Comparison of Two Rapid Assays for the Detection of BRAF V600 Mutations in Metastatic Melanoma including Positive Sentinel Lymph Nodes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Assessment of the BRAF Mutation Status with the Idylla TM Method
2.3. Assessment of the BRAF V600E Status with Immunohistochemistry
2.4. Literature Search
3. Results
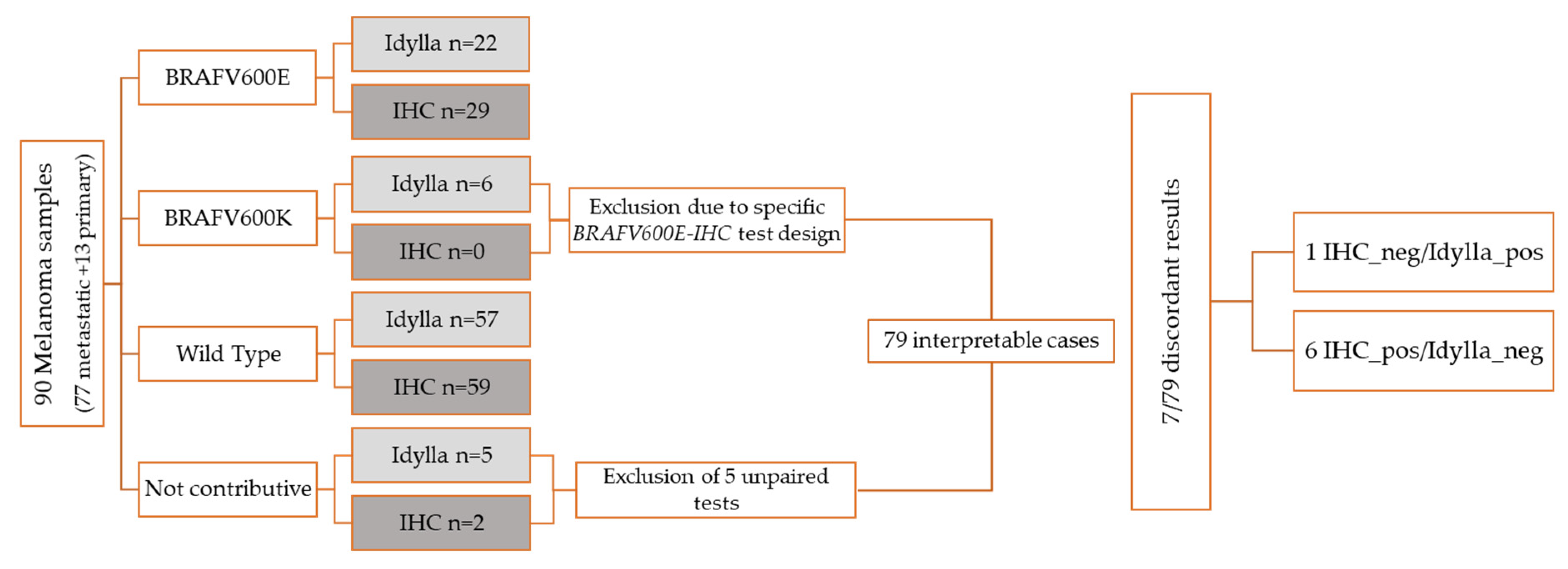
3.1. Samples
3.2. Molecular Testing
3.3. Immunohistochemistry
3.4. Performance Comparison of Both Methods
3.5. Interpretation of the Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Bastholt, L.; Bataille, V.; Del Marmol, V.; Dréno, B.; Fargnoli, M.C.; et al. European Consensus-Based Interdisciplinary Guideline for Melanoma. Part 1: Diagnostics—Update 2019. Eur. J. Cancer 2020, 126, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Prieto, V.G. Sentinel Lymph Nodes in Cutaneous Melanoma. Clin. Lab. Med. 2017, 37, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Bastholt, L.; Bataille, V.; Del Marmol, V.; Dréno, B.; Fargnoli, M.C.; et al. European Consensus-Based Interdisciplinary Guideline for Melanoma. Part 2: Treatment—Update 2019. Eur. J. Cancer 2020, 126, 159–177. [Google Scholar] [CrossRef]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted Agents and Immunotherapies: Optimizing Outcomes in Melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Platz, A.; Egyhazi, S.; Ringborg, U.; Hansson, J. Human Cutaneous Melanoma; a Review of NRAS and BRAF Mutation Frequencies in Relation to Histogenetic Subclass and Body Site. Mol. Oncol. 2008, 1, 395–405. [Google Scholar] [CrossRef]
- Bergdorf, K.N.; Lee, L.A.; Weiss, V.L. BRAF Molecular Testing in Cytopathology: Implications for Diagnosis, Prognosis, and Targeted Therapeutics. Cancer Cytopathol. 2020, 128, 9–11. [Google Scholar] [CrossRef]
- Cheng, L.; Lopez-Beltran, A.; Massari, F.; MacLennan, G.T.; Montironi, R. Molecular Testing for BRAF Mutations to Inform Melanoma Treatment Decisions: A Move toward Precision Medicine. Mod. Pathol. 2018, 31, 24–38. [Google Scholar] [CrossRef]
- Long, G.V.; Wilmott, J.S.; Capper, D.; Preusser, M.; Zhang, Y.E.; Thompson, J.F.; Kefford, R.F.; von Deimling, A.; Scolyer, R.A. Immunohistochemistry Is Highly Sensitive and Specific for the Detection of V600E BRAF Mutation in Melanoma. Am. J. Surg. Pathol. 2013, 37, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Ritterhouse, L.L.; Barletta, J.A. BRAF V600E Mutation-Specific Antibody: A Review. Semin. Diagn. Pathol. 2015, 32, 400–408. [Google Scholar] [CrossRef]
- Long, E.; Ilie, M.; Lassalle, S.; Butori, C.; Poissonnet, G.; Washetine, K.; Mouroux, J.; Lespinet, V.; Lacour, J.P.; Taly, V.; et al. Why and How Immunohistochemistry Should Now Be Used to Screen for the BRAFV600E Status in Metastatic Melanoma? The Experience of a Single Institution (LCEP, Nice, France). J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2436–2443. [Google Scholar] [CrossRef] [PubMed]
- Long-Mira, E.; Washetine, K.; Hofman, P. Sense and Nonsense in the Process of Accreditation of a Pathology Laboratory. Virchows Arch. 2016, 468, 43–49. [Google Scholar] [CrossRef]
- Lhermitte, B.; Egele, C.; Weingertner, N.; Ambrosetti, D.; Dadone, B.; Kubiniek, V.; Burel-Vandenbos, F.; Coyne, J.; Michiels, J.-F.; Chenard, M.-P.; et al. Adequately Defining Tumor Cell Proportion in Tissue Samples for Molecular Testing Improves Interobserver Reproducibility of Its Assessment. Virchows Arch. 2017, 470, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Claes, B.; Huang, H.J.; Falchook, G.S.; Devogelaere, B.; Kockx, M.; Bempt, I.V.; Reijans, M.; Naing, A.; Fu, S.; et al. BRAF Mutation Testing with a Rapid, Fully Integrated Molecular Diagnostics System. Oncotarget 2015, 6, 26886–26894. [Google Scholar] [CrossRef] [PubMed]
- Schiefer, A.-I.; Parlow, L.; Gabler, L.; Mesteri, I.; Koperek, O.; von Deimling, A.; Streubel, B.; Preusser, M.; Lehmann, A.; Kellner, U.; et al. Multicenter Evaluation of a Novel Automated Rapid Detection System of BRAF Status in Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn. 2016, 18, 370–377. [Google Scholar] [CrossRef]
- Ilie, M.I.; Lassalle, S.; Long-Mira, E.; Bonnetaud, C.; Bordone, O.; Lespinet, V.; Lamy, A.; Sabourin, J.-C.; Haudebourg, J.; Butori, C.; et al. Diagnostic Value of Immunohistochemistry for the Detection of the BRAF(V600E) Mutation in Papillary Thyroid Carcinoma: Comparative Analysis with Three DNA-Based Assays. Thyroid 2014, 24, 858–866. [Google Scholar] [CrossRef]
- Van Akkooi, A.C.J.; De Wilt, J.H.W.; Verhoef, C.; Graveland, W.J.; Van Geel, A.N.; Kliffen, M.; Eggermont, A.M.M. High Positive Sentinel Node Identification Rate by EORTC Melanoma Group Protocol. Prognostic Indicators of Metastatic Patterns after Sentinel Node Biopsy in Melanoma. Eur. J. Cancer 2006, 42, 372–380. [Google Scholar] [CrossRef]
- Dewar, D.J.; Newell, B.; Green, M.A.; Topping, A.P.; Powell, B.W.E.M.; Cook, M.G. The Microanatomic Location of Metastatic Melanoma in Sentinel Lymph Nodes Predicts Nonsentinel Lymph Node Involvement. J. Clin. Oncol. 2004, 22, 3345–3349. [Google Scholar] [CrossRef]
- Lito, P.; Rosen, N.; Solit, D.B. Tumor Adaptation and Resistance to RAF Inhibitors. Nat. Med. 2013, 19, 1401–1409. [Google Scholar] [CrossRef]
- Yancovitz, M.; Litterman, A.; Yoon, J.; Ng, E.; Shapiro, R.L.; Berman, R.S.; Pavlick, A.C.; Darvishian, F.; Christos, P.; Mazumdar, M.; et al. Intra- and Inter-Tumor Heterogeneity of BRAF(V600E)) Mutations in Primary and Metastatic Melanoma. PLoS ONE 2012, 7, e29336. [Google Scholar] [CrossRef] [PubMed]
- Satzger, I.; Marks, L.; Kerick, M.; Klages, S.; Berking, C.; Herbst, R.; Völker, B.; Schacht, V.; Timmermann, B.; Gutzmer, R. Allele Frequencies of BRAFV600 Mutations in Primary Melanomas and Matched Metastases and Their Relevance for BRAF Inhibitor Therapy in Metastatic Melanoma. Oncotarget 2015, 6, 37895–37905. [Google Scholar] [CrossRef] [PubMed]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.-P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A Landscape of Driver Mutations in Melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Melchior, L.; Grauslund, M.; Bellosillo, B.; Montagut, C.; Torres, E.; Moragón, E.; Micalessi, I.; Frans, J.; Noten, V.; Bourgain, C.; et al. Multi-Center Evaluation of the Novel Fully-Automated PCR-Based IdyllaTM BRAF Mutation Test on Formalin-Fixed Paraffin-Embedded Tissue of Malignant Melanoma. Exp. Mol. Pathol. 2015, 99, 485–491. [Google Scholar] [CrossRef]
- Janku, F.; Huang, H.J.; Claes, B.; Falchook, G.S.; Fu, S.; Hong, D.; Ramzanali, N.M.; Nitti, G.; Cabrilo, G.; Tsimberidou, A.M.; et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol. Cancer Ther. 2016, 15, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Harlé, A.; Salleron, J.; Franczak, C.; Dubois, C.; Filhine-Tressarieu, P.; Leroux, A.; Merlin, J.-L. Detection of BRAF Mutations Using a Fully Automated Platform and Comparison with High Resolution Melting, Real-Time Allele Specific Amplification, Immunohistochemistry and Next Generation Sequencing Assays, for Patients with Metastatic Melanoma. PLoS ONE 2016, 11, e0153576. [Google Scholar] [CrossRef]
- Barel, F.; Guibourg, B.; Lambros, L.; Flahec, G.; Marcorelles, P.; Uguen, A. Evaluation of a Rapid, Fully Automated Platform for Detection of BRAF and NRAS Mutations in Melanoma. Acta Derm. Venerol. 2018, 98, 44–49. [Google Scholar] [CrossRef]
- Bisschop, C.; Ter Elst, A.; Bosman, L.J.; Platteel, I.; Jalving, M.; Van den Berg, A.; Diepstra, A.; Van Hemel, B.; Diercks, G.F.H.; Hospers, G.A.P.; et al. Rapid BRAF Mutation Tests in Patients with Advanced Melanoma: Comparison of Immunohistochemistry, Droplet Digital PCR, and the Idylla Mutation Platform. Melanoma Res. 2018, 28, 96–104. [Google Scholar] [CrossRef]
- Long-Mira, E.; Ilie, M.; Chamorey, E.; Leduff-Blanc, F.; Montaudié, H.; Tanga, V.; Allégra, M.; Lespinet-Fabre, V.; Bordone, O.; Bonnetaud, C.; et al. Monitoring BRAF and NRAS Mutations with Cell-Free Circulating Tumor DNA from Metastatic Melanoma Patients. Oncotarget 2018, 9, 36238–36249. [Google Scholar] [CrossRef]
- Seremet, T.; Planken, S.; Schreuer, M.; Jansen, Y.; Delaunoy, M.; El Housni, H.; Lienard, D.; Del Marmol, V.; Heimann, P.; Neyns, B. Illustrative Cases for Monitoring by Quantitative Analysis of BRAF/NRAS CtDNA Mutations in Liquid Biopsies of Metastatic Melanoma Patients Who Gained Clinical Benefits from Anti-PD1 Antibody Therapy. Melanoma Res. 2018, 28, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Serre, D.; Salleron, J.; Husson, M.; Leroux, A.; Gilson, P.; Merlin, J.-L.; Geoffrois, L.; Harlé, A. Accelerated BRAF Mutation Analysis Using a Fully Automated PCR Platform Improves the Management of Patients with Metastatic Melanoma. Oncotarget 2018, 9, 32232–32237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vallée, A.; Denis-Musquer, M.; Herbreteau, G.; Théoleyre, S.; Bossard, C.; Denis, M.G. Prospective Evaluation of Two Screening Methods for Molecular Testing of Metastatic Melanoma: Diagnostic Performance of BRAF V600E Immunohistochemistry and of a NRAS-BRAF Fully Automated Real-Time PCR-Based Assay. PLoS ONE 2019, 14, e0221123. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Springborn, S.; Haug, K.; Bartow, K.; Samra, H.; Menon, S.; Mackinnon, A.C. Evaluation, Validation, and Implementation of the Idylla System as Rapid Molecular Testing for Precision Medicine. J. Mol. Diagn. 2019, 21, 862–872. [Google Scholar] [CrossRef]
- Bourhis, A.; Le Flahec, G.; Uguen, A. Decalcification Can Cause the Failure of BRAF Molecular Analyses and Anti-BRAFV600E VE1 Immunohistochemistry. Pathol. Int. 2019, 69, 219–223. [Google Scholar] [CrossRef]
- Van Haele, M.; Vander Borght, S.; Ceulemans, A.; Wieërs, M.; Metsu, S.; Sagaert, X.; Weynand, B. Rapid Clinical Mutational Testing of KRAS, BRAF and EGFR: A Prospective Comparative Analysis of the Idylla Technique with High-Throughput next-Generation Sequencing. J. Clin. Pathol. 2020, 73, 35–41. [Google Scholar] [CrossRef]
- Petty, D.R.; Hassan, O.A.; Barker, C.S.; O’Neill, S.S. Rapid BRAF Mutation Testing in Pigmented Melanomas. Am. J. Dermatopathol. 2020, 42, 343–348. [Google Scholar] [CrossRef]
- Colombino, M.; Rozzo, C.; Paliogiannis, P.; Casula, M.; Manca, A.; Doneddu, V.; Fedeli, M.A.; Sini, M.C.; Palomba, G.; Pisano, M.; et al. Comparison of BRAF Mutation Screening Strategies in a Large Real-Life Series of Advanced Melanoma Patients. J. Clin. Med. 2020, 9, 2430. [Google Scholar] [CrossRef]
- Durślewicz, J.; Klimaszewska-Wiśniewska, A.; Antosik, P.; Kasperska, A.; Grzanka, D.; Szylberg, T.; Szylberg, Ł. Detection of BRAF V600E Mutation in Ganglioglioma and Pilocytic Astrocytoma by Immunohistochemistry and Real-Time PCR-Based Idylla Test. Dis. Markers 2020, 2020, 8880548. [Google Scholar] [CrossRef]
- Sadlecki, P.; Walentowicz, P.; Bodnar, M.; Marszalek, A.; Grabiec, M.; Walentowicz-Sadlecka, M. Determination of BRAF V600E (VE1) Protein Expression and BRAF Gene Mutation Status in Codon 600 in Borderline and Low-Grade Ovarian Cancers. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef]
- Colling, R.; Wang, L.M.; Soilleux, E. Validating a Fully Automated Real-Time PCR-Based System for Use in the Molecular Diagnostic Analysis of Colorectal Carcinoma: A Comparison with NGS and IHC. J. Clin. Pathol. 2017, 70, 610–614. [Google Scholar] [CrossRef]
- Bodnar, M.; Burduk, P.; Antosik, P.; Jarmuz-Szymczak, M.; Wierzbicka, M.; Marszalek, A. Assessment of BRAF V600E (VE1) Protein Expression and BRAF Gene Mutation Status in Codon 600 in Benign and Malignant Salivary Gland Neoplasms. J. Oral. Pathol. Med. 2017, 46, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Cardus, B.; Colling, R.; Hamblin, A.; Soilleux, E. Comparison of Methodologies for the Detection of BRAF Mutations in Bone Marrow Trephine Specimens. J. Clin. Pathol. 2019, 72, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Viray, H.; Li, K.; Long, T.A.; Vasalos, P.; Bridge, J.A.; Jennings, L.J.; Halling, K.C.; Hameed, M.; Rimm, D.L. A Prospective, Multi-Institutional Diagnostic Trial to Determine Pathologist Accuracy in Estimation of Percentage of Malignant Cells. Arch. Pathol. Lab. Med. 2013, 137, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Smits, A.J.J.; Kummer, J.A.; Dde Bruin, P.C.; Bol, M.; Van den Tweel, J.G.; Seldenrijk, K.A.; Willems, S.M.; Offerhaus, G.J.A.; De Weger, R.A.; Van Diest, P.J.; et al. The Estimation of Tumor Cell Percentage for Molecular Testing by Pathologists Is Not Accurate. Mod. Pathol. 2014, 27, 168–174. [Google Scholar] [CrossRef]
- Chen, G.; Dudley, J.; Tseng, L.-H.; Smith, K.; Gurda, G.T.; Gocke, C.D.; Eshleman, J.R.; Lin, M.-T. Lymph Node Metastases of Melanoma: Challenges for BRAF Mutation Detection. Hum. Pathol. 2015, 46, 113–119. [Google Scholar] [CrossRef]
- Dudley, J.C.; Gurda, G.T.; Tseng, L.-H.; Anderson, D.A.; Chen, G.; Taube, J.M.; Gocke, C.D.; Eshleman, J.R.; Lin, M.-T. Tumor Cellularity as a Quality Assurance Measure for Accurate Clinical Detection of BRAF Mutations in Melanoma. Mol. Diagn. Ther. 2014, 18, 409–418. [Google Scholar] [CrossRef]
- Long, G.V.; Hauschild, A.; Santinami, M.; Atkinson, V.; Mandalà, M.; Chiarion-Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; Haydon, A.; et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N. Engl. J. Med. 2017, 377, 1813–1823. [Google Scholar] [CrossRef]
- Maio, M.; Lewis, K.; Demidov, L.; Mandalà, M.; Bondarenko, I.; Ascierto, P.A.; Herbert, C.; Mackiewicz, A.; Rutkowski, P.; Guminski, A.; et al. Adjuvant Vemurafenib in Resected, BRAFV600 Mutation-Positive Melanoma (BRIM8): A Randomised, Double-Blind, Placebo-Controlled, Multicentre, Phase 3 Trial. Lancet Oncol. 2018, 19, 510–520. [Google Scholar] [CrossRef]
- Varada, S.; Mahalingam, M. Mutation Stability in Primary and Metastatic Melanoma: What We Know and What We Don’t. Histol. Histopathol. 2015, 30, 763–770. [Google Scholar] [CrossRef]
- Ito, T.; Tanaka, Y.; Murata, M.; Kaku-Ito, Y.; Furue, K.; Furue, M. BRAF Heterogeneity in Melanoma. Curr. Treat. Options Oncol. 2021, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.A.; Wiggins, J.M.; Corless, B.C.; Syeda, M.M.; Tadepalli, J.S.; Blake, S.; Fleming, N.; Darvishian, F.; Pavlick, A.; Berman, R.; et al. TERT, BRAF, and NRAS Mutational Heterogeneity between Paired Primary and Metastatic Melanoma Tumors. J. Investig. Dermatol. 2020, 140, 1609–1618.e7. [Google Scholar] [CrossRef] [PubMed]
- Heinzerling, L.; Baiter, M.; Kühnapfel, S.; Schuler, G.; Keikavoussi, P.; Agaimy, A.; Kiesewetter, F.; Hartmann, A.; Schneider-Stock, R. Mutation Landscape in Melanoma Patients Clinical Implications of Heterogeneity of BRAF Mutations. Br. J. Cancer 2013, 109, 2833–2841. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, L.; Meyer, N.; Tournier, E.; Grand, D.; Uro-Coste, E.; Rochaix, P.; Brousset, P.; Lamant, L. Highly Concordant Results Between Immunohistochemistry and Molecular Testing of Mutated V600E BRAF in Primary and Metastatic Melanoma. Acta Derm. Venereol. 2016, 96, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Guadarrama-Orozco, J.A.; Ortega-Gómez, A.; Ruiz-García, E.B.; Astudillo-de la Vega, H.; Meneses-García, A.; Lopez-Camarillo, C. Braf V600E Mutation in Melanoma: Translational Current Scenario. Clin. Transl. Oncol. 2016, 18, 863–871. [Google Scholar] [CrossRef]
- Cormican, D.; Kennedy, C.; Murphy, S.; Werner, R.; Power, D.G.; Heffron, C.C.B.B. High Concordance of BRAF Mutational Status in Matched Primary and Metastatic Melanoma. J. Cutan. Pathol. 2019, 46, 117–122. [Google Scholar] [CrossRef]
- Nielsen, L.B.; Dabrosin, N.; Sloth, K.; Bønnelykke-Behrndtz, M.L.; Steiniche, T.; Lade-Keller, J. Concordance in BRAF V600E Status over Time in Malignant Melanoma and Corresponding Metastases. Histopathology 2018, 72, 814–825. [Google Scholar] [CrossRef]
- Parakh, S.; Murphy, C.; Lau, D.; Cebon, J.S.; Andrews, M.C. Response to MAPK Pathway Inhibitors in BRAF V600M-Mutated Metastatic Melanoma. J. Clin. Pharm. Ther. 2015, 40, 121–123. [Google Scholar] [CrossRef]
- Popescu, A.; Haidar, A.; Anghel, R.M. Treating Malignant Melanoma When a Rare BRAF V600M Mutation Is Present: Case Report and Literature Review. Rom. J. Intern. Med. 2018, 56, 122–126. [Google Scholar] [CrossRef]
- Eckhart, L.; Bach, J.; Ban, J.; Tschachler, E. Melanin Binds Reversibly to Thermostable DNA Polymerase and Inhibits Its Activity. Biochem. Biophys. Res. Commun. 2000, 271, 726–730. [Google Scholar] [CrossRef]
- Cook, M.G.; Green, M.A.; Anderson, B.; Eggermont, A.M.M.; Ruiter, D.J.; Spatz, A.; Kissin, M.W.; Powell, B.W.E.M.; EORTC Melanoma Group. The Development of Optimal Pathological Assessment of Sentinel Lymph Nodes for Melanoma. J. Pathol. 2003, 200, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.G.; Massi, D.; Szumera-Ciećkiewicz, A.; Van den Oord, J.; Blokx, W.; Van Kempen, L.C.; Balamurugan, T.; Bosisio, F.; Koljenović, S.; Portelli, F.; et al. An Updated European Organisation for Research and Treatment of Cancer (EORTC) Protocol for Pathological Evaluation of Sentinel Lymph Nodes for Melanoma. Eur. J. Cancer 2019, 114, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.B.; Moore, H.M. Effects of Preanalytical Variables on the Detection of Proteins by Immunohistochemistry in Formalin-Fixed, Paraffin-Embedded Tissue. Arch. Pathol. Lab. Med. 2011, 135, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Menzies, A.M.; Lum, T.; Wilmott, J.S.; Hyman, J.; Kefford, R.F.; Thompson, J.F.; O’Toole, S.; Long, G.V.; Scolyer, R.A. Intrapatient Homogeneity of BRAFV600E Expression in Melanoma. Am. J. Surg. Pathol. 2014, 38, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Hauschild, A.; Santinami, M.; Atkinson, V.; Mandalà, M.; Kirkwood, J.M.; Chiarion Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; et al. Five-Year Analysis of Adjuvant Dabrafenib plus Trametinib in Stage III Melanoma. N. Engl. J. Med. 2020, 383, 1139–1148. [Google Scholar] [CrossRef]
- Dummer, R.; Lebbé, C.; Atkinson, V.; Mandalà, M.; Nathan, P.D.; Arance, A.; Richtig, E.; Yamazaki, N.; Robert, C.; Schadendorf, D.; et al. Combined PD-1, BRAF and MEK Inhibition in Advanced BRAF-Mutant Melanoma: Safety Run-in and Biomarker Cohorts of COMBI-i. Nat. Med. 2020, 26, 1557–1563. [Google Scholar] [CrossRef]
- Hauschild, A.; Dummer, R.; Schadendorf, D.; Santinami, M.; Atkinson, V.; Mandalà, M.; Chiarion-Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; et al. Longer Follow-Up Confirms Relapse-Free Survival Benefit with Adjuvant Dabrafenib Plus Trametinib in Patients with Resected BRAF V600-Mutant Stage III Melanoma. J. Clin. Oncol. 2018, 36, 3441–3449. [Google Scholar] [CrossRef]
- Bellomo, D.; Arias-Mejias, S.M.; Ramana, C.; Heim, J.B.; Quattrocchi, E.; Sominidi-Damodaran, S.; Bridges, A.G.; Lehman, J.S.; Hieken, T.J.; Jakub, J.W.; et al. Model Combining Tumor Molecular and Clinicopathologic Risk Factors Predicts Sentinel Lymph Node Metastasis in Primary Cutaneous Melanoma. JCO Precis. Oncol. 2020, 4, 319–334. [Google Scholar] [CrossRef]
- Yousaf, A.; Tjien-Fooh, F.J.; Rentroia-Pacheco, B.; Quattrocchi, E.; Kobic, A.; Tempel, D.; Kolodney, M.; Meves, A. Validation of CP-GEP (Merlin Assay) for Predicting Sentinel Lymph Node Metastasis in Primary Cutaneous Melanoma Patients: A U.S. Cohort Study. Int. J. Dermatol. 2021, 60, 851–856. [Google Scholar] [CrossRef]
- Salvianti, F.; Massi, D.; De Giorgi, V.; Gori, A.; Pazzagli, M.; Pinzani, P. Evaluation of the Liquid Biopsy for the Detection of BRAFV600E Mutation in Metastatic Melanoma Patients. Cancer Biomark. 2019, 26, 271–279. [Google Scholar] [CrossRef]
| Exon | Results Given with Idylla | Mutation Detected |
|---|---|---|
| 15 | V600E/E2/D | c.1799T > A |
| c.1799_1800TG > AA | ||
| c.1799_1800TG > AT | ||
| c.1799_1800TG > AC | ||
| V600K/R/M | c.1798_1799GT > AA | |
| c.1798_1799GT > AG | ||
| c.1798G > A | ||
| Wild Type | c.1799T |
| (A) | ||
|---|---|---|
| Clinical and Pathological Characteristics | N (%) | |
| 90 (100) | ||
| Age at diagnostic | Median | 63 |
| Range | 10–93 | |
| Gender | Male | 42 (47) |
| Female | 48 (53) | |
| Tissue sample | Metastasis | 77 (85) |
| Primary | 13 (15) | |
| Metastatic site | Lymph node | 54 (70) |
| Including SLN | 32 (60) | |
| Sub-cutaneous | 14 (18) | |
| Pulmonary | 7 (9) | |
| Other | 2 (3) | |
| Percentage of tumor cells (TC) | ≤1% | 6 (7) |
| 1 < TC < 10% | 12 (13) | |
| 10 ≤ TC < 50% | 15 (17) | |
| ≥50 | 57 (63) | |
| Mean percentage of tumor cell in SLN | 9.3% | |
| Molecular analysis | Assessed | 90 (100) |
| Not contributive | 5 (5) | |
| Molecular status | mutated BRAF | 28 (33) |
| BRAF V600E | 22 (79) | |
| BRAF V600K | 6 (21) | |
| wt-BRAF | 57 (67) | |
| IHC analysis | Assessed | 90 (100) |
| Not contributive | 2 (2) | |
| IHC status | BRAF V600E | 29 (33) |
| wt-BRAF V600E | 59 (67) | |
| (B) | ||
| Pathological Characteristics of Metastatic SLN | N (%) | |
| 90 (100) | ||
| SLN | Number | 32 (35) |
| SLN tumor size | <0.1 mm | 1 (3) |
| 0.1–1 mm | 14 (43) | |
| >1 mm | 17 (53) | |
| SLN tumor microanatomical localization | Sub-capsular | 8 (25) |
| Parenchymal | 6 (19) | |
| Combined | 10 (31) | |
| Multifocal | 6 (19) | |
| Extensive | 2 (6) | |
| Cases | Diagnostic | Location | Stage | TC % | Idylla | Idylla Explore | ΔCQ (Idylla) | BRAF VE1 IHC | Other Sample Available | TC (%) | Molecular Methods | Results | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
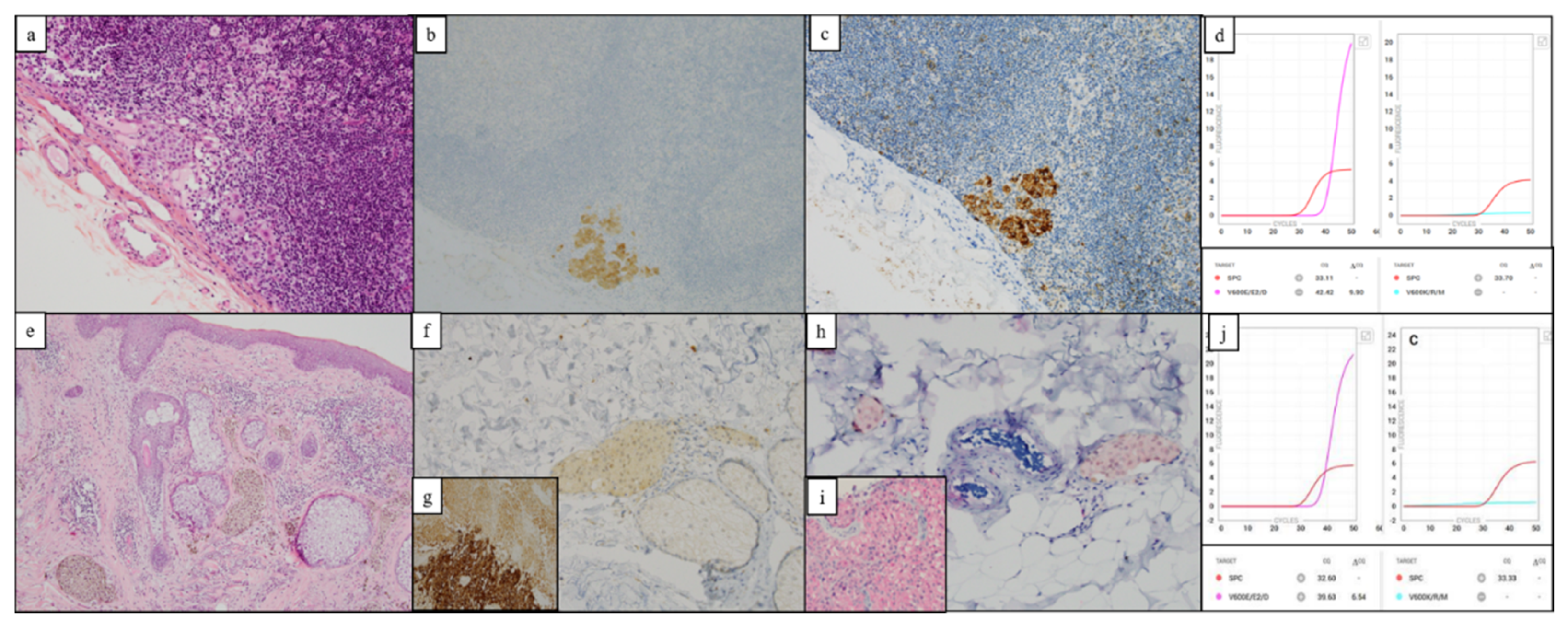
| Positive Idylla/Negative IHC | #62 | Metastatic melanoma | Sub-cutaneous | IV | 20 | V600E/E2/D | Amplification | 6.54 | Negative | Metastatic melanoma | 20 | Idylla and NGS and IHC | Wild type |
| Negative Idylla/Positive IHC | #2 | Metastatic melanoma | Sentinel lymph node | IIIA | 5 | Wild type | Delayed amplification | 10.07 | Positive | Metastatic melanoma | 50 | PS ** | V600E |
| #17 | Metastatic melanoma | Sentinel lymph node | IIIA | 5 | Wild type | Delayed amplification | 8.69 | Positive | Metastatic melanoma | 80 | Idylla | V600E/E2/D | |
| #22 | Metastatic melanoma | Lymph node | IIIA | 5 | Wild type | Delayed amplification | 11.39 | Positive | Metastatic melanoma | 70 | NGS * | V600E | |
| #23 | Metastatic melanoma | Sentinel lymph node | IIIA | ≤1 | Wild type | No amplification | Not applicable | Positive | Primitive melanoma | 30 | Idylla | V600E/E2/D | |
| #27 | Metastatic melanoma | Sentinel lymph node | IIIA | 1 < CT < 5 | Wild type | Delayed amplification | 10.08 | Positive | Metastatic melanoma | 50 | PS ** | V600E | |
| #28 | Metastatic melanoma | Sentinel lymph node | IIIA | ≤1 | Wild type | Delayed amplification | 9.9 | Positive | Metastatic melanoma | 80 | PS ** | V600E |
| Cases | SLN | Stage | % TC | Idylla Result | Delayed Amplification | Idylla Explore Tool (ΔCQ) | IHC BRAF | Re-Test/Other Sample |
|---|---|---|---|---|---|---|---|---|
| #3 | yes | IIIA | 1 | Wild type | No | Not applicable | Negative | Not applicable |
| #5 | yes | IIIA | <1 | Wild type | Yes | ΔCQ = 10.08 | Negative | Wild type (primitive) |
| #10 | yes | IIIA | <1 | Wild type | No | Not applicable | Negative | Wild type (metastasis) |
| #11 | yes | IIIA | <1 | Wild type | Yes | ΔCQ = 10.98 | Negative | Not applicable |
| #23 | yes | IIIA | <1 | Wild type | No | Not applicable | Positive | BRAF V600E (primitif) |
| #28 | yes | IIIA | <1 | Wild type | Yes | ΔCQ = 9.90 | Positive | BRAF V600E (metastasis) |
| Ref. | Study | Gene | Idylla Test | CE-IVD | Mutation Detected for BRAF | Sample | Number of Samples | Type of Tumor | Sample Origin | TAT min | Reference Method | Concordance for BRAF % | Sens % | Spe % | PPV % | NPV % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Melchior et al. 2015 [25] | Multicenter retrospective | BRAF | Idylla BRAF Mutation Test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue | 139 | Melanoma | NS | 90 | SS, RT-PCR, ddPCR, HRM | 97.84 | NS | NS | NS | NS |
| Janku et al. 2015 [16] | Retrospective | BRAF | Idylla BRAF Mutation Test | NS | V600E/E2/D V600K/R/M | FFPE tissue | 60 | Melanoma, CRC, PTC and others | NS | 90 | RT-PCR, NGS | 97 (RT-PCR) 100 (NGS) | 95 | 97 | 98 | 92 |
| Janku et al. 2016 [26] | Prospective | BRAF | BRAF Mutation Test prototype | RUO | Not specified | Cell-free DNA vs FFPE | 160 | CRC, Melanoma, NSCLCC and others | Blood | 90 | PCR-based method, mass spectrometry, NGS | 88 | 73 | 98 | 96 | 85 |
| Schiefer et al. 2016 [17] | Multicenter Retrospective | BRAF | Idylla BRAF Mutation test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue | 419 | Melanoma, PTC, CRC and others | Primary and Metastases | 90 | SS, PS, NGS | 96.2 (SS); 97.5 (PS) | NS | NS | NS | NS |
| Harlé et al. 2016 [27] | Retrospective | BRAF | Idylla BRAF Mutation Test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue | 59 | Melanoma | NS | 90 | HRM, real-time PCR, NGS, IHC | NS | 93.5 (NGS) | 100 | 100 | 93.3 |
| Barel et al. 2018 [28] | Retrospective | NRAS-BRAF-EGFR | Idylla NRAS-BRAF-EGFR S492R Mutation Test | RUO | V600 E/E2/D V600 K/R | FFPE tissue | 36 | Melanoma | Primary and metastases | 110 | NGS, IHC | 97.2 (overall) | NS | NS | NS | NS |
| Bisschop et al. 2018 [29] | Retrospective | BRAF | Idylla BRAF Mutation Test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue | 37 | Melanoma | Primary and metastases | 90 | HRM, SS, IHC, NGS | 97.3 (overall) | 100 | 0.94 | 100 | 100 |
| Long-Mira et al. 2018 [30] | Prospective | BRAF—NRAS | ctNRAS-BRAF Mutation Test | RUO | V600E/E2/D V600 K/R/M | Cell-free DNA vs FFPE | 19 | Melanoma | Blood | 90 | PS, NGS | 84 | 80 | 89 | NS | NS |
| Seremet et al. 2018 [31] | Prospective short communication | BRAF-NRAS | NS | NS | NS | cell-free DNA | 7 | Melanoma | Blood | NS | No | NS | NS | NS | NS | NS |
| Serre et al. 2018 [32] | Prospective and retrospective | BRAF | Idylla BRAF Mutation Test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue | 37 | Melanoma | Metastases | 90 | No | NS | NS | NS | NS | NS |
| Vallée et al. 2019 [33] | Prospective | NRAS-BRAF-EGFR | Idylla NRAS-BRAF-EGFR S492R Mutation Test | RUO | V600 E/E2/D V600 K/R | FFPE tissue | 65 | Melanoma | Primary and metastases | 120 | IHC, ASA, SS, ddPCR | 92.1 (overall) | 100 | 100 | 100 | 100 |
| Huang et al. 2019 [34] | Retrospective | BRAF | Idylla NRAS-BRAF and Idylla BRAF Mutation Test | NS | V600 E/E2/D V600 K/R | FFPE tissue | 210 | CRC, Melanoma, NSCLCC and others | NS | NS | NGS, SS | 100 | NS | NS | NS | NS |
| Bourhis et al. 2019 [35] | Retrospective | BRAF | Idylla BRAF Mutation Test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue and decalcified tissue | 11 samples (paired) | Melanoma, Hairy cell leukemia | Metastases | 90 | IHC | 100 (except decalcified samples) | NS | NS | NS | NS |
| Van Haele et al. 2020 [36] | Prospective | BRAF | Idylla BRAF Mutation Test | CE-IVD | V600E/E2/D V600K/R/M | FFPE tissue and cell block | 48 | Melanoma, NSCLCC, CRC | Metastases | 90 | NGS, Cobas | 100 (NGS) | NS | NS | NS | NS |
| Petty et al. 2020 [37] | Retrospective | BRAF | Idylla BRAF Mutation Test | NS | V600E/E2/D V600K/R/M | FFPE tissue and cell block | 23 | Melanoma | Primary and metastases | 90 | SS, ARMS | 100 | 100 | 100 | 100 | 100 |
| Colombino et al. 2020 [38] | Retrospective | BRAF | NS | NS | V600E/E2/D V600K/R/M | DNA | 319 | Melanoma | Primary and metastases | 120 | SS, PS, NGS | 98.4 (BRAF+) | NS | NS | NS | NS |
| Reference | Tumor Type | Number of Samples | Type Antibody | BRAF Mutation | CE-IVD | Immunostaining System | Sample | Idylla Method | BRAF Mutation with Idylla | Concordance Rate (%) | IHC Sensitivity (%) | IHC Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Durślewicz et al. 2020 [39] | CNS tumor | 22 | Clone VE1 (Ventana Medical System) | BRAF V600E | Yes | Ventana BenchMark ULTRA stainer | FFPE tissue | Idylla BRAF mutation Test | V600E/E2/D V600K/R/M | 86 | NS | NS | NS | NS |
| Sadlecki et al. 2017 [40] | Ovarian tumor | 42 | Clone VE1 (Ventana Medical System) | BRAF V600E | Yes | Ventana BenchMark GX | FFPE tissue | Idylla BRAF mutation Test | V600E/E2/D V600K/R/M | 100 | NS | NS | NS | NS |
| Bourhis et al. 2019 [35] | Metastatic melanoma and hairy cell leukemia | 11 | Clone VE1 | BRAF V600E | No | Ventana Benchmark XT | FFPE tissue and decalcified | Idylla BRAF mutation Test | V600E/E2/D V600K/R/M | 100 | NS | NS | NS | NS |
| Bisschop et al. 2018 [29] | Metastatic melanoma | 37 | Clone VE1 (Ventana Medical System) | BRAF V600E | Yes | Ventana BenchMark ULTRA stainer | FFPE tissue | Idylla NRAS-BRAF-EGFR S492R Mutation Test | V600 E/E2/D V600 K/R | 97.3 (overall) | 94 | 95 | NS | NS |
| Barel et al. 2018 [28] | Melanoma (metastatic and primary) | 36 | Clone VE1 | BRAF V600E | No | Ventana Benchmark XT | FFPE tissue | Idylla NRAS-BRAF-EGFR S492R Mutation Test | V600 E/E2/D V600 K/R | 100 | NS | NS | NS | NS |
| Colling et al. 2017 [41] | CRC | 20 | Clone VE1 (Ventana Medical System) | BRAF V600E | Yes | Ventana Benchmark Immunostainer | FFPE tissue | Idylla NRAS-BRAF-EGFR S492R Mutation Test | V600 E/E2/D V600 K/R | 90 | NS | NS | NS | NS |
| Vallée et al. 2019 [33] | Melanoma (metastatic and primary) | 65 | Clone VE1 (Eurobio) | BRAF V600E | No | NS | FFPE tissue | Idylla NRAS-BRAF-EGFR S492R Mutation Test | V600 E/E2/D V600 K/R | 89 (overall) | 82,3 | 100 | 100 | 93 |
| Bodnar et al. 2017 [42] | Salivary gland tumor | 95 | Clone VE1 (Ventana Medical System) | BRAF V600E | Yes | Ventana BenchMark GX | FFPE tissue | Idylla BRAF Mutation Test | V600E/E2/D V600K/R/M | 97 | NS | NS | NS | NS |
| Cardus et al. 2019 [43] | Hairy cell leukemia and B/T cell neoplasm | 218 | Clone VE1 (Ventana Medical System) | BRAF V600E | Yes | Ventana BenchMark ULTRA stainer | FFPE tissue and decalcified | Idylla BRAF Mutation Test | V600E/E2/D V600K/R/M | 100 | NS | NS | NS | NS |
| Idylla (Biocartis, Belgium) | IHC BRAFV600E (Clone VE1, Roche Ventana) | |
|---|---|---|
| Principles of the Technology | DNA, RT-PCR | Protein expression, Antigen-Antibody |
| Mutations | Detection of a Group of Mutant Only V600E/E2/D; V600K/R/M | V600E |
| Cost/Patient * | 140 € | 54 € |
| Duration run | 90 mn | 255 mn |
| Hands-on time | 20 mn Including block selection, cutting section or macrodissection, insertion in the cartridge | 70 mn Including cutting slide, drying time, preparation of the instrument and mounting of the slide |
| Total duration time | 110 mn | 325 mn |
| Competence of the operator | Not required | Trained technician |
| Ease of interpretation | Very easy—No specific skills | Easy—Trained Pathologist |
| Analytical sensibility | Very high (1%) | Very high (single cell-level resolution) |
| Minimal amount of material | 50% tumor cell and 250 mm3 of tissue are recommended | Few cells, methods independant of the percentage of tumor cell |
| Preanalytic parameter | Robust (formalin fixative) | Delicate (formalin fixative, cold ischemia) |
| Major advantage | Easy to use | Single cell-level |
| Major limitation | Impossibility to collect DNA from the cartridge after test completion for NGS | Limited to the detection of the BRAF V600E mutant protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long-Mira, E.; Picard-Gauci, A.; Lassalle, S.; Hofman, V.; Lalvée, S.; Tanga, V.; Zahaf, K.; Bonnetaud, C.; Lespinet, V.; Camuzard, O.; et al. Comparison of Two Rapid Assays for the Detection of BRAF V600 Mutations in Metastatic Melanoma including Positive Sentinel Lymph Nodes. Diagnostics 2022, 12, 751. https://doi.org/10.3390/diagnostics12030751
Long-Mira E, Picard-Gauci A, Lassalle S, Hofman V, Lalvée S, Tanga V, Zahaf K, Bonnetaud C, Lespinet V, Camuzard O, et al. Comparison of Two Rapid Assays for the Detection of BRAF V600 Mutations in Metastatic Melanoma including Positive Sentinel Lymph Nodes. Diagnostics. 2022; 12(3):751. https://doi.org/10.3390/diagnostics12030751
Chicago/Turabian StyleLong-Mira, Elodie, Alexandra Picard-Gauci, Sandra Lassalle, Véronique Hofman, Salomé Lalvée, Virginie Tanga, Katia Zahaf, Christelle Bonnetaud, Virginie Lespinet, Olivier Camuzard, and et al. 2022. "Comparison of Two Rapid Assays for the Detection of BRAF V600 Mutations in Metastatic Melanoma including Positive Sentinel Lymph Nodes" Diagnostics 12, no. 3: 751. https://doi.org/10.3390/diagnostics12030751
APA StyleLong-Mira, E., Picard-Gauci, A., Lassalle, S., Hofman, V., Lalvée, S., Tanga, V., Zahaf, K., Bonnetaud, C., Lespinet, V., Camuzard, O., Montaudié, H., Poissonnet, G., Passeron, T., Ilié, M., & Hofman, P. (2022). Comparison of Two Rapid Assays for the Detection of BRAF V600 Mutations in Metastatic Melanoma including Positive Sentinel Lymph Nodes. Diagnostics, 12(3), 751. https://doi.org/10.3390/diagnostics12030751








