightly lower than that of the Dartmouth dataset due to the technical and the pathological point of view. The main technical reason arises from ambiguous data that are found in the set of patches for each class generated from the ICIAR2018 data by our WSI pre-processing system. We generated patches automatically using the label information provided. According to a pathologist, there are a number of patches that our system has made from areas designated as invasive, which should be classified as normal or benign. This is because we created a patch with a size of 224 224, which occupies more of the normal or benign than the invasive. However, in the Dartmouth dataset, such patches are extremely rare.
A pathologist said that the ICIAR2018 dataset has morphologic heterogeneity. This means the ICIAR2018 dataset comprises various spectrums of normal, benign, in situ, and invasive carcinoma. Even more, breast cancer is also morphologically heterogeneous in tumor types, such as invasive ductal, invasive lobular, mucinous, micropapillary, or invasive papillary carcinoma, and various histological grading scales, such as well-differentiated (more tubular formation) to poorly differentiated (more solid). In other words, each cluster of the ICIAR2018 dataset has a diverse spectrum of various diseases. The Dartmouth dataset includes only invasive carcinoma with a single histological subtype. Therefore, each cluster is a more homogenous composition compared to that of the ICIAR2018 dataset.
From the confusion matrices shown in Figure 5, the ICIAR2018 dataset showed 18.4% differences of in situ carcinoma, which was interpreted as invasive carcinoma. This is because small patches of a training set of invasive carcinoma could contain scattered in situ lesions in between invasive areas, resulting in the deep learning being able to learn small patches of the in situ carcinoma as invasive carcinoma. In the real world, much invasive breast carcinoma, especially the luminal molecular subtype, has scattered in situ carcinoma adjacent to and mixed with an invasive component. However, carcinoma of the lung consists of purely invasive components with a very low proportion of in situ carcinoma. In lung cancer, in situ carcinoma itself is not a more common lesion than breast cancer.
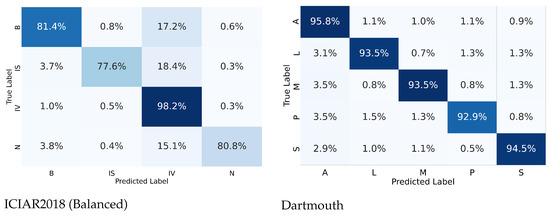
Figure 6 shows a two-dimensional visual representation of the activations of the last fully connected layer of the classifier stage. These representations result from the application of t-SNE [52], which is an efficient parametric embedding technique for dimensionality reduction that preserves the distance between samples. In these representations, each point corresponds to a sample instance, and the 2D distance between points is an approximation of the original Euclidean distance in a multidimensional space. In each row, the test datasets are represented by a cross symbol. The images appear in more organized and well-separated clusters for each class, indicating a good differentiation between images with different labels after the two fully connected layers of the model.
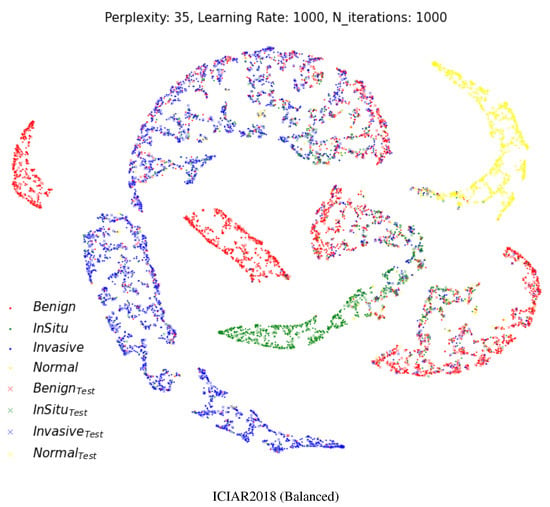
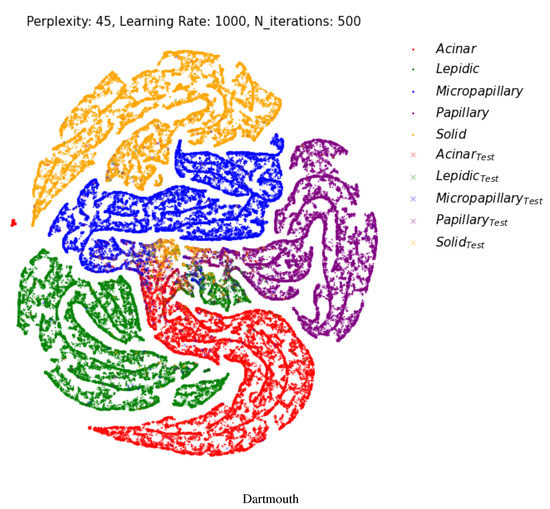
Figure 6.
Two-dimensional visualization of two different layers using t-SNE [52] for the multi-class classification. Projection of the last fully connected layer. Cross shapes represent test samples. (Best viewed in color).
The visualization result shows that our model makes a very accurate and meaningful classification even from a pathological point of view. In Figure 6, the ICIAR2018 (Balanced) dataset has benign lesions (red) of the breast with four clusters. That is why benign breast lesions are not a single disease entity. Benign breast lesions encompass adenosis, ductal hyperplasia, cystic change, columnar cell change, columnar cell hyperplasia, papilloma, fibroadenoma, even atypical ductal hyperplasia, etc. Benign lesions composed of microglandular adenosis could resemble invasive carcinoma, or benign lesions composed of atypical ductal hyperplasia could have a morphologic similarity to in situ carcinoma. Compared to the various spectrum of benign lesions, normal, in situ carcinoma and invasive carcinoma are relatively homogenous disease entities.
In the Dartmouth dataset, two small isolated clusters (green and yellow) separated from the main clusters were identified, which were lepidic and solid, as shown in Figure 6. A small lepidic cluster was present in between acinar and papillary clusters. There could be two reasons for this. First, tumor type sub-classification is decided based on the main component of the invasive carcinoma, usually more than 90% of one subtype. In the case of more than 90% of acinar-type carcinoma and less than 10% of lepidic-type carcinoma, the main classification could be the acinar type. A smaller proportion of the lepidic type is really present in acinar-type carcinoma. The other reason is the morphologic similarities of each tumor subtype. Some patches of acinar-type carcinoma or papillary carcinoma share a resemblance with the microscopic features of lepidic-type carcinoma.
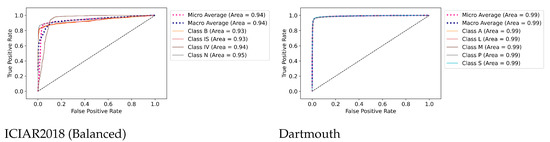
7.3. ROC Curves’ Visualizations
Figure 7 shows the AUC (ROC) results for the ICIAR and Dartmouth datasets. The AUCs of benign (B) and in situ (IS) of the ICIAR2018 dataset are 0.93, respectively, and the average is 0.94. The average AUC of the Dartmouth dataset is 0.99. This confirms that the proposed model can be used clinically for cancer diagnosis.
8. Conclusions
In this paper, we proposed an unsupervised deep learning model for WSI diagnosis using stacked autoencoders that learn and generalize different cellular heterogeneity representations from multiple-image descriptors, along with the original patches extracted from WSIs. Our model can use a set of N independent feature descriptors (i.e., currently, we used N = 3). We showed additional inputs help the model learn and classify multiple variants’ cellular information robustly on two publicly available WSI benchmark datasets. Our model outperformed existing state-of-the-art models by providing the best score for binary-class and multi-class classification using four metrics. The visualization results showed that the breast cancer and lung cancer classification are similar from the point of view of a pathologist.
The primary limitation of the model is deciding the size of patches that can learn more robust representations in the bottleneck space to separate the morphologically heterogeneous regions in sub-multi-level tumor types. We observed that this trend is due to the limited and unbalanced data, which affect the feature fusion stage. In future studies, we intend to make a more robust WSI processing toolbox to generate an accurate patch dataset for studying different feature descriptors to learn more about the WSI patch dataset. We will investigate the performance of the model on other feature descriptors that provide various variant comparisons and other datasets that provide diverse cancer cases. The proposed system can be adapted for diverse tasks associated with the domain of WSI-based diagnosis with relevance to clinical settings.
Author Contributions
Conceptualization, T.S.S. and M.C.; methodology, T.S.S.; software, T.S.S.; validation, T.S.S., J.-Y.K. and J.S.; formal analysis, T.S.S., J.-Y.K. and J.S.; investigation, J.-Y.K. and J.S.; resources, M.C.; data curation, T.S.S.; writing—original draft preparation, T.S.S. and M.C.; writing—review and editing, T.S.S., M.C., J.-Y.K. and J.S.; visualization, T.S.S. and J.-Y.K.; supervision, M.C., J.-Y.K. and J.S.; project administration, M.C.; funding acquisition, M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1A2B5B01001789).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not Applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shamshirband, S.; Fathi, M.; Dehzangi, A.; Chronopoulos, A.T.; Alinejad-Rokny, H. A review on deep learning approaches in healthcare systems: Taxonomies, challenges, and open issues. J. Biomed. Inform. 2021, 113, 103627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Y.; Wu, Q.; Zhao, P.; Niu, S.; Huang, J.; Tan, M. Collaborative unsupervised domain adaptation for medical image diagnosis. IEEE Trans. Image Process. 2020, 29, 7834–7844. [Google Scholar] [CrossRef]
- Lee, J.; Warner, E.; Shaikhouni, S.; Bitzer, M.; Kretzler, M.; Gipson, D.; Pennathur, S.; Bellovich, K.; Bhat, Z.; Gadegbeku, C.; et al. Unsupervised machine learning for identifying important visual features through bag-of-words using histopathology data from chronic kidney disease. Sci. Rep. 2022, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, H.; Li, X.; Yao, J. Deep contrastive learning based tissue clustering for annotation-free histopathology image analysis. Comput. Med. Imaging Graph. 2022, 97, 102053. [Google Scholar] [CrossRef] [PubMed]
- Chikontwe, P.; Kim, M.; Nam, S.J.; Go, H.; Park, S.H. Multiple instance learning with center embeddings for histopathology classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 519–528. [Google Scholar]
- Wang, X.; Han, T.X.; Yan, S. An HOG-LBP human detector with partial occlusion handling. In Proceedings of the 2009 IEEE 12th International Conference on Computer Vision, Kyoto, Japan, 27 September–4 October 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 32–39. [Google Scholar]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; IEEE: Piscataway, NJ, USA, 2005; Volume 1, pp. 886–893. [Google Scholar]
- Ahonen, T.; Hadid, A.; Pietikainen, M. Face description with local binary patterns: Application to face recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 2037–2041. [Google Scholar] [CrossRef]
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Ré, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef]
- Balazsi, M.; Blanco, P.; Zoroquiain, P.; Levine, M.D.; Burnier, M.N., Jr. Invasive ductal breast carcinoma detector that is robust to image magnification in whole digital slides. J. Med. Imaging 2016, 3, 027501. [Google Scholar] [CrossRef][Green Version]
- Barker, J.; Hoogi, A.; Depeursinge, A.; Rubin, D.L. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med. Image Anal. 2016, 30, 60–71. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using convolutional neural networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 2560–2567. [Google Scholar]
- Zhu, X.; Yao, J.; Zhu, F.; Huang, J. Wsisa: Making survival prediction from whole slide histopathological images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7234–7242. [Google Scholar]
- Whitney, J.; Corredor, G.; Janowczyk, A.; Ganesan, S.; Doyle, S.; Tomaszewski, J.; Feldman, M.; Gilmore, H.; Madabhushi, A. Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer 2018, 18, 610. [Google Scholar] [CrossRef]
- Bahlmann, C.; Patel, A.; Johnson, J.; Ni, J.; Chekkoury, A.; Khurd, P.; Kamen, A.; Grady, L.; Krupinski, E.; Graham, A.; et al. Automated detection of diagnostically relevant regions in H&E stained digital pathology slides. In Proceedings of the Medical Imaging 2012: Computer-Aided Diagnosis; San Diego, CA, USA, 4–9 February 2012, International Society for Optics and Photonics: Bellingham, WA, USA, 2012; Volume 8315, p. 831504. [Google Scholar]
- Bejnordi, B.E.; Litjens, G.; Hermsen, M.; Karssemeijer, N.; van der Laak, J.A. A multi-scale superpixel classification approach to the detection of regions of interest in whole slide histopathology images. In Proceedings of the Medical Imaging 2015: Digital Pathology, Orlando, FL, USA, 25–26 February 2015; International Society for Optics and Photonics: Bellingham, WA, USA, 2015; Volume 9420, p. 94200H. [Google Scholar]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van De Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Zuidhof, G.; Balkenhol, M.; Hermsen, M.; Bult, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J. Med. Imaging 2017, 4, 044504. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chen, H.; Dou, Q.; Wang, L.; Qin, J.; Heng, P.A. Scannet: A fast and dense scanning framework for metastastic breast cancer detection from whole-slide image. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 539–546. [Google Scholar]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A.; González, F. High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: Application to invasive breast cancer detection. PLoS ONE 2018, 13, e0196828. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. MB-AI-His: Histopathological diagnosis of pediatric medulloblastoma and its subtypes via AI. Diagnostics 2021, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.; Zaghlool, S. AI-Based Pipeline for Classifying Pediatric Medulloblastoma Using Histopathological and Textural Images. Life 2022, 12, 232. [Google Scholar] [CrossRef]
- Anwar, F.; Attallah, O.; Ghanem, N.; Ismail, M.A. Automatic breast cancer classification from histopathological images. In Proceedings of the 2019 International Conference on Advances in the Emerging Computing Technologies (AECT), Al Madinah Al Munawwarah, Saudi Arabia, 10 February 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Attallah, O.; Anwar, F.; Ghanem, N.M.; Ismail, M.A. Histo-CADx: Duo cascaded fusion stages for breast cancer diagnosis from histopathological images. PeerJ Comput. Sci. 2021, 7, e493. [Google Scholar] [CrossRef]
- Dundar, M.M.; Badve, S.; Bilgin, G.; Raykar, V.; Jain, R.; Sertel, O.; Gurcan, M.N. Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans. Biomed. Eng. 2011, 58, 1977–1984. [Google Scholar] [CrossRef]
- Sudharshan, P.; Petitjean, C.; Spanhol, F.; Oliveira, L.E.; Heutte, L.; Honeine, P. Multiple instance learning for histopathological breast cancer image classification. Expert Syst. Appl. 2019, 117, 103–111. [Google Scholar] [CrossRef]
- Mercan, C.; Aksoy, S.; Mercan, E.; Shapiro, L.G.; Weaver, D.L.; Elmore, J.G. Multi-instance multi-label learning for multi-class classification of whole slide breast histopathology images. IEEE Trans. Med. Imaging 2017, 37, 316–325. [Google Scholar] [CrossRef]
- Xia, Y.; Nie, L.; Zhang, L.; Yang, Y.; Hong, R.; Li, X. Weakly supervised multilabel clustering and its applications in computer vision. IEEE Trans. Cybern. 2016, 46, 3220–3232. [Google Scholar] [CrossRef]
- Doyle, S.; Feldman, M.; Tomaszewski, J.; Madabhushi, A. A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans. Biomed. Eng. 2010, 59, 1205–1218. [Google Scholar] [CrossRef]
- Basavanhally, A.; Ganesan, S.; Feldman, M.; Shih, N.; Mies, C.; Tomaszewski, J.; Madabhushi, A. Multi-field-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Trans. Biomed. Eng. 2013, 60, 2089–2099. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gadepalli, K.; Norouzi, M.; Dahl, G.E.; Kohlberger, T.; Boyko, A.; Venugopalan, S.; Timofeev, A.; Nelson, P.Q.; Corrado, G.S.; et al. Detecting cancer metastases on gigapixel pathology images. arXiv 2017, arXiv:1703.02442. [Google Scholar]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-based convolutional neural network for whole slide tissue image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar]
- Mahmood, T.; Kim, S.G.; Koo, J.H.; Park, K.R. Artificial Intelligence-Based Tissue Phenotyping in Colorectal Cancer Histopathology Using Visual and Semantic Features Aggregation. Mathematics 2022, 10, 1909. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Vega, J.E.V.; Brat, D.J.; Cooper, L.A. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef]
- Guo, W.; Liang, W.; Deng, Q.; Zou, X. A Multimodal Affinity Fusion Network for Predicting the Survival of Breast Cancer Patients. Front. Genet. 2021, 1323. [Google Scholar] [CrossRef]
- Tong, L.; Sha, Y.; Wang, M.D. Improving classification of breast cancer by utilizing the image pyramids of whole-slide imaging and multi-scale convolutional neural networks. In Proceedings of the 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC), Milwaukee, WI, USA, 15–19 July 2019; IEEE: Piscataway, NJ, USA, 2019; Volume 1, pp. 696–703. [Google Scholar]
- Vincent, P.; Larochelle, H.; Bengio, Y.; Manzagol, P.A. Extracting and composing robust features with denoising autoencoders. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008; pp. 1096–1103. [Google Scholar]
- Roy, A.G.; Conjeti, S.; Carlier, S.G.; Houissa, K.; König, A.; Dutta, P.K.; Laine, A.F.; Navab, N.; Katouzian, A.; Sheet, D. Multiscale distribution preserving autoencoders for plaque detection in intravascular optical coherence tomography. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1359–1362. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press Cambridge: Cambridge, MA, USA, 2016; Volume 1. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. Bach: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Wetteland, R.; Engan, K.; Eftestøl, T.; Kvikstad, V.; Janssen, E.A. Multiclass Tissue Classification of Whole-Slide Histological Images using Convolutional Neural Networks. ICPRAM 2019, 1, 320–327. [Google Scholar]
- Toğaçar, M.; Özkurt, K.B.; Ergen, B.; Cömert, Z. BreastNet: A novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phys. A Stat. Mech. Appl. 2020, 545, 123592. [Google Scholar] [CrossRef]
- Aatresh, A.A.; Alabhya, K.; Lal, S.; Kini, J.; Saxena, P. LiverNet: Efficient and robust deep learning model for automatic diagnosis of sub-types of liver hepatocellular carcinoma cancer from H&E stained liver histopathology images. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1549–1563. [Google Scholar]
- Burçak, K.C.; Baykan, Ö.K.; Uğuz, H. A new deep convolutional neural network model for classifying breast cancer histopathological images and the hyperparameter optimisation of the proposed model. J. Supercomput. 2021, 77, 973–989. [Google Scholar] [CrossRef]
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).