1. Introduction
A chest X-ray (CXR), which is generated by exposing the chest to a small dose of ionizing radiation, is a projection radiograph of the chest used for imaging subtle lesions and the density of human tissues. It is commonly used for visualizing the condition of the thoracic cage, chest cavity, lung tissue, mediastinum, and heart. It can thus facilitate the diagnosis of common thorax diseases, including aortic sclerosis or calcification, arterial curvature, abnormal lung fields, anomalous lung patterns, spinal lesions, intercostal pleural thickening, and cardiac hypertrophy.
Computer vision technology and hardware computing capabilities have progressed considerably. Considering the overload of medical resources and the high demand for medical image analysis, the development of computer-aided diagnosis systems with a high diagnostic efficiency and accuracy is warranted. CXR images containing large amounts of physiological data can aid data-hungry deep learning paradigms in the construction of valuable intelligent auxiliary systems. Deep learning is a branch of machine learning; through linear or non-linear transform from multiple layers, deep learning can automatically extract sufficient and representative features from a data set. In traditional machine learning, features are usually extracted using handcrafted rules, which are created by relevant domain experts. After the data characteristics are understood, useful and effective features can be produced. However, the ability to automatically extract features from deep learning can reduce the time spent by experts in feature engineering. Therefore, deep learning may afford an excellent performance in applications where machines may have failed in the past.
Several studies have used deep learning for CXR analysis, particularly for image classification. Most of these studies have trained deep learning models by using well-designed convolutional neural network (CNN) architectures such as VGG [
1], GoogleNet [
2], ResNet [
3], and DenseNet [
4]. CNN architecture depth, data augmentation, input preprocessing methods, image size, and pretraining schemes can affect the performance of a deep learning model [
5]. No standardized design methodology for improving deep learning model performance has been reported thus far. Most of the relevant studies have focused on comparing the performance of multiple design methodologies for a specific task, rather than reporting novel methodologies [
6,
7,
8]. Some studies have achieved methodological novelty by utilizing methods that can aid in improving model performance. For example, Hu et al. [
9] and Wu et al. [
10] used an extreme learning machine (ELM) to replace the conventional fully connected layer in a deep CNN for real-time analysis and applied a Chimp optimization algorithm or sine-cosine algorithm to ameliorate the ELM’s ill-conditioning and nonoptimal problems. Wang et al. [
11] trained CNN models by using the whale optimization algorithm, which can resolve difficulties related to requiring a considerable amount of manual parameter tuning and parallelizing the training process of traditional gradient descent-based approaches. Khishe et al. [
12] proposed an efficient biogeography-based optimization approach for automatically finetuning model hyperparameters (e.g., number of output channels, convolution kernel size, layer type, training algorithm’s learning rate, epoch, and batch size), which are typically selected manually. CXR images are commonly classified as normal or abnormal in the literature [
7]. Although CXR can be used to detect multiple diseases of the thorax, few methods have been proposed for classifying multiple disease labels [
13].
Applying deep learning methods to CXR image analysis may have promising applications. However, the advancement of automatic image analysis in hindered by several underlying limitations. The main limitation is the lack of large-scale CXR datasets. Although the number of parameters required for deep learning models is considerably large, CXR image training data are limited; this can cause model overfitting. Compared with ordinary images, collecting and labeling CXR images can be difficult and cost intensive. CXR images generated using various instruments with different settings and in different environments cannot be naively analyzed together because this can lead to various errors. Transfer learning [
14], which uses the knowledge learned from one task as a starting point for related tasks, can aid in making the best use of different CXR databases. Transfer learning mainly involves using a large amount of open-source data to make up for target data shortages so as to achieve improved performance in model fitting.
In this study, we used one private target data set—1630 chest radiographs provided by the E-Da Hospital, I-Shou University, Taiwan—and three open-source datasets—the ImageNet dataset [
15], ChestX-ray dataset [
16,
17] (with >100,000 chest radiograph images provided by the National Institutes of Health (NIH)), and CheXpert dataset [
18] (collected by the Stanford ML Group, comprising nearly 220,000 chest radiographs). The images from the private target data set were labeled by radiology specialists as either being disease-free or containing multiple sites of abnormalities—representing a typical multilabel classification problem. Although ImageNet is sufficiently large for deep learning, most of its 14 million images were dissimilar to those in our private data set; therefore, this data set may not have been able to provide accurate feature representations to classify images in our private data set. ChestX-ray and CheXpert, although modest in size, are more similar to our private data set, and thus, they may be able to support target data training more efficiently.
Transfer learning has been widely applied in CXR deep learning analysis. Most studies have first trained deep CNN models on the large ImageNet dataset for natural image classification, followed by the use of trained weights for initialization to retrain all layers or only retrain the final (fully connected) layer for target CXR image classification [
8]. The performance of transfer learning might be affected by several factors such as the sizes of the source and target data set, similarity between these data sets, retraining of all or partial layers in the target task, and CNN architecture [
19]. For natural image classification, Azizpour et al. [
19] and Cui et al. [
20] have reported best practices on how these factors should be set in generic and domain-specific tasks, respectively. However, only a few studies have focused on the factors that affect the transferability of medical image analysis approaches. Tajbakhsh et al. [
21] considered four different medical imaging applications and demonstrated that CNNs pretrained on ImageNet performed better and were more robust to the size of the target training data than the CNNs trained from scratch. Gozes and Greenspan [
22] pretrained their model on the ChestX-ray data set and demonstrated that the pretrained weights enabled the model to exhibit improved predictions on small-scale CXR data sets compared with the performance of a model pretrained on ImageNet.
To maximize the performance of transfer learning for CXR image classification, the effects of different transfer learning characteristics in medical image analysis must be systematically investigated. Because of the substantial differences between natural and medical images, we could not apply the knowledge learned for natural images in previous studies [
19,
20] to our current CXR analysis. Accordingly, our study focused on the multilabel classification of CXR images, which is a crucial topic that warrants research. We thoroughly investigated the aforementioned transferability factors and included the ImageNet, ChestX-ray, and CheXpert data sets as the source data. Previous studies have typically used one type of source data at a time for transfer learning. In this study, we developed new approaches for integrating different source data sets practically to eventually obtain novel powerful source data sets. Our results may aid in devising best practices for the efficient use of different types of data sets to alleviate the insufficiency of training data and enhance the performance of deep learning models in the medical field.
3. Methods
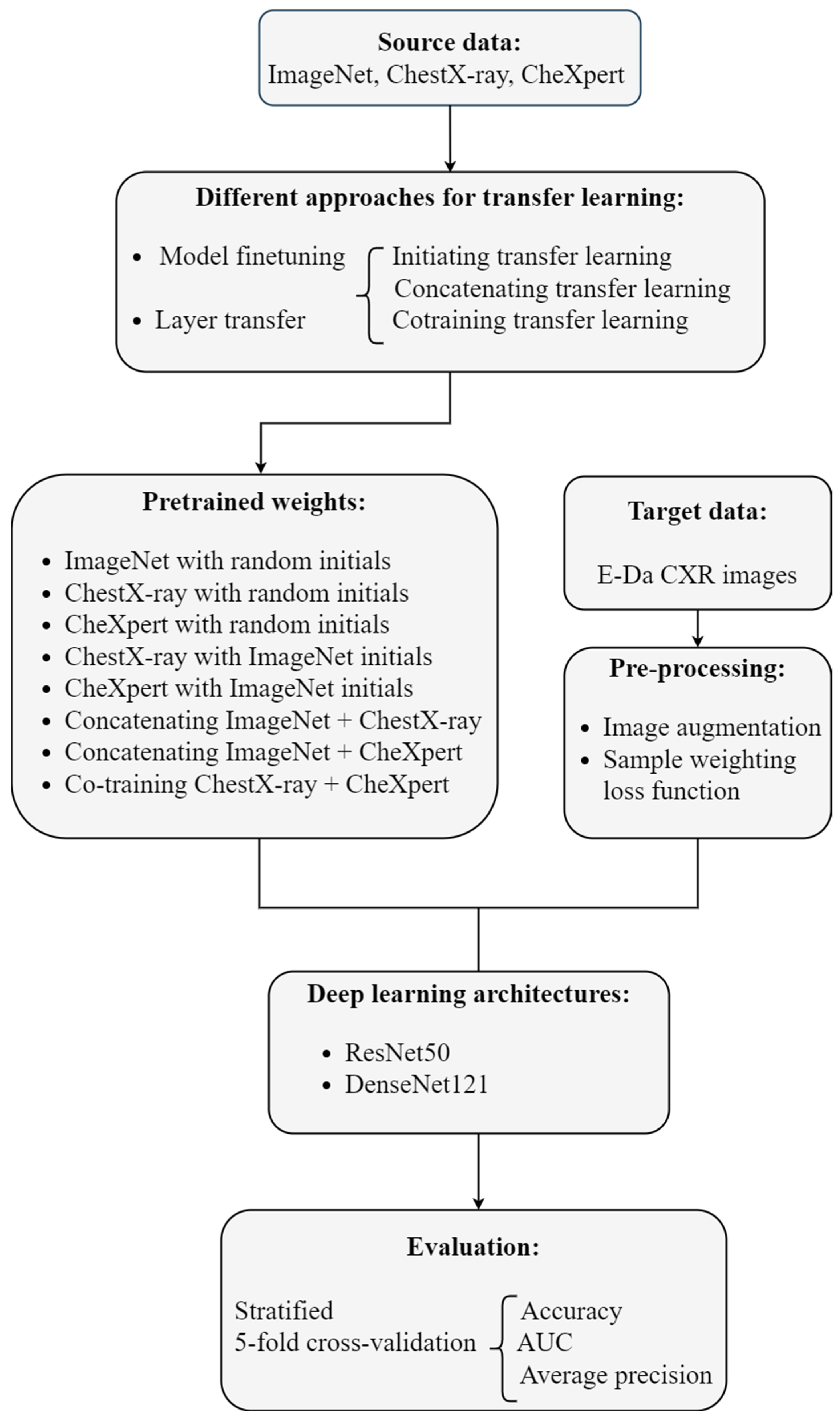
This analysis aimed to predict common thorax diseases in patients through their CXR images. The images were labeled as being disease-free or containing multiple sites of abnormalities, representing a typical multilabel classification problem. Deep learning architectures were used to automatically extract features and build a classifier, and transfer learning was conducted to combine information from different datasets to improve model performance. We trained deep learning models using the well-designed convolutional neural network (CNN) architectures ResNet50 [
3] and DenseNet121 [
4]. Implementing transfer learning in the CNN involved reusing the first several layers of the network for classifying images in the open-source data sets ImageNet, CheXpert, and ChestX-ray (the source task). Pretrained weights from these layers served as the starting points for classifying the CXR images from E-Da Hospital (the target task). By combining three source data sets in various manners, we obtained several different sets of pretrained weights. To transfer the pretrained weights to the target task, we either used the pretrained weights as the initial values and retrained the model from the scratch or fixed the weights of some early layers at the corresponding pretrained weights and reconstructed the others.
Image augmentation techniques were applied to artificially create variations in the existing images; this expanded the training data set to represent a comprehensive set of possible images. Our target dataset was imbalanced: the number of images containing each disease label was unequal. Deep learning algorithms can be biased toward the majority labels and fail to detect the minority labels. To prevent imbalanced data, sample weighting in the loss function was used. Model performance was evaluated using various metrics and stratified five-fold cross-validation. Our analytic approach is illustrated in
Figure 1. The programming language Python [
23] was used to implement these methods.
3.1. Multilabel Classification
Image classification aims at building a model that maps the input of the th image to a label vector , where is the indicator for the th disease class. Multiclass classification involves classifying an image into one of multiple classes; in other words, the label vector of multiclass classification has only a single element equal to 1. However, in multilabel classification, more than one class may be assigned to the image, with the label vector possibly having 0 or 1 in each element.
CXR images in our target data set were labeled for seven diseases, and multiple diseases were often identified in one image. The target task was multilabel classification and defined a seven-dimensional label vector with an all-zero vector representing a normal status (where none of the seven diseases was detected). We treated multilabel classification as a multiple binary classification problem; thus, the loss function was the sum of multiple binary cross-entropies. However, our data were seriously imbalanced: the elements of the label vectors were almost equal to 0. The number of 0 s was much larger than that of 1 s; this could have misled our model toward predicting 0 s if the usual loss function was used. Therefore, we adjusted the common binary cross-entropy with weights that considered the proportions of 0 s and 1 s in the same sampling batch.
Suppose that the entire training set is divided into
batches, with each batch size being
. Let
be the input for the
th image in the
th batch and
be its label vector. Then, the proposed weighted binary cross-entropy (WBCE) loss is defined as:
where
is
’s
th input for the final fully-connected layer,
is the sigmoid function, and
is set to
, whereas
is set to
, with
and
being the number of 1 s and 0 s in the label vectors of the
th batch, respectively.
3.2. Image Augmentation
Training deep CNNs requires a considerable amount of data, but the sample size of medical images is typically not sufficiently large. Image augmentation is a powerful technique for generating images using a combination of affine transformations—such as shift, zoom in-zoom out, rotate, flip, distort, and shade with a hue—which can feed more images into the neural networks and exploit information in the original data more fully. We augmented our training images by using the ImageDataGenerator function in the Keras Python library. “Online” augmentation was applied; here, we applied the image augmentation techniques in mini-batches and then fed them to the model. The model with online augmentation was presented with different images at each epoch; this aided the model in generalizing, and the model did not need to save the augmented images on the disk, which reduced the computing burden.
3.3. Deep CNNs
Image classification using deep CNNs has an outstanding performance compared with traditional machine learning approaches. CNN is a deep learning method in which a series of layers is constructed to progressively extract higher-level features from the raw input. In a CNN, the typical layers comprise the input layer (which imports image data for model training), the convolution layer (which uses various filters to automatically learn representation of features at different levels), the pooling layer (which selects the most prominent features to reduce the dimension of subsequent layers), and the fully connected layer (which flattens the matrix feature maps into a single vector for label prediction).
The CNN architecture refers to the overall structure of the network: the types of layers it should have, the number of units each layer type should contain, and the manner in which these units should be connected to each other. CNN architectures such as VGG [
1], GoogleNet [
2], ResNet [
3], and DenseNet [
4] have been widely used. We included two high-performing CNN models: 50-layer ResNet (ResNet50) and 121-layer DenseNet (DenseNet121). ResNet50 has fewer filters and a lower complexity than VGG nets, and DenseNet121 requires fewer parameters than ResNet50 does.
3.3.1. ResNet
In theory, the deeper the network model, the better its results. Nevertheless, the degradation problem may arise: as the network becomes deeper, the model’s accuracy may become saturated or even decrease. This problem is different from overfitting because of increased training errors. ResNet made a historical breakthrough in deep neural networks by solving the degradation problem through residual learning.
In this study, we established a deeper model by stacking new layers on a shallower architecture. Let denote the output of the shallow part of the model and denote the output of the deeper model. No higher training error should be obtained if the added layers are for identity mapping. Rather than expecting the stack of layers to learn identity mapping, ResNet argues that it is easier to let these layers fit a residual mapping of to zero and recast the output of the deeper model as , where is the fitted residual. Generally, will not be zero; therefore, the stacking layers can still learn new features and demonstrate an improved performance.
The formulation of
can be realized using feedforward neural networks with a shortcut connection, which skips some of the layers in the neural network and feeds the output of one layer as the input to the next layers. A series of shortcut connections (residual blocks) forms ResNet. This study applied the deeper ResNet50 that contained 50 layers and used a stack of three layers with
,
, and
convolutions as the building residual block. This three-layer residual block adopted a bottleneck design to improve computational efficiency, where the
layers were responsible for reducing and then increasing (restoring) the dimensions, leaving the
layer as a bottleneck with smaller input and output dimensions [
3]. In ResNet50, batch normalization (BN) [
24] is adopted immediately after each convolution and before ReLU activation, and global average pooling (GAP) [
25] is performed to form the final fully connected layer.
3.3.2. DenseNet
As CNNs become increasingly deep, information about the input passes through many layers and can vanish by the time it reaches the end of the network. Different approaches, which vary in network topology and training procedure, create short paths from early layers to later layers to address this problem. DenseNet proposes an architecture that distills this insight into a simple connectivity pattern of dense blocks and transition layers. A dense block is a module containing many layers connected densely with feature maps of the same size. In a dense block, each layer obtains additional inputs from all preceding layers, and it passes on its own feature maps to all the subsequent layers. The transition layer connects two adjacent dense blocks, and it reduces the size of the feature map through pooling. Compared with ResNet that connects layers through element-level addition, layers in DenseNet are connected by concatenating them at the channel level.
In a dense block, the convolution for each layer produces feature maps, which denote channels in the output. is a hyperparameter known as the growth rate of DenseNet, which is usually set to be small. Under the assumption that the initial number of channels is , the number of input channels in the th layer is . As the number of layers increases, the input can be extremely large even if is small. Because the input of the latter layer grows quickly, the bottleneck design is introduced into the dense block to reduce the burden of calculation, where a convolution and then a convolution are applied to each layer to generate the output. Similar to ResNet, DenseNet uses a composite of three consecutive operations for each convolution: .
3.3.3. Implementation
We used the Python package Keras to implement ResNet50 and DenseNet121. Optimizer Adam [
26] with a mini-batch size of 16 and an epoch number of 30 was used. The learning rate started from 0.0001 and was divided by 10 when the validation loss did not decrease in 10 epochs. DenseNet121 set the growth rate to
.
3.4. Transfer Learning
In practice, we usually do not have sufficient data to train a deep and complex network such as ResNet or DenseNet. Techniques such as image argumentation are insufficient for resolving this problem. The lack of data can cause overfitting, which can make our trained model overly rely on a particular data set and then fail to fit to additional data. This problem can be resolved using transfer learning [
14]. The main concepts of transfer learning are to first train the network with sufficiently large source data and then transfer the structure and weights from this trained network to predict the target data.
In the current study, we used the data from the ImageNet [
15], ChestX-ray [
16,
17], and CheXpert [
18] data sets as the source data and the CXR images from the E-Da Hospital as the target data. ImageNet contains more than 14 million natural images and thus is sufficiently large for a deep learning application; however, it does not contain images with similarity to medical images, and therefore, it may fail to provide useful feature representations to classify our target data. ChestX-ray and CheXpert—consisting of about 100,000 and 220,000 CXR images, respectively—are modest in size but are more similar to our target data set.
Implementing transfer learning in the CNN involves reusing the parameter estimators pretrained on the source data when fitting the target data. Model finetuning is a method in which these pretrained parameter estimators are used as the initial values and finetuned to fit the target data. In the layer transfer approach, target data fitting preserves some of the layers from a pretrained model and reconstructs the others. We thus adopted model finetuning or layer transfer to reuse the parameter estimators pretrained on the source data (from ImageNet, ChestX-ray, or CheXpert).
We next tried to combine these source data sets to obtain new powerful source data sets for transfer learning.
3.4.1. Initiating Transfer Learning
In Keras, we can implement transfer learning with initial weights selected randomly or from ImageNet pretraining. ImageNet pretraining may result in robust parameter estimation due to the diversity and richness of the data set, and the pretraining of the similar data sets ChestX-ray and CheXpert may accelerate the convergence of the model. Therefore, we adopted five different pretrained weights in modeling our target data: ImageNet pretraining with randomly selected initials, ChestX-ray pretraining with random or ImageNet pretraining initials, and CheXpert pretraining with random or ImageNet pretraining initials.
3.4.2. Concatenating Transfer Learning
In this approach, we first constructed two ResNet backbone models by using different pretrained weights: one from ImageNet pretraining with random initials (IR) and the other from ChestX-ray pretraining with random initials (CR). Thereafter, we concatenated the outputs of the final convolution layer from two models and then used the GAP to reduce the dimension, and finally applied the fully connected layer to generate the final prediction (
Figure 2). The ResNet model concatenating IR and CheXpert pretraining with random initials (XR) was obtained in a similar manner. The DenseNet backbone models concatenating IR and CR and concatenating IR and XR could also be obtained. Models using different pretrained weights may extract different features. In contrast to the first approach aimed at extracting features from a single domain, this approach could expand features from two different domains. However, compared with the first approach, this approach used twice the memory and time to store and upgrade parameters.
3.4.3. Co-Training Transfer Learning
In this approach, we aimed to combine two source data sets, namely ChestX-ray and CheXpert, to a larger CXR data set to serve as source data for medical image classification tasks. ChestX-ray and CheXpert cannot be combined directly: although both the data sets contain only CXR images, they use different class definitions. To resolve this issue, we applied a co-training approach. First, we fed images from two source data sets to jointly train convolutional layers but to also connect to different fully connected layers to predict their corresponding classes. In other words, they shared the parameters from the convolution layers but not those after the convolution layers. Finally, we reserved these shared convolutional weights as the pretrained weights in target data model fitting. The reason that we did not cotrain ImageNet with ChestX-ray (or CheXpert) was that merging different domain data may mislead the model and reduce its efficiency. The co-training approach is illustrated in
Figure 3.
3.5. Evaluation
3.5.1. Stratified -Fold Cross-Validation
In -fold cross-validation, first, the data are shuffled to ensure the randomness when the data are split. Second, whole data are split into groups, where is usually set to be 5 or 10 based on the sample size. Third, one of groups is considered “test data,” and the others are considered “training data” with a total of different test-training combinations ( cross-validation rounds). Finally, the “training data” are further divided into “training” and “validation” sets, which are used to train the model parameters and to instantly evaluate the performance of various hyper-parameters, respectively.
In stratified
-fold cross-validation, every group must have the same class distribution. This method has great applicability for multiclass classification, where each sample belongs to only one class. In a multilabel task, every sample may have multiple class categories. To appropriately perform stratified
-fold cross-validation for a multilabel classification task, performing iterative stratification is essential [
27]; it can be implemented using Python’s scikit-learn-compatible package MultilabelStratifiedKFold.
3.5.2. Metrics
For an imbalanced data set, good accuracy does not indicate that a classifier is reliable. A receiver operating characteristic (ROC) curve is a graphical plot that illustrates the performance of a binary classifier (with vs. without the label) at different thresholds, where the x-axis represents the false positive rate, and the y-axis represents the true positive rate. The area under the ROC curve (AUC) is used to summarize the ROC curve. A precision (PR) curve is another type of plot that evaluates the performance of a model, with the x-axis being the recall and the y-axis being the precision. The area under the PR curve is the average precision (AP). Moreover, training accuracy represents the average accuracy of the training sets during the five cross-validation rounds. Test accuracy, AUC, and AP refer to the accuracy, AUC, and AP of the five test groups, respectively. In multilabel classification, we can treat the problem as a combination of multiple binary classifications; thus, we can evaluate every label individually with binary metrics and obtain the average of these binary metrics (i.e., the mean metric). In this study, we calculated the training accuracy, test accuracy, test AUC, and test AP for each individual label and the mean training accuracy, test accuracy, test AUC, and test AP for all labels.
5. Discussion
When reusing pretrained weights in transfer learning, the approach that re-trains the whole model with pretrained weights as initial values (i.e., the model finetuning approach) typically afforded excellent results but required many more computational resources compared with the other approaches. The layer transfer approach, which freezes some layers on pretrained weights, demonstrated advantages over the model finetuning approach by allowing larger batch sizes and requiring a shorter run time and less GPU/CPU memory. For building the most cost-effective model by using the layer transfer approach, the number of frozen layers should be determined accurately. Our results demonstrated that the higher the similarity between the source and target data, the larger the allowed number of frozen layers should be. However, none of the included approaches were universally applicable; selecting the most appropriate approach would require the assessment of its benefits and costs on the basis of specific goals and available resources.
When adopting only one single-source dataset, ChestX-ray demonstrated a better performance than CheXpert. ChestX-ray is only half the size of CheXpert; nonetheless, its accurate labeling was found to make up for its smaller size. However, after ImageNet initials were attached, CheXpert outperformed ChestX-ray. As such, ImageNet appeared to enhance the data volume and variety of these data sets.
ResNet50 was suitable for initiating transfer learning, whereas DenseNet121 performed better in concatenating and co-training transfer learning. Transfer learning combined with various source data sets was also preferable with the use of a single-source data set; however, DenseNet121 led to greater benefits than ResNet50 did. Compound weights from several source data sets may be superior to single weights because they contain additional information offered by another data set. Nevertheless, a more complex transfer process may produce more noise. Compared with ResNet50, DenseNet121 was more complex, and its dense block mechanism could process more data and absorb more information; consequently, DenseNet121 is more suitable than BesNet50 for integrating source data sets.
Few studies have focused on the factors that affect the performance of transfer learning in medical image analysis. The strength of the present study lies in its systematic approach to investigating the transferability of CXR image analytic approaches. Nevertheless, this study had several limitations that should be resolved in future studies. First, our experiments were based on the 50-layer ResNet and 121-layer DenseNet architectures, and the derived weights were estimated using the gradient descent-based optimizer Adam [
26] under the consideration of manually selected hyperparameter values. ResNet and DenseNet are both widely used for conducting deep learning analyses on CXR images; nevertheless, they differ in several aspects, and such differences can be leveraged to investigate the impact of the CNN architecture on transfer learning. Alternatively, new architectures—such as EfficientNet [
28] and CoAtNet [
29], which have shown a high performance in challenging computer vision tasks—could be used for analysis. Moreover, to enhance the efficiency of model parameter estimation, Adam may be replaced with recent optimizers such as Chimp [
9] and Whale [
11], and biogeography-based optimization can be applied to automatically finetune model hyperparameters [
12]. Second, CXR images can be taken in posteroanterior, anteroposterior, and lateral views. In this study, we included only one target data set in which all CXR images were in the posteroanterior view; this may have limited the real-world applicability of our findings. Moreover, we included only image classification as the target task. However, CXR images can be used for several other types of deep learning tasks such as segmentation, localization, and image generation [
5]. Accordingly, future studies could use CXR images taken in different positions and could consider a wide range of deep learning tasks.