Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Fundus Examination
2.3. Grading of Fundoscopy Images and Subject Exclusion
2.4. Coronary Computed Tomography Angiography (CCTA) Acquisition and Examination
2.5. CAD-RADS Score
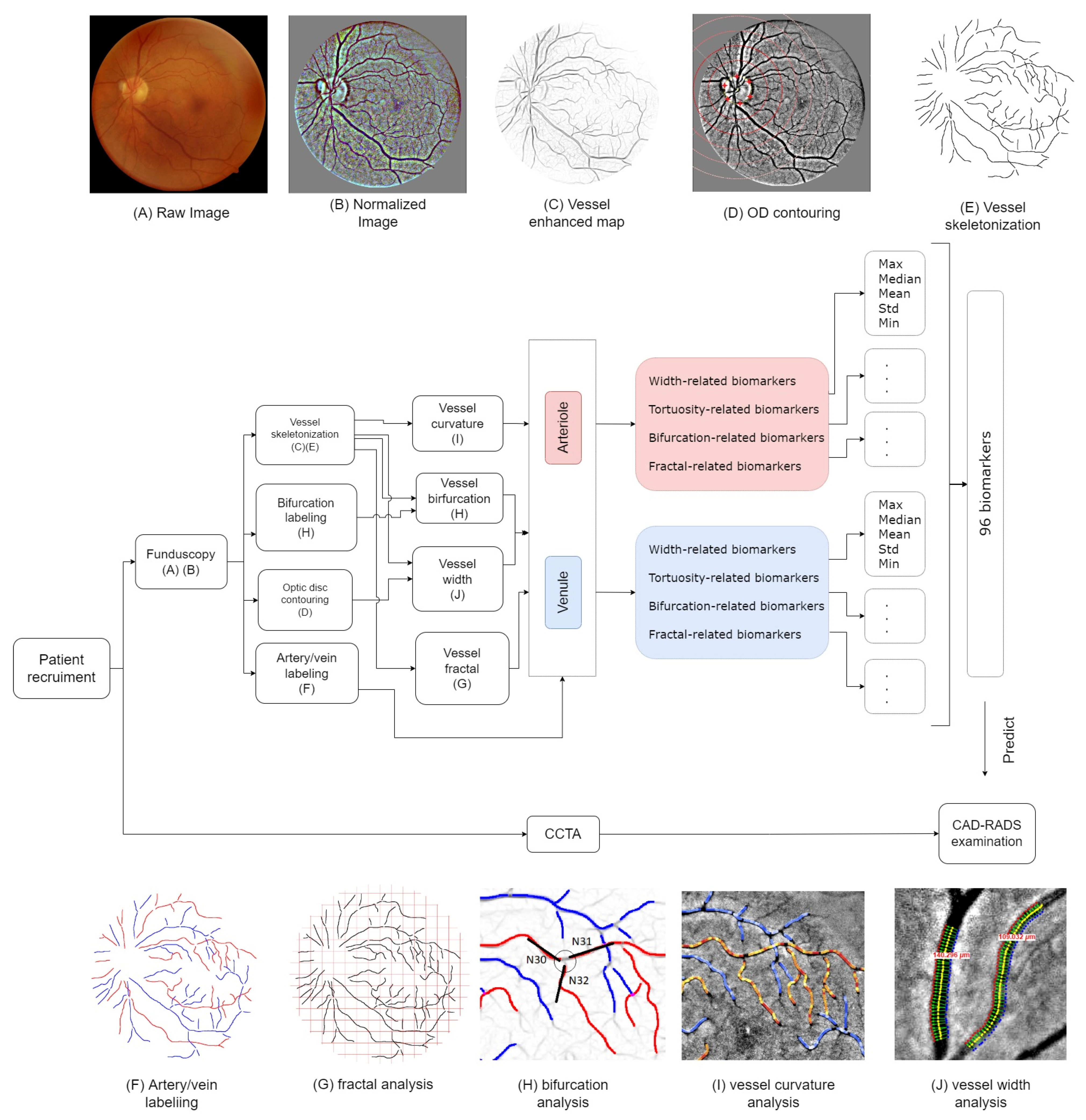
2.6. Fundus Biomarkers
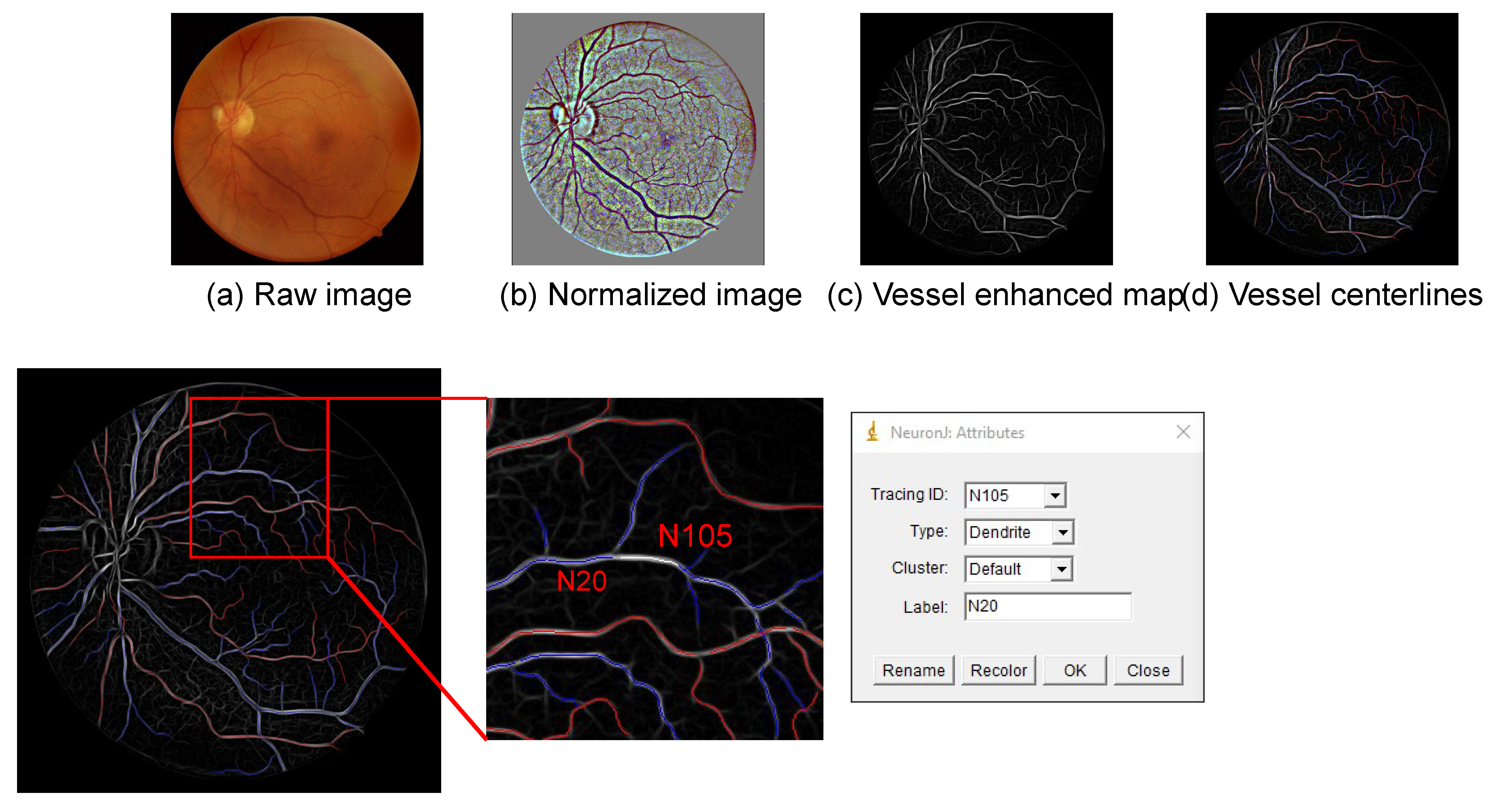
2.7. Vessel Manual Modelling
2.8. Optic Disc Labelling
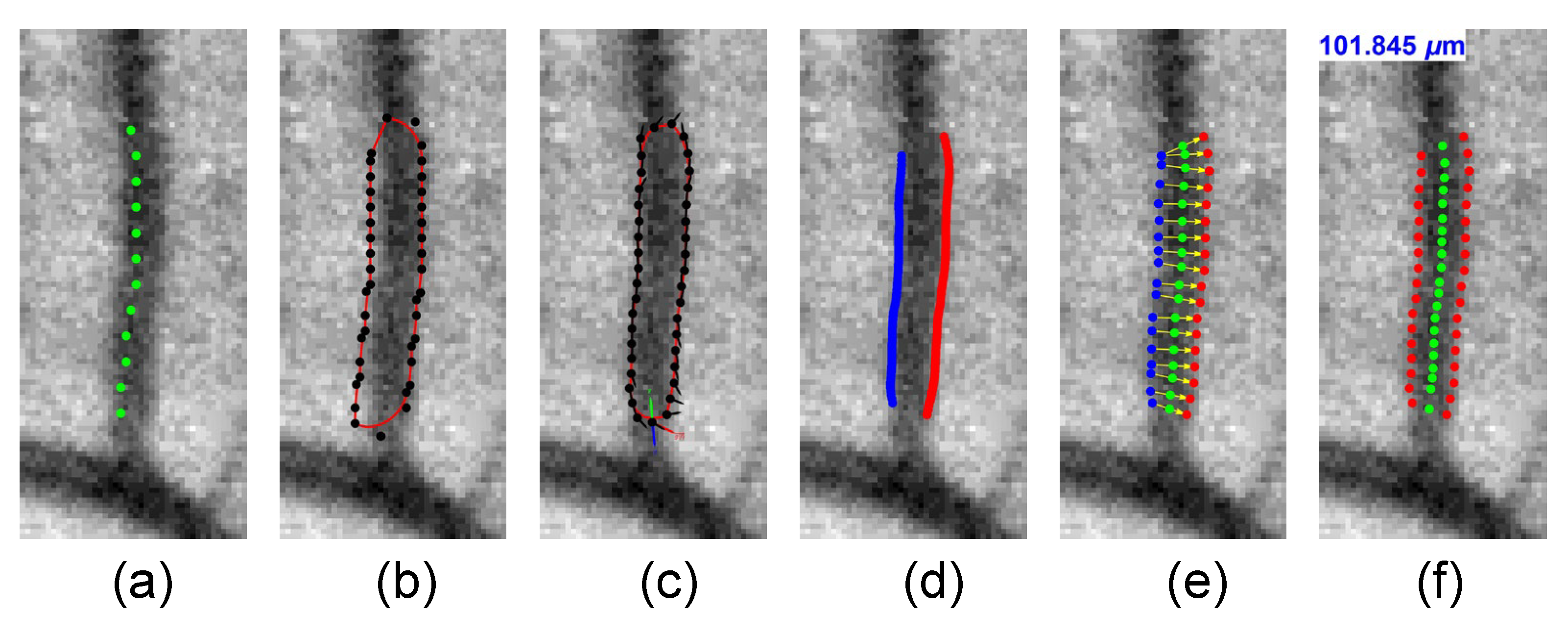
2.9. Vessel Width
2.10. Vessel Tortuosity
2.11. Bifurcation Junction Parameters
2.12. Vessel Fractal Dimensions
2.13. The GraphSAGE Model
2.14. Traditional Machine Learning Models
2.15. Feature Selection and Dimensionality Reduction
2.16. Statistical Study
3. Results
3.1. Association Analysis of CAD-RADS Scores with Patient Characteristics, Retinal Diseases, and Quantitative Vascular Biomarkers
3.2. GNN and Traditional Machine Learning Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix B
| Layers | Input Features | Output Features | Parameters |
|---|---|---|---|
| Input = G | 96 | - | - |
| SAGEConv | 96 | 128 | Aggregator = mean |
| ReLU | - | - | - |
| Dropout layers | - | - | Probability = 0.5 |
| SAGEConv | 128 | 2 | Aggregator = mean |
| Softmax layer | 2 | 2 | - |
| Loss | - | - | Cross-entropy loss |
References
- Marcus, M.L.; Chilian, W.M.; Kanatsuka, H.; Dellsperger, K.C.; Eastham, C.L.; Lamping, K.G. Understanding the coronary circulation through studies at the microvas cular level. Circulation 1990, 82, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Sharrett, A.R.; Hubbard, L.D.; Cooper, L.S.; Sorlie, P.D.; Brothers, R.J.; Nieto, F.J.; Pinsky, J.L.; Klein, R. Retinal arteriolar diameters and elevated blood pressure: The Atherosc lerosis Risk in Communities Study. Am. J. Epidemiol. 1999, 150, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wong, T.Y.; Sharrett, A.R.; Klein, R.; Folsom, A.R.; Jerosch-Herold, M. Relationship between retinal arteriolar narrowing and myocardial perfu sion: Multi-ethnic study of atherosclerosis. Hypertension 2008, 51, 119–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillum, R.F. Retinal arteriolar findings and coronary heart disease. Am. Heart J. 1991, 122, 262–263. [Google Scholar] [CrossRef]
- Wong, T.Y.; Klein, R.; Klein, B.E.; Tielsch, J.M.; Hubbard, L.; Nieto, F.J. Retinal microvascular abnormalities and their relationship with hypertension, cardiovascular disease, and mortality. Surv. Ophthalmol. 2001, 46, 59–80. [Google Scholar] [CrossRef]
- Tedeschi-Reiner, E.; Strozzi, M.; Skoric, B.; Reiner, Z. Relation of atherosclerotic changes in retinal arteries to the extent of coronary artery disease. Am. J. Cardiol. 2005, 96, 1107–1109. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Garvin, M.K.; Sonka, M. Retinal imaging and image analysis. IEEE Rev. Biomed. Eng. 2010, 3, 169–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, R.; Sharrett, A.R.; Klein, B.E.; Chambless, L.E.; Cooper, L.S.; Hubbard, L.D.; Evans, G. Are retinal arteriolar abnormalities related to atherosclerosis? The A therosclerosis Risk in Communities Study. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1644–1650. [Google Scholar] [CrossRef] [Green Version]
- Hubbard, L.D.; Brothers, R.J.; King, W.N.; Clegg, L.X.; Klein, R.; Cooper, L.S.; Sharrett, A.R.; Davis, M.D.; Cai, J. Methods for evaluation of retinal microvascular abnormalities associat ed with hypertension/sclerosis in the Atherosclerosis Risk in Communit ies Study. Ophthalmology 1999, 106, 2269–2280. [Google Scholar] [CrossRef]
- Ikram, M.K.; de Jong, F.J.; Vingerling, J.R.; Witteman, J.C.; Hofman, A.; Breteler, M.M.; de Jong, P.T. Are retinal arteriolar or venular diameters associated with markers fo r cardiovascular disorders? The Rotterdam Study. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2129–2134. [Google Scholar] [CrossRef]
- Lyu, M.; Lee, Y.; Kim, B.S.; Kim, H.; Hong, R.; Shin, Y.U.; Cho, H.; Shin, J. Clinical significance of subclinical atherosclerosis in retinal vein o cclusion. Sci. Rep. 2021, 11, 11905. [Google Scholar] [CrossRef]
- Wong, T.Y.; Cheung, N.; Islam, F.M.; Klein, R.; Criqui, M.H.; Cotch, M.F.; Carr, J.J.; Klein, B.E.; Sharrett, A.R. Relation of retinopathy to coronary artery calcification: The multi-ethnic study of atherosclerosis. Am. J. Epidemiol. 2008, 167, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cury, R.C.; Abbara, S.; Achenbach, S.; Agatston, A.; Berman, D.S.; Budoff, M.J.; Dill, K.E.; Jacobs, J.E.; Maroules, C.D.; Rubin, G.D.; et al. CAD-RADSTM coronary artery disease–reporting and data system. An exper t consensus document of the Society of Cardiovascular Computed Tomogra phy (SCCT), the American College of Radiology (ACR) and the North Amer ican Society for Cardiovascular Imaging (NASCI). Endorsed by the American College of Cardiology. J. Cardiovasc. Comput. Tomogr. 2016, 10, 269–281. [Google Scholar]
- Zhang, J.; Dashtbozorg, B.; Bekkers, E.; Pluim, J.P.W.; Duits, R.; ter Haar Romeny, B.M. Robust retinal vessel segmentation via locally adaptive derivative fra mes in orientation scores. IEEE Trans. Med. Imaging 2016, 35, 2631–2644. [Google Scholar] [CrossRef] [Green Version]
- Meijering, E.; Jacob, M.; Sarria, J.C.; Steiner, P.; Hirling, H.; Unser, M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. A 2004, 58, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Dashtbozorg, B.; Zhang, J.; Huang, F.; ter Haar Romeny, B.M. Automatic Optic Disc and Fovea Detection in Retinal Images Using Super-Elliptical Convergence Index Filters. In Proceedings of the International Conference on Image Analysis and Recognition, Póvoa de Varzim, Portugal, 13–15 July 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 697–706. [Google Scholar]
- Cheung, C.Y.; Tay, W.T.; Mitchell, P.; Wang, J.J.; Hsu, W.; Lee, M.L.; Lau, Q.P.; Zhu, A.L.; Klein, R.; Saw, S.M.; et al. Quantitative and qualitative retinal microvascular characteristics and blood pressure. J. Hypertens. 2011, 29, 1380–1391. [Google Scholar] [CrossRef]
- Sasongko, M.B.; Wong, T.Y.; Nguyen, T.T.; Cheung, C.Y.; Shaw, J.E.; Kawasaki, R.; Lamoureux, E.L.; Wang, J.J. Retinal vessel tortuosity and its relation to traditional and novel va scular risk markers in persons with diabetes. Curr. Eye Res. 2003, 41, 551–557. [Google Scholar]
- Zhang, J.; Dashtbozorg, B.; Huang, F.; Berendschot, T.T.J.M.; ter Haar Romeny, B.M. Analysis of Retinal Vascular Biomarkers for Early Detection of Diabetes. In Proceedings of the European Congress on Computational Methods in Applied Sciences and Engineering, Porto, Portugal, 18–20 October 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 811–817. [Google Scholar]
- Huang, F.; Dashtbozorg, B.; Yeung, A.K.S.; Zhang, J.; Berendschot, T.T.J.M.; ter Haar Romeny, B.M. A Comparative Study Towards the Establishment of an Automatic Retinal Vessel Width Measurement Technique. In Proceedings of the Fetal, Infant and Ophthalmic Medical Image Analysis, Québec City, QC, Canada, 14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 227–234. [Google Scholar]
- Huang, F.; Dashtbozorg, B.; Yeung, A.K.S.; Zhang, J.; Berendschot, T.T.J.M.; ter Haar Romeny, B.M. Validation Study on Retinal Vessel Caliber Measurement Technique. In Proceedings of the European Congress on Computational Methods in Applied Sciences and Engineering, Porto, Portugal, 18–20 October 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 818–826. [Google Scholar]
- Caselles, V.; Kimmel, R.; Sapiro, G. Geodesic active contours. Int. J. Comput. Vis. 1997, 22, 61–79. [Google Scholar] [CrossRef]
- Kalitzeos, A.A.; Lip, G.Y.H.; Heitmar, R. Retinal vessel tortuosity measures and their applications. Exp. Eye Res. 2013, 106, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Al-Diri, B.; Hunter, A. Automated Measurements of Retinal Bifurcations. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering, Munich, Germany, 7–12 September 2009; Springer: Berlin/Heidelberg, Germany, 2009; pp. 205–208. [Google Scholar]
- Rényi, A. On the dimension and entropy of probability distributions. Acta Math. Acad. Sci. Hung. 1959, 10, 193–215. [Google Scholar] [CrossRef]
- Liebovitch, L.S.; Toth, T. A fast algorithm to determine fractal dimensions by box counting. Phys. Lett. A 1989, 141, 386–390. [Google Scholar] [CrossRef]
- Harte, D. Multifractals: Theory and Applications; Chapman and Hall/CRC: New York, NY, USA, 2001. [Google Scholar]
- Ktena, S.I.; Ferrante, E.; Lee, M.; Moreno, R.G.; Glocker, B.; Rueckert, D. Spectral Graph Convolutions for Population-Based Disease Prediction. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 177–185. [Google Scholar]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive representation learning on large graphs. Adv. Neural Inf. Process. Syst. 2017, 30, 1025–1035. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature selection: A data perspective. ACM Comput. Surv. 2017, 50, 1–45. [Google Scholar] [CrossRef]
- Hall, M.A.; Smith, L.A. Feature selection for machine learning: Comparing a correlation-based filter approach to the wrapper. In Proceedings of the FLAIRS Conference, Orlando, FL, USA, 1–5 May 1999; pp. 235–239. [Google Scholar]
- Vidal-Naquet, M.; Ullman, S. Object Recognition with Informative Features and Linear Classification. In Proceedings of the ICCV, Nice, France, 14–17 October 2003; p. 281. [Google Scholar]
- Meyer, P.E.; Bontempi, G. On the use of variable complementarity for feature selection in cancer classification. In Proceedings of the Workshops on Applications of Evolutionary Computation, Budapest, Hungary, 10–12 April 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 91–102. [Google Scholar]
- Jakulin, A. Machine Learning Based on Attribute Interactions. Ph.D. Thesis, Univerza v Ljubljani, Ljubljana, Slovenia, 2005. [Google Scholar]
- He, X.; Cai, D.; Niyogi, P. Laplacian score for feature selection. In Proceedings of the 18th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 5–8 December 2005; pp. 507–514. [Google Scholar]
- George, D.; Mallery, P. IBM SPSS Statistics 26 Step by Step: A Simple Guide and Reference; Routledge: Oxfordshire, UK, 2019. [Google Scholar]
- Cheung, N.; Wang, J.J.; Klein, R.; Couper, D.J.; Sharrett, A.R.; Wong, T.Y. Diabetic retinopathy and the risk of coronary heart disease: The Ather osclerosis Risk in Communities Study. Diabetes Care 2007, 30, 1742–1746. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, V.P.; Joshi, C.K.; Luu, A.T.; Laurent, T.; Bengio, Y.; Bresson, X. Benchmarking graph neural networks. arXiv 2020, arXiv:2003.00982. [Google Scholar]
- Hemelings, R.; Elen, B.; Stalmans, I.; van Keer, K.; de Boever, P.; Blaschko, M.B. Artery-vein segmentation in fundus images using a fully convolutional network. Comput. Med. Imaging Graph. 2019, 76, 101636. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Artif. Intell. Med. 2020, 103, 167–175. [Google Scholar] [CrossRef] [Green Version]
- Sharafeldeen, A.; Elsharkawy, M.; Khalifa, F.; Soliman, A.; Ghazal, M.; AlHalabi, M.; Yaghi, M.; Alrahmawy, M.; Elmougy, S.; Sandhu, H.S.; et al. Precise higher-order reflectivity and morphology models for early diagnosis of diabetic retinopathy using OCT images. Sci. Rep. 2021, 11, 4730. [Google Scholar] [CrossRef] [PubMed]


| 0 | 1 | 2 | 3 | 4 | 5 | Model 1 | Model 2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAD-RADS * ≤ 1 | CAD-RADS ≥ 2 | CAT ** = 0 | CAT = 1 | |||||||||||||||||
| Number of participants | 55 | 15 | 37 | 20 | 13 | 5 | 70 | 75 | 108 | 37 | ||||||||||
| No. | (%) | No. | (%) | No. | (%) | No. | (%) | No. | (%) | No. | (%) | No. | (%) | No. | (%) | No. | (%) | No. | (%) | |
| Gender | ||||||||||||||||||||
| Male | 28 | (50.91) | 7 | (46.67) | 19 | (51.35) | 17 | (85.0) | 10 | (76.92) | 3 | (60.0) | 35 | (50.0) | 49 | (65.33) | 57 | (52.78) | 27 | (72.97) |
| Female | 27 | (49.09) | 8 | (53.33) | 18 | (48.65) | 3 | (15.0) | 3 | (23.08) | 2 | (40.0) | 35 | (50.0) | 26 | (34.67) | 51 | (47.22) | 10 | (27.03) |
| Tobacco use | ||||||||||||||||||||
| Non-smoker | 42 | (76.36) | 11 | (73.33) | 21 | (56.76) | 16 | (80.0) | 7 | (53.85) | 4 | (80.0) | 53 | (75.71) | 48 | (64.0) | 76 | (70.37) | 25 | (67.57) |
| Current smoker | 3 | (5.45) | 3 | (20.0) | 5 | (13.51) | 2 | (10.0) | 2 | (15.38) | 0 | (0) | 6 | (8.57) | 9 | (12.0) | 12 | (11.11) | 3 | (8.11) |
| Ex-smoker | 10 | (18.18) | 1 | (6.67) | 11 | (29.73) | 2 | (10.0) | 4 | (30.77) | 1 | (20.0) | 11 | (15.71) | 18 | (24.0) | 20 | (18.52) | 9 | (24.32) |
| Retinopathy | ||||||||||||||||||||
| Non-retinopathy | 35 | (63.64) | 10 | (66.67) | 22 | (59.46) | 12 | (60.0) | 6 | (46.15) | 3 | (60.0) | 45 | (64.29) | 43 | (57.33) | 64 | (59.26) | 24 | (64.86) |
| Tessellated retina | 12 | (21.82) | 3 | (20.0) | 9 | (24.32) | 5 | (25.0) | 3 | (23.08) | 1 | (20.0) | 15 | (21.43) | 18 | (24.0) | 27 | (25.0) | 6 | (16.22) |
| DM-related retinopathy | 2 | (3.64) | 0 | (0) | 2 | (5.41) | 1 | (5.0) | 1 | (7.69) | 0 | (0) | 2 | (2.86) | 4 | (5.33) | 4 | (3.7) | 2 | (5.41) |
| AMD | 6 | (10.91) | 2 | (13.33) | 5 | (13.51) | 1 | (5.0) | 2 | (15.38) | 1 | (20.0) | 8 | (11.43) | 9 | (12.0) | 13 | (12.04) | 4 | (10.81) |
| Pathologic myopia | 1 | (1.82) | 0 | (0) | 2 | (5.41) | 1 | (5.0) | 0 | (0) | 0 | (0) | 1 | (1.43) | 3 | (4.0) | 4 | (3.7) | 0 | (0) |
| Comorbidities | ||||||||||||||||||||
| Heart failure | 2 | (3.64) | 1 | (6.67) | 1 | (2.7) | 2 | (10) | 1 | (7.69) | 0 | (0) | 3 | (4.29) | 4 | (5.33) | 5 | (4.63) | 2 | (5.41) |
| Ischemic heart disease | 12 | (21.82) | 3 | (20) | 5 | (13.51) | 8 | (40) | 2 | (15.38) | 1 | (20) | 15 | (21.43) | 16 | (21.33) | 10 | (9.26) | 21 | (56.76) |
| Hyperlipidemia | 17 | (30.91) | 10 | (66.67) | 15 | (40.54) | 15 | (75) | 8 | (61.54) | 4 | (80) | 27 | (38.57) | 42 | (56) | 40 | (37.04) | 29 | (78.97) |
| Hypertension | 25 | (45.45) | 7 | (46.67) | 18 | (48.65) | 10 | (50) | 10 | (76.92) | 4 | (80) | 32 | (45.71) | 42 | (56) | 47 | (43.52) | 27 | (72.97) |
| Diabetes mellitus | 8 | (14.55) | 2 | (13.33) | 2 | (5.41) | 9 | (45) | 3 | (23.08) | 1 | (20) | 10 | (14.29) | 15 | (20) | 15 | (13.89) | 10 | (27.03) |
| mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | |||||||||||
| Age | 54.35 ± 12.33 | 59.73 ± 10.31 | 62.86 ± 12.3 | 61.25 ± 12.51 | 65.0 ± 8.56 | 59.2 ± 12.3 | 55.5 ± 12.06 | 62.56 ± 11.67 | 58.48 ± 12.92 | 61.11 ± 10.37 | ||||||||||
| BMI (kg/m2) | 24.52 ± 5.5 | 25.38 ± 3.16 | 26.04 ± 4.51 | 25.68 ± 5.37 | 25.07 ± 3.14 | 25.72 ± 1.91 | 24.7 ± 5.08 | 25.75 ± 4.38 | 25.35 ± 5.19 | 24.94 ± 3.14 | ||||||||||
| Blood pressure (mmHg) | ||||||||||||||||||||
| Systolic | 129.31 ± 19.83 | 134.8 ± 16.89 | 135.11 ± 18.47 | 123.65 ± 16.5 | 134.69 ± 17.95 | 130.2 ± 19.51 | 130.49 ± 19.26 | 131.65 ± 18.27 | 131.16 ± 18.54 | 130.89 ± 19.41 | ||||||||||
| Diastolic | 79.73 ± 13.38 | 80.47 ± 10.6 | 81.72 ± 10.52 | 75.85 ± 10.44 | 81.62 ± 11.12 | 79.4 ± 6.58 | 79.89 ± 12.77 | 79.98 ± 10.53 | 79.69 ± 11.7 | 80.65 ± 11.53 | ||||||||||
| Heart rate (BPM) | 74.82 ± 11.46 | 70.27 ± 14. | 71.11 ± 10.56 | 71.3 ± 12.84 | 68.23 ± 6.02 | 71.8 ± 18.47 | 73.84 ± 12.09 | 70.71 ± 11.05 | 73.12 ± 11.83 | 69.59 ± 10.75 | ||||||||||
| CAD-RADS Model 1 | CAD-RADS Model 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| CAD-RADS ≤ 1 | CAD-RADS ≥ 2 | CAT = 0 | CAT = 1 | |||||
| Tessellated retina | OR | 95%CI | p-value | OR | 95%CI | p-value | ||
| OR-Model 1 * | 1.00 | 2.139 | (0.188, 24.345) | 0.54 | 1.00 | - | (-, -) | - |
| OR-Model 2 † | 1.00 | 2.257 | (0.182, 27.949) | 0.526 | 1.00 | - | (-, -) | - |
| DM-related retinopathy | ||||||||
| OR-Model 1 | 1.00 | 1.481 | (0.24, 9.119) | 0.672 | 1.00 | 1.64 | (0.249, 10.805) | 0.607 |
| OR-Model 2 | 1.00 | 2.112 | (0.3, 14.881) | 0.453 | 1.00 | 1.542 | (0.205, 11.594) | 0.674 |
| AMD | ||||||||
| OR-Model 1 | 1.00 | 1.09 | (0.45, 2.636) | 0.849 | 1.00 | 0.628 | (0.225, 1.753) | 0.375 |
| OR-Model 2 | 1.00 | 1.361 | (0.524, 3.532) | 0.527 | 1.00 | 0.733 | (0.245, 2.193) | 0.578 |
| Pathologic myopia | ||||||||
| OR-Model 1 | 1.00 | 1.02 | (0.34, 3.057) | 0.972 | 1.00 | 1.006 | (0.284, 3.561) | 0.993 |
| OR-Model 2 | 1.00 | 1.071 | (0.33, 3.476) | 0.909 | 1.00 | 1.334 | (0.344, 5.169) | 0.677 |
| Methods ** | Feature Selection | Sens. | Spec. | Accu. | AUC | F1-Score | Precision | p-Value * |
|---|---|---|---|---|---|---|---|---|
| CAD-RADS Model 1 (class 0: CAD-RADS ≤ 1; class 1: CAD-RADS ≥ 2) for image-wise classification | ||||||||
| GraphSAGE | all | 0.711 (0.621, 0.786) | 0.697 (0.605, 0.776) | 0.704 (0.644, 0.764) | 0.739 (0.675, 0.804) | 0.711 (0.672, 0.746) | 0.711 (0.621, 0.786) | - |
| LR | CFS | 0.509 (0.418, 0.599) | 0.541 (0.448, 0.632) | 0.525 (0.459, 0.59) | 0.521 (0.445, 0.596) | 0.514 (0.473, 0.555) | 0.537 (0.443, 0.628) | <0.01 |
| LDA | DISR | 0.553 (0.461, 0.641) | 0.468 (0.377, 0.561) | 0.511 (0.446, 0.577) | 0.507 (0.431, 0.583) | 0.546 (0.505, 0.586) | 0.521 (0.432, 0.608) | <0.05 |
| KNN | CFS | 0.158 (0.102, 0.236) | 0.862 (0.785, 0.915) | 0.502 (0.437, 0.568) | 0.527 (0.451, 0.603) | 0.184 (0.152, 0.221) | 0.545 (0.38, 0.702) | <0.01 |
| NB | CFS | 0.491 (0.401, 0.582) | 0.495 (0.403, 0.588) | 0.493 (0.428, 0.559) | 0.52 (0.444, 0.596) | 0.494 (0.453, 0.535) | 0.505 (0.413, 0.596) | <0.01 |
| SVM | all | 0.535 (0.444, 0.624) | 0.569 (0.475, 0.658) | 0.552 (0.486, 0.617) | 0.604 (0.53, 0.678) | 0.541 (0.5, 0.581) | 0.565 (0.471, 0.654) | <0.01 |
| CAD-RADS Model 1 (class 0: CAD-RADS ≤ 1; class 1: CAD-RADS ≥ 2) for subject-wise classification | ||||||||
| GraphSAGE | LAP | 0.747 (0.638, 0.831) | 0.571 (0.455, 0.681) | 0.662 (0.585, 0.739) | 0.769 (0.708, 0.831) | 0.725 (0.679, 0.768) | 0.651 (0.546, 0.743) | - |
| LR | CFS | 0.507 (0.396, 0.617) | 0.543 (0.427, 0.654) | 0.524 (0.443, 0.605) | 0.512 (0.436, 0.588) | 0.514 (0.463, 0.564) | 0.543 (0.427, 0.654) | < 0.01 |
| LDA | DISR | 0.453 (0.346, 0.566) | 0.5 (0.386, 0.614) | 0.476 (0.395, 0.557) | 0.526 (0.45, 0.601) | 0.461 (0.411, 0.512) | 0.493 (0.378, 0.608) | <0.05 |
| KNN | CFS | 0.387 (0.285, 0.5) | 0.657 (0.54, 0.758) | 0.517 (0.436, 0.599) | 0.531 (0.455, 0.607) | 0.411 (0.361, 0.463) | 0.547 (0.415, 0.673) | <0.01 |
| NB | CFS | 0.453 (0.346, 0.566) | 0.514 (0.4, 0.628) | 0.483 (0.401, 0.564) | 0.492 (0.416, 0.568) | 0.462 (0.412, 0.513) | 0.5 (0.384, 0.616) | <0.01 |
| SVM | SVMB | 0.653 (0.541, 0.751) | 0.614 (0.497, 0.72) | 0.634 (0.556, 0.713) | 0.697 (0.629, 0.765) | 0.652 (0.602, 0.698) | 0.645 (0.533, 0.743) | <0.05 |
| CAD-RADS Model 2 (class 0: CAT = 0; class 1: CAT = 1) for image-wise classification | ||||||||
| GraphSAGE | all | 0.544 (0.416, 0.666) | 0.681 (0.606, 0.747) | 0.646 (0.583, 0.709) | 0.692 (0.608, 0.776) | 0.497 (0.442, 0.552) | 0.369 (0.274, 0.476) | - |
| LR | CFS | 0.561 (0.433, 0.682) | 0.5 (0.425, 0.575) | 0.516 (0.45, 0.581) | 0.513 (0.426, 0.601) | 0.466 (0.414, 0.519) | 0.278 (0.205, 0.366) | >0.05 |
| LDA | CFS | 0.544 (0.416, 0.666) | 0.428 (0.355, 0.504) | 0.457 (0.392, 0.523) | 0.497 (0.41, 0.584) | 0.438 (0.387, 0.49) | 0.246 (0.179, 0.328) | >0.05 |
| KNN | CFS | 0.228 (0.138, 0.352) | 0.819 (0.754, 0.87) | 0.668 (0.606, 0.73) | 0.561 (0.473, 0.649) | 0.24 (0.193, 0.294) | 0.302 (0.186, 0.451) | >0.05 |
| NB | LAP | 0.544 (0.416, 0.666) | 0.422 (0.349, 0.498) | 0.453 (0.388, 0.518) | 0.498 (0.411, 0.585) | 0.437 (0.386, 0.489) | 0.244 (0.178, 0.326) | >0.05 |
| SVM | LAP | 0.544 (0.416, 0.666) | 0.488 (0.413, 0.563) | 0.502 (0.437, 0.568) | 0.514 (0.426, 0.601) | 0.451 (0.399, 0.503) | 0.267 (0.195, 0.354) | >0.05 |
| CAD-RADS Model 2 (class 0: CAT = 0; class 1: CAT = 1) for subject-wise classification | ||||||||
| GraphSAGE | CFS | 0.649 (0.488, 782) | 0.75 (0.661, 0.822) | 0.724 (0.651, 0.797) | 0.753 (0.674, 0.832) | 0.603 (0.534, 0.668) | 0.471 (0.341, 0.605) | - |
| LR | CFS | 0.568 (0.409, 0.713) | 0.444 (0.354, 0.538) | 0.476 (0.395, 0.557) | 0.501 (0.414, 0.588) | 0.459 (0.395, 0.523) | 0.259 (0.176, 0.364) | >0.05 |
| LDA | CFS | 0.541 (0.384, 0.69) | 0.463 (0.372, 0.557) | 0.483 (0.401, 0.564) | 0.501 (0.414, 0.588) | 0.442 (0.379, 0.508) | 0.256 (0.173, 0.363) | >0.05 |
| KNN | CFS | 0.243 (0.134, 0.401) | 0.759 (0.671, 0.83) | 0.628 (0.549, 0.706) | 0.572 (0.485, 0.66) | 0.246 (0.189, 0.313) | 0.257 (0.142, 0.421) | >0.05 |
| NB | CMIM | 0.568 (0.409, 0.713) | 0.417 (0.328, 0.511) | 0.455 (0.374, 0.536) | 0.52 (0.432, 0.607) | 0.453 (0.39, 0.517) | 0.25 (0.17, 0.352) | <0.05 |
| SVM | SVMB | 0.595 (0.435, 0.737) | 0.556 (0.462, 0.646) | 0.566 (0.485, 0.646) | 0.565 (0.477, 0.653) | 0.505 (0.439, 0.57) | 0.314 (0.218, 0.43) | >0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, F.; Lian, J.; Ng, K.-S.; Shih, K.; Vardhanabhuti, V. Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network. Diagnostics 2022, 12, 1390. https://doi.org/10.3390/diagnostics12061390
Huang F, Lian J, Ng K-S, Shih K, Vardhanabhuti V. Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network. Diagnostics. 2022; 12(6):1390. https://doi.org/10.3390/diagnostics12061390
Chicago/Turabian StyleHuang, Fan, Jie Lian, Kei-Shing Ng, Kendrick Shih, and Varut Vardhanabhuti. 2022. "Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network" Diagnostics 12, no. 6: 1390. https://doi.org/10.3390/diagnostics12061390
APA StyleHuang, F., Lian, J., Ng, K.-S., Shih, K., & Vardhanabhuti, V. (2022). Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network. Diagnostics, 12(6), 1390. https://doi.org/10.3390/diagnostics12061390







