A Comprehensive Survey on Bone Segmentation Techniques in Knee Osteoarthritis Research: From Conventional Methods to Deep Learning
Abstract
:1. Introduction
- This paper provides a comprehensive survey and analysis of a wide range of state-of-the-art recent methodologies for knee bone segmentation. Moreover, we present quantitative results and the findings of other studies, in order to evaluate their potential and limitations;
- We perform an extended analysis of knee bone segmentation methods, taking it to the next level of depth by breaking the approaches down into their building pieces and emphasizing the algorithmic aspects;
- Unlike other studies, we not only investigate the existing methods, but also provide recommendations and future directions to enhance them;
- Finally, we highlight deep learning’s diagnostic value as the key to future computer-aided diagnosis applications to conclude this review.
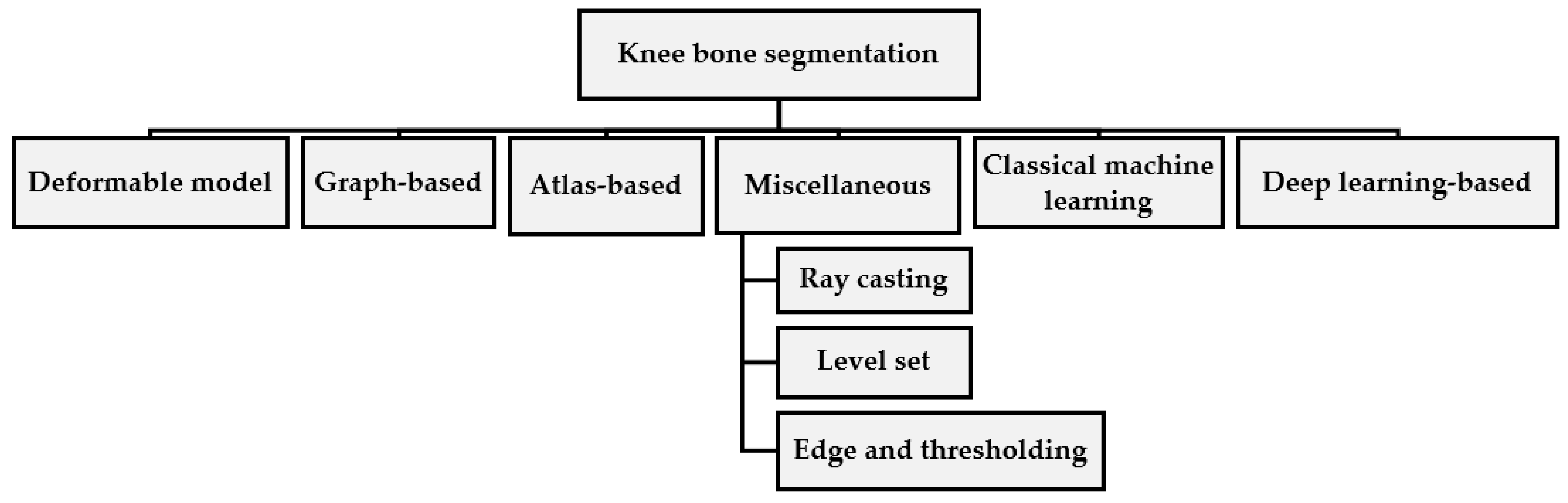
2. Knee Bone Segmentation
2.1. Deformable Model-Based
| Active Shape Model Algorithm |
|
2.2. Graph-Based Methods
- Pre-segmentation of bones;
- Mesh generation and optimization by Gcuts;
- Co-segmentation of knee bone and cartilage surfaces.
2.3. Atlas-Based Methods
- Single atlas: Utilizes a separate segmented image; also, the selection might be random or based on particular criteria, for example, image quality;
- Probabilistic atlas (average shape atlas): Plots all of the original individual images on a common reference to produce a median image. Then, the original images are correlated with the first average to produce a new average. The mapping process occurs frequently until convergence;
- Best atlas: Used to determine the optimal segmentation from the results of the different atlases; one can check the similarity of the image using standardized mutual information and the size of the distortion after registration.
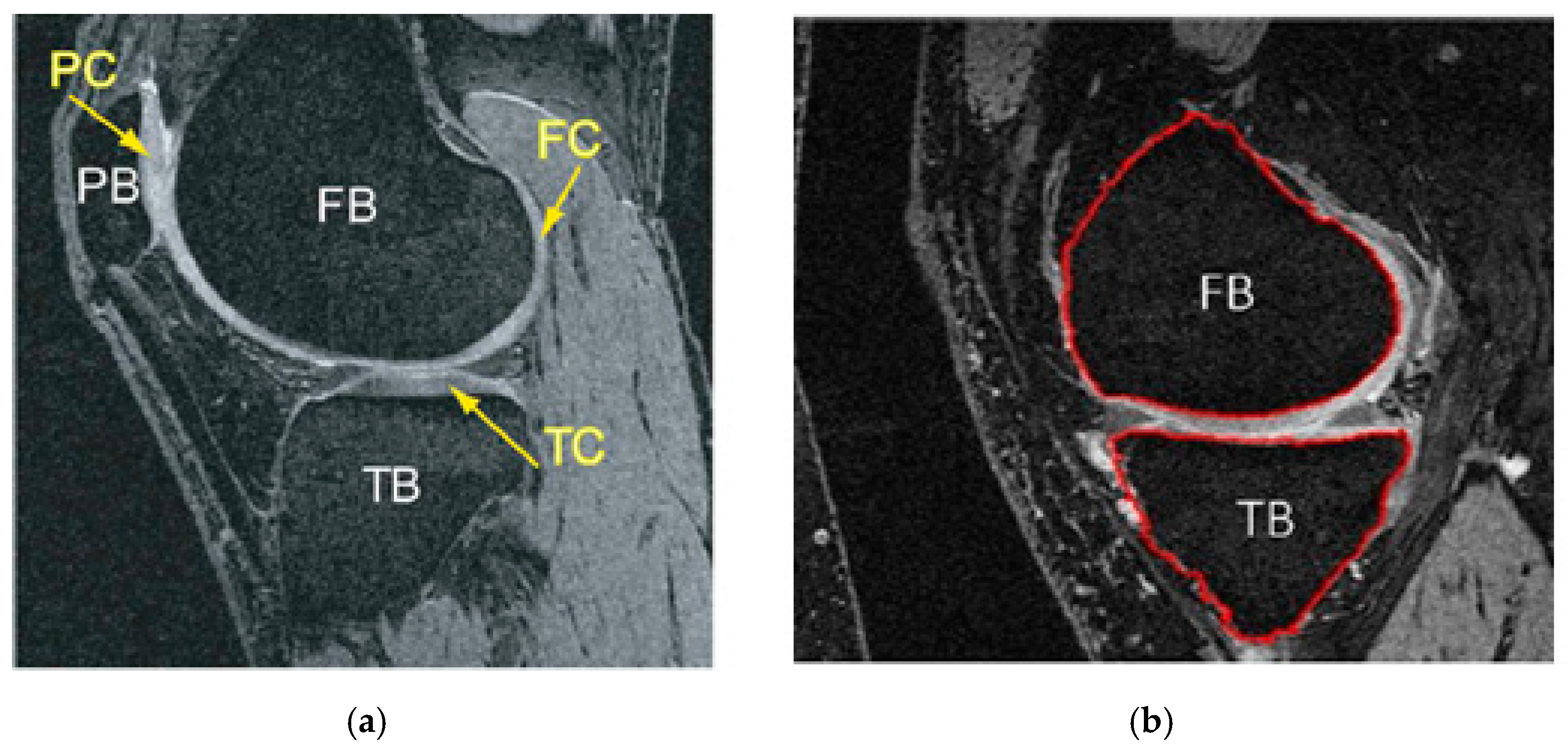
- Multiple atlases: This method applies various atlases to a raw image. Then, the segments are combined into a final hash based on the merging of the “voting rule” decision. This method applies various atlases to a raw image. Then, the segments are combined into a final hash based on the merging of the “voting rule” decision. This can be implemented as labeling cost C in Equation (4) per label l in {FB; BG. TB} (“FB”,”BG” and “TB” stand for femur, background and tibia bone in succession) is determined by the probability of recording each of the labels given image I in voxel Y site:
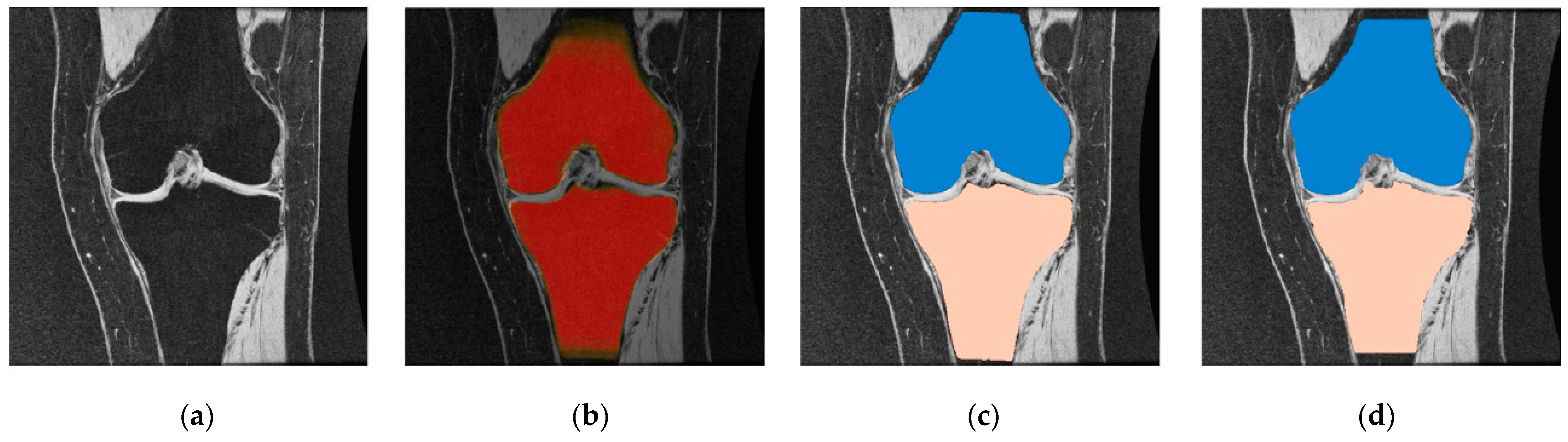
2.4. Miscellaneous Segmentation Approaches
- The region r in Yj is connected to the region Yi if there exists a sequence (Yj … Yi). for instance, Yt and Yt+1 are connected to R;
- R is a continuous region if x and R are connected;
- Whole image,
- 4.
- Equation (10)
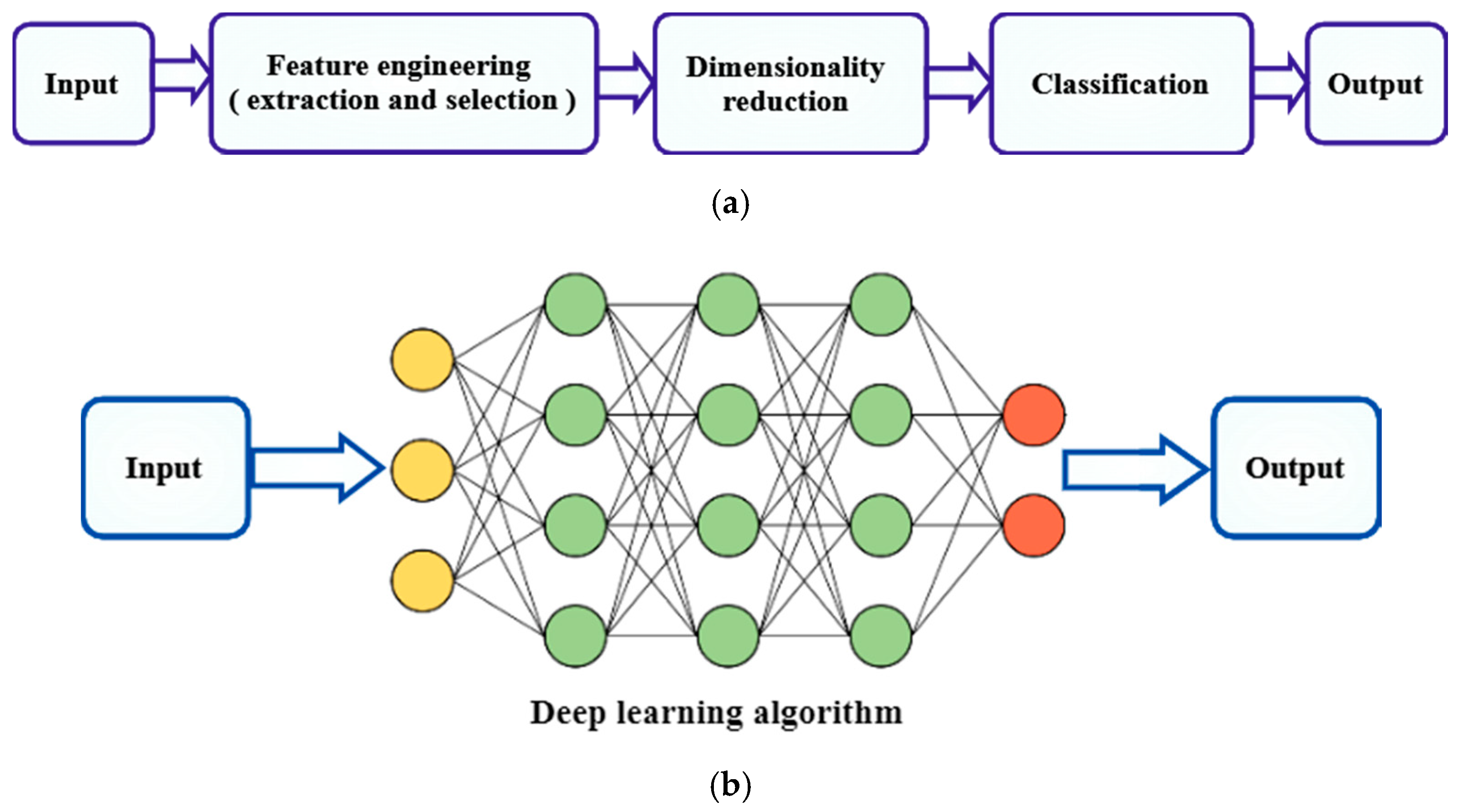
2.5. Machine Learning Based
- Decision Tree: The algorithm is structured like a tree, with branches and nodes. Each branch indicates the outcome, whereas each leaf node represents a class label. The method will sort characteristics in a hierarchical order from the root of the tree to the leaf node [62];
- Naïve Bayes: The technique is based on the Bayes theorem, which assumes characteristics are statistically independent. The classification is based on the conditional likelihood that a result is produced from the probabilities imposed by the input variables [63];
- Support Vector Machine: The algorithm aims to draw the most appropriate margins in which the distance between each category is maximized to the nearest margin. A margin is defined as the distance between two hyperplane support vectors. A bigger margin involves minor mistakes in categorization [64];
- Ensemble Learning: A method of grouping multiple weak classifiers to build a strong classifier. It is known that aggregation methods can be used to improve prediction performance. Boosting and bagging are important ensemble learning techniques [65].
- K-Means: This algorithm groups data into k-clusters based on their homogeneity, where the center of each cluster is an individual mean value. Moreover, the data values are allocated based on their closeness to the nearest average with the least possible error function during implementation [66];
- Principal Component Analysis: This method aims to reduce the dimensionality of the data by finding a set of uncorrelated low dimensional linear data representations that have greater variance. This linear dimensional technique is useful for exploring the latent interaction of a variable in an unsupervised environment [67].
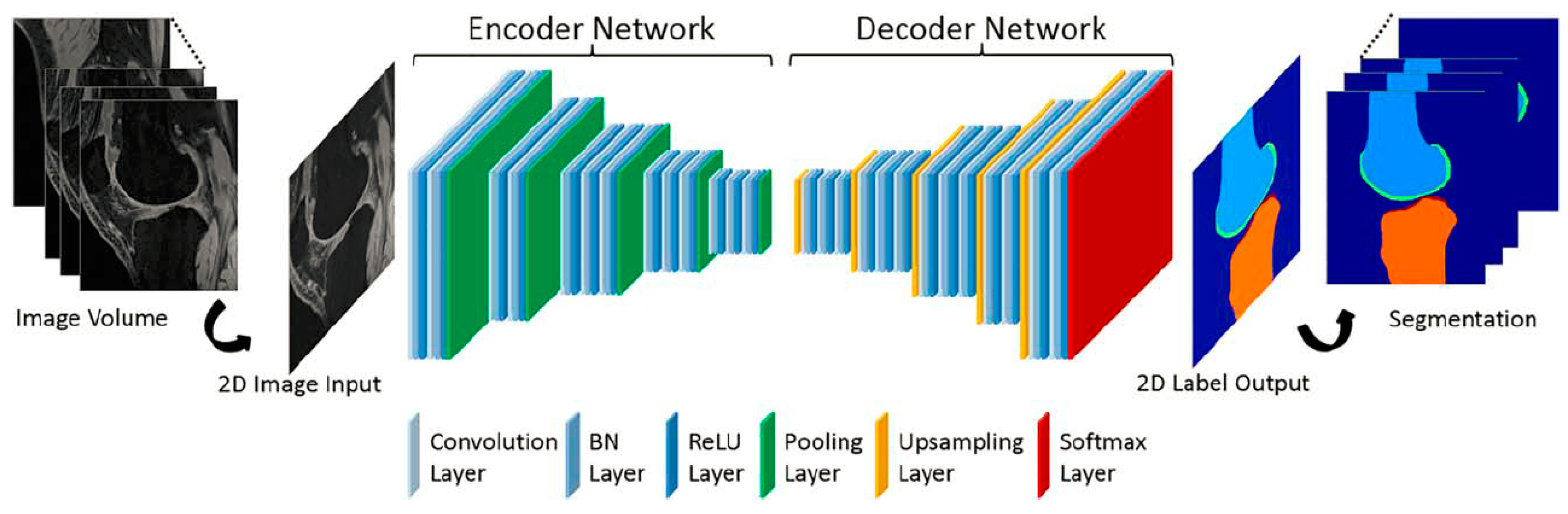
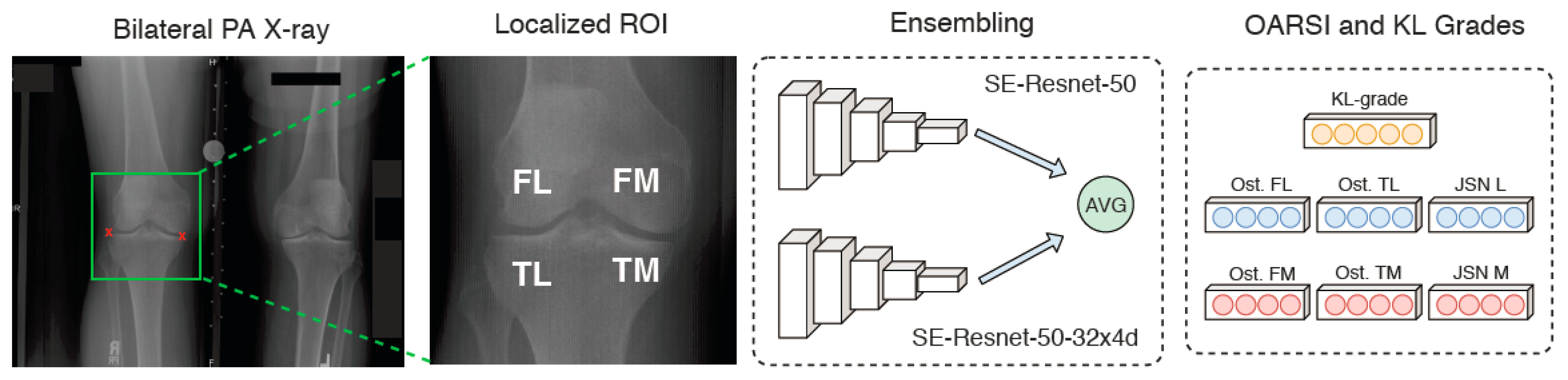
2.6. Deep Learning-Based
3. Approaches
3.1. Research Approach to Literature
3.2. Estimated Results
3.3. Data Sources
4. Discussion and Recommendations
- The development of a useful tool based on CNNs for assessing morphological and structural changes in the musculoskeletal system could be an interesting research field for assisting clinical applications, particularly for longitudinal assessments;
- More research is needed to improve current methods to address issues such as a lack of full assessment for intensity inhomogeneity and clinical practices;
- Combining DL strategies with other machine learning approaches such as KNN, SVM, and so on, can achieve an acceptable result;
- The design and development of a 3D CNN learning-based framework for a graph representation of knee joints that can accommodate both edge and shape information for the graph.
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, P.; Gao, L.; Shi, X.; Allen, K.; Yang, L. Fully Automatic Knee Osteoarthritis Severity Grading Using Deep Neural Networks with a Novel Ordinal Loss. Comput. Med. Imaging Graph. 2019, 75, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Liu, F.; Mizaian, A.H.; Demehri, S.; Samsonov, A.; Guermazi, A.; Kijowski, R. Deep Learning Approach to Predict Pain Progression in Knee Osteoarthritis. Skelet. Radiol. 2022, 51, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Neogi, T. The Epidemiology and Impact of Pain in Osteoarthritis. Osteoarthr. Cartil. 2013, 21, 1145–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaul, E.; Barron, J. Age-Related Diseases and Clinical and Public Health Implications for the 85 Years Old and over Population. Front. Public Health 2017, 5, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briggs, A.M.; Shiffman, J.; Shawar, Y.R.; Åkesson, K.; Ali, N.; Woolf, A.D. Global Health Policy in the 21st Century: Challenges and Opportunities to Arrest the Global Disability Burden from Musculoskeletal Health Conditions. Best Pract. Res. Clin. Rheumatol. 2020, 34, 101549. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.; Smith, E.; Hoy, D.; Carmona, L.; Wolfe, F.; Vos, T.; Williams, B.; Gabriel, S.; Lassere, M.; Johns, N. The Global Burden of Rheumatoid Arthritis: Estimates from the Global Burden of Disease 2010 Study. Ann. Rheum. Dis. 2014, 73, 1316–1322. [Google Scholar] [CrossRef]
- Migliore, A.; Gigliucci, G.; Alekseeva, L.; Avasthi, S.; Bannuru, R.R.; Chevalier, X.; Conrozier, T.; Crimaldi, S.; Damjanov, N.; de Campos, G.C. Treat-to-Target Strategy for Knee Osteoarthritis. International Technical Expert Panel Consensus and Good Clinical Practice Statements. Ther. Adv. Musculoskelet. Dis. 2019, 11, 1759720X19893800. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, X.; Gao, T.; Du, L.; Liu, W. An Automatic Knee Osteoarthritis Diagnosis Method Based on Deep Learning: Data from the Osteoarthritis Initiative. J. Healthc. Eng. 2021, 2021, 5586529. [Google Scholar] [CrossRef]
- Hayashi, D.; Roemer, F.W.; Jarraya, M.; Guermazi, A. Imaging in Osteoarthritis. Radiol. Clin. N. Am. 2017, 55, 1085–1102. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An Overview of Deep Learning in Medical Imaging Focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Aprovitola, A.; Gallo, L. Knee Bone Segmentation from MRI: A Classification and Literature Review. Biocybern. Biomed. Eng. 2016, 36, 437–449. [Google Scholar] [CrossRef]
- Goldring, S.R. Cross-Talk between Subchondral Bone and Articular Cartilage in Osteoarthritis. Arthritis Res. Ther. 2012, 14, A7. [Google Scholar] [CrossRef] [Green Version]
- Hunter, D.J.; Zhang, Y.; Niu, J.; Goggins, J.; Amin, S.; LaValley, M.P.; Guermazi, A.; Genant, H.; Gale, D.; Felson, D.T. Increase in Bone Marrow Lesions Associated with Cartilage Loss: A Longitudinal Magnetic Resonance Imaging Study of Knee Osteoarthritis. Arthritis Rheum. 2006, 54, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Neogi, T.; Bowes, M.A.; Niu, J.; De Souza, K.M.; Vincent, G.R.; Goggins, J.; Zhang, Y.; Felson, D.T. Magnetic Resonance Imaging-Based Three-Dimensional Bone Shape of the Knee Predicts Onset of Knee Osteoarthritis: Data from the Osteoarthritis Initiative. Arthritis Rheum. 2013, 65, 2048–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies-Tuck, M.L.; Wluka, A.E.; Forbes, A.; Wang, Y.; English, D.R.; Giles, G.G.; O’Sullivan, R.; Cicuttini, F.M. Development of Bone Marrow Lesions Is Associated with Adverse Effects on Knee Cartilage While Resolution Is Associated with Improvement—A Potential Target for Prevention of Knee Osteoarthritis: A Longitudinal Study. Arthritis Res. Ther. 2010, 12, R10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgeat, P.; Fripp, J.; Stanwell, P.; Ramadan, S.; Ourselin, S. MR Image Segmentation of the Knee Bone Using Phase Information. Med. Image Anal. 2007, 11, 325–335. [Google Scholar] [CrossRef]
- Kashyap, S.; Zhang, H.; Rao, K.; Sonka, M. Learning-Based Cost Functions for 3-D and 4-D Multi-Surface Multi-Object Segmentation of Knee MRI: Data from the Osteoarthritis Initiative. IEEE Trans. Med. Imaging 2017, 37, 1103–1113. [Google Scholar] [CrossRef]
- Yin, Y.; Zhang, X.; Williams, R.; Wu, X.; Anderson, D.D.; Sonka, M. LOGISMOS-Layered Optimal Graph Image Segmentation of Multiple Objects and Surfaces: Cartilage Segmentation in the Knee Joint. IEEE Trans. Med. Imaging 2010, 29, 2023–2037. [Google Scholar] [CrossRef] [Green Version]
- Becker, M.; Magnenat-Thalmann, N. Deformable Models in Medical Image Segmentation. In 3D Multiscale Physiological Human; Springer: London, UK, 2014; pp. 81–106. [Google Scholar] [CrossRef]
- Mcinerney, T.; Terzopoulos, D. Deformable models in medical image analysis: A survey. Med. Image Anal. 1996, 1, 91–108. [Google Scholar] [CrossRef]
- Cootes, T.F.; Taylor, C.J. Active Shape Models—‘Smart Snakes’ BT—BMVC92; Hogg, D., Boyle, R., Eds.; Springer: London, UK, 1992; pp. 266–275. [Google Scholar]
- Heimann, T.; Meinzer, H.P. Statistical Shape Models for 3D Medical Image Segmentation: A Review. Med. Image Anal. 2009, 13, 543–563. [Google Scholar] [CrossRef]
- Sarkalkan, N.; Weinans, H.; Zadpoor, A.A. Statistical Shape and Appearance Models of Bones. Bone 2014, 60, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Terzopoulos, D. On Matching Deformable Models to Images. Top. Meet. Mach. Vis. Tech 1987, 12, 160–167. [Google Scholar]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active Contour Models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Cootes, T.; Baldock, E.R.; Graham, J. An Introduction to Active Shape Models. Image Processing Anal. 2000, 243657, 223–248. [Google Scholar]
- Cootes, T.; Taylor, C.; Cooper, D.; Graham, J. Active Shape Models-Their Training and Application. Comput. Vis. Image Underst. 1995, 61, 38–59. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Jiang, J.; Hao, S.; Zhan, S. Distribution-Based Active Contour Model for Medical Image Segmentation. In Proceedings of the 6th International Conference on Image and Graphics, ICIG 2011, Hefei, China, 12–15 August 2011; pp. 61–65. [Google Scholar] [CrossRef]
- Lorigo, L.M.; Faugeras, O.; Grimson, W.E.L.; Antipolis, S. Segmentation of Bone in Clinical Knee MRI Using Texture—Bas Ed Geodesic Active Contours. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Cambridge, MA, USA, 11–13 October 1998. [Google Scholar]
- Cheng, R.; Roth, H.R.; Lu, L.; Wang, S.; Turkbey, B.; Gandler, W.; McCreedy, E.S.; Agarwal, H.K.; Choyke, P.; Summers, R.M.; et al. Active Appearance Model and Deep Learning for More Accurate Prostate Segmentation on MRI. In Medical Imaging 2016: Image Processing; SPIE: Bellingham, WA, USA, 2016; Volume 9784, p. 97842I. [Google Scholar] [CrossRef]
- Fripp, J.; Crozier, S.; Warfield, S.K.; Ourselin, S. Automatic Segmentation of the Bone and Extraction of the Bone-Cartilage Interface from Magnetic Resonance Images of the Knee. Phys. Med. Biol. 2007, 52, 1617–1631. [Google Scholar] [CrossRef] [PubMed]
- Vincent, G.; Wolstenholme, C.; Scott, I.; Bowes, M. Fully Automatic Segmentation of the Knee Joint Using Active Appearance Models. Med. Image Anal. Clin. A Grand Chall. 2010, 1, 224–230. [Google Scholar]
- Seim, H.; Kainmueller, D.; Lamecker, H.; Bindernagel, M.; Malinowski, J.; Zachow, S. Model-Based Auto-Segmentation of Knee Bones and Cartilage in MRI Data. In Proceedings of the 13th International Conference on Medical Image Computing and Computer Assisted Intervention, Beijing, China, 24 September 2010; pp. 215–223. [Google Scholar]
- Bindernagel, M.; Kainmueller, D.; Seim, H.; Lamecker, H.; Zachow, S.; Hege, H.C. An Articulated Statistical Shape Model of the Human Knee. In Bildverarbeitung für die Medizin; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar] [CrossRef]
- Tamez-Pena, J.G.; Farber, J.; Gonzalez, P.C.; Schreyer, E.; Schneider, E.; Totterman, S. Unsupervised Segmentation and Quantification of Anatomical Knee Features: Data from the Osteoarthritis Initiative. IEEE Trans. Biomed. Eng. 2012, 59, 1177–1186. [Google Scholar] [CrossRef]
- Boykov, Y.Y.; Jolly, M.P. Interactive Graph Cuts for Optimal Boundary & Region Segmentation of Objects in N-D Images. In Proceedings of the IEEE International Conference on Computer Vision, Vancouver, BC, Canada, 7–14 July 2001; Volume 1, pp. 105–112. [Google Scholar] [CrossRef]
- Bourmaud, G.; Mégret, R.; Giremus, A.; Berthoumieu, Y. Global Motion Estimation from Relative Measurements Using Iterated Extended Kalman Filter on Matrix LIE Groups. In Proceedings of the 2014 IEEE International Conference on Image Processing, ICIP 2014, Paris, France, 27–30 October 2014; Volume 22, pp. 3362–3366. [Google Scholar] [CrossRef]
- Toennies, K.D. Guide to Medical Image Analysis; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Camilus, K.S.; Govindan, V.K. A Review on Graph Based Segmentation. Int. J. Image Graph. Signal Process. 2012, 4, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Peng, B.; Zhang, L.; Zhang, D. A Survey of Graph Theoretical Approaches to Image Segmentation. Pattern Recognit. 2013, 46, 1020–1038. [Google Scholar] [CrossRef] [Green Version]
- Leahy, Z.W. and R. An Optimal Graph Theoretic Approach to Data Clustering: Theory and Its Application to Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 1101–1113. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Malik, J. Normalized Cuts and Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 888–905. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Lee, S.; Shim, H.; Yun, I.D.; Lee, S.U.; Lee, K.H.; Kang, H.S.; Han, J.K. Fully Automatic 3-D Segmentation of Knee Bone Compartments by Iterative Local Branch-And-Mincut on Mr Images from Osteoarthritis Initiative (OAI). In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 3381–3384. [Google Scholar]
- Ababneh, S.Y.; Prescott, J.W.; Gurcan, M.N. Automatic Graph-Cut Based Segmentation of Bones from Knee Magnetic Resonance Images for Osteoarthritis Research. Med. Image Anal. 2011, 15, 438–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ababneh, S.Y.; Gurcan, M.N. An Efficient Graph-Cut Segmentation for Knee Bone Osteoarthritis Medical Images. In Proceedings of the 2010 IEEE International Conference on Electro/Information Technology, EIT2010, Normal, IL, USA, 20–22 May 2010. [Google Scholar] [CrossRef]
- Somasundar, M.K.A.; Somashekar, B.H.; Somasundar, R.A. Segmentation of Tibia Femoral Bone Using Graph Cut Method and 3D Rendering for FEA. Int. J. Inf. Technol. 2020, 12, 1435–1441. [Google Scholar] [CrossRef]
- Rohlfing, T.; Brandt, R.; Menzel, R.; Russakoff, D.B.; Maurer, C.R. Quo Vadis, Atlas-Based Segmentation? In Handbook of Biomedical Image Analysis; Springer: Boston, MA, USA, 2005; pp. 435–486. [Google Scholar]
- Shan, L.; Zach, C.; Charles, C.; Niethammer, M. Automatic Atlas-Based Three-Label Cartilage Segmentation from MR Knee Images. Med. Image Anal. 2014, 18, 1233–1246. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.G.; Gumus, S.; Moon, C.H.; Kwoh, C.K.; Bae, K.T. Fully Automated Segmentation of Cartilage from the MR Images of Knee Using a Multi-Atlas and Local Structural Analysis Method. Med. Phys. 2014, 41, 092303. [Google Scholar] [CrossRef]
- Dam, E.B.; Lillholm, M.; Marques, J.; Nielsen, M. Automatic Segmentation of High-and Low-Field Knee MRIs Using Knee Image Quantification with Data from the Osteoarthritis Initiative. J. Med. Imaging 2015, 2, 24001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anshad, P.Y.M.; Kumar, S.S.; Shahudheen, S. Segmentation of Chondroblastoma from Medical Images Using Modified Region Growing Algorithm. Clust. Comput. 2019, 22, 13437–13444. [Google Scholar] [CrossRef]
- Pan, Z.; Lu, J. A Bayes-Based Region-Growing Algorithm for Medical Image Segmentation. Comput. Sci. Eng. 2007, 9, 32–38. [Google Scholar] [CrossRef]
- Lee, J.-S.; Chung, Y.-N. Integrating edge detection and thresholding approaches to segmenting femora and patellae from magnetic resonance images. Biomed. Eng. Appl. Basis Commun. 2005, 17, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Dodin, P.; Martel-Pelletier, J.; Pelletier, J.-P.; Abram, F. A Fully Automated Human Knee 3D MRI Bone Segmentation Using the Ray Casting Technique. Med. Biol. Eng. Comput. 2011, 49, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Dalvi, R.; Abugharbieh, R.; Wilson, D.C.; Wilson, D.R. Multi-Contrast MR for Enhanced Bone Imaging and Segmentation. In Proceedings of the 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; IEEE: Piscataway, NJ, USA, 2007; pp. 5620–5623. [Google Scholar]
- Gandhamal, A.; Talbar, S.; Gajre, S.; Razak, R.; Hani, A.F.M.; Kumar, D. Fully Automated Subchondral Bone Segmentation from Knee MR Images: Data from the Osteoarthritis Initiative. Comput. Biol. Med. 2017, 88, 110–125. [Google Scholar] [CrossRef]
- Cabitza, F.; Locoro, A.; Banfi, G. Machine Learning in Orthopedics: A Literature Review. Front. Bioeng. Biotechnol. 2018, 6, 75. [Google Scholar] [CrossRef] [Green Version]
- Jamshidi, A.; Pelletier, J.-P.; Martel-Pelletier, J. Machine-Learning-Based Patient-Specific Prediction Models for Knee Osteoarthritis. Nat. Rev. Rheumatol. 2019, 15, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Kluzek, S.; Mattei, T.A. Machine-Learning for Osteoarthritis Research. Osteoarthr. Cartil. 2019, 27, 977–978. [Google Scholar] [CrossRef] [PubMed]
- Zheng, A.; Casari, A. Feature Engineering for Machine Learning: Principles and Techniques for Data Scientists; O’Reilly Media, Inc.: Newton, MA, USA, 2018. [Google Scholar]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Rish, I. An Empirical Study of the Naive Bayes Classifier. In IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence; IBM: New York, NY, USA, 2001; Volume 3, pp. 41–46. [Google Scholar]
- Farhat, N.H. Photonic Neural Networks and Learning Machines. IEEE Expert 1992, 7, 63–72. [Google Scholar] [CrossRef]
- Rokach, L. Ensemble-Based Classifiers. Artif. Intell. Rev. 2010, 33, 1–39. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967; Volume 1, pp. 281–297. [Google Scholar]
- Jolliffe, I.T. Generalizations and Adaptations of Principal Component Analysis. In Principal Component Analysis; Springer: New York, NY, USA, 2002; pp. 373–405. [Google Scholar]
- Zhang, K.; Lu, W.; Marziliano, P. Automatic Knee Cartilage Segmentation from Multi-Contrast MR Images Using Support Vector Machine Classification with Spatial Dependencies. Magn. Reson. Imaging 2013, 31, 1731–1743. [Google Scholar] [CrossRef]
- Brahim, A.; Jennane, R.; Riad, R.; Janvier, T.; Khedher, L.; Toumi, H.; Lespessailles, E. A Decision Support Tool for Early Detection of Knee OsteoArthritis Using X-Ray Imaging and Machine Learning: Data from the OsteoArthritis Initiative. Comput. Med. Imaging Graph. 2019, 73, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Kubkaddi, S.; Ravikumar, K.M. Early Detection of Knee Osteoarthritis Using SVM Classifier. IJSEAT 2017, 5, 259–262. [Google Scholar]
- Du, Y.; Almajalid, R.; Shan, J.; Zhang, M. A Novel Method to Predict Knee Osteoarthritis Progression on MRI Using Machine Learning Methods. IEEE Trans. Nanobiosci. 2018, 17, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S.; Oguz, I.; Zhang, H.; Sonka, M. Automated segmentation of knee MRI using hierarchical classifiers and just enough interaction based learning: Data from osteoarthritis initiative. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2016; pp. 344–351. [Google Scholar] [CrossRef] [Green Version]
- Halilaj, E.; Le, Y.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Modeling and Predicting Osteoarthritis Progression: Data from the Osteoarthritis Initiative. Osteoarthr. Cartil. 2018, 26, 1643–1650. [Google Scholar] [CrossRef] [Green Version]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Geetharamani, G.; Pandian, A. Identification of Plant Leaf Diseases Using a Nine-Layer Deep Convolutional Neural Network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar] [CrossRef]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Harapanahalli, S.; Hernandez, G.V.; Krpalkova, L.; Riordan, D.; Walsh, J. Deep Learning vs. Traditional Computer Vision. In Science and Information Conference; Springer: Cham, Switzerland, 2019; pp. 128–144. [Google Scholar]
- Wang, Z. Deep Learning for Image Segmentation: Veritable or Overhyped? arXiv 2019, arXiv:1904.08483. [Google Scholar]
- Wang, G.; Ye, J.C.; Mueller, K.; Fessler, J.A. Image Reconstruction Is a New Frontier of Machine Learning. IEEE Trans. Med. Imaging 2018, 37, 1289–1296. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, J.; Liu, B.; Zhang, Z.; Yan, X.; Wei, M. A Review on Deep Learning Applications in Prognostics and Health Management. IEEE Access 2019, 7, 162415–162438. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT press: Cambridge, MA, USA, 2016. [Google Scholar]
- Jozefowicz, R.; Zaremba, W.; Sutskever, I. An Empirical Exploration of Recurrent Network Architectures. In Proceedings of the 32nd International Conference on Machine Learning, ICML 2015, Lille, France, 7–9 July 2015; Volume 3, pp. 2332–2340. [Google Scholar]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.A. Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Liu, F.; Zhou, Z.; Jang, H.; Samsonov, A.; Zhao, G.; Kijowski, R. Deep Convolutional Neural Network and 3D Deformable Approach for Tissue Segmentation in Musculoskeletal Magnetic Resonance Imaging. Magn. Reson. Med. 2018, 79, 2379–2391. [Google Scholar] [CrossRef] [PubMed]
- Ambellan, F.; Tack, A.; Ehlke, M.; Zachow, S. Automated Segmentation of Knee Bone and Cartilage Combining Statistical Shape Knowledge and Convolutional Neural Networks: Data from the Osteoarthritis Initiative. Med. Image Anal. 2019, 52, 109–118. [Google Scholar] [CrossRef]
- Cheng, R.; Alexandridi, N.A.; Smith, R.M.; Shen, A.; Gandler, W.; McCreedy, E.; McAuliffe, M.J.; Sheehan, F.T. Fully Automated Patellofemoral MRI Segmentation Using Holistically Nested Networks: Implications for Evaluating Patellofemoral Osteoarthritis, Pain, Injury, Pathology, and Adolescent Development. Magn. Reson. Med. 2020, 83, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, J.; Cheon, S. A Deep Neural Network-Based Method for Early Detection of Osteoarthritis Using Statistical Data. Int. J. Environ. Res. Public Health 2019, 16, 1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiulpin, A.; Saarakkala, S. Automatic Grading of Individual Knee Osteoarthritis Features in Plain Radiographs Using Deep Convolutional Neural Networks. Diagnostics 2020, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; McGuinness, K.; Moran, K.; O’Connor, N.E. Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity Using Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning and Data Mining in Pattern Recognition, New York, NY, USA, 15–20 July 2017; Springer: Cham, Switzerland, 2017; pp. 376–390. [Google Scholar]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci. Rep. 2018, 8, 1727. [Google Scholar] [CrossRef]
- Christodoulou, E.; Moustakidis, S.; Papandrianos, N.; Tsaopoulos, D.; Papageorgiou, E. Exploring Deep Learning Capabilities in Knee Osteoarthritis Case Study for Classification. In Proceedings of the 2019 10th International Conference on Information, Intelligence, Systems and Applications (IISA), Patras, Greece, 15–17 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–6. [Google Scholar]
- Tiulpin, A.; Klein, S.; Bierma-Zeinstra, S.M.A.; Thevenot, J.; Rahtu, E.; van Meurs, J.; Oei, E.H.G.; Saarakkala, S. Multimodal Machine Learning-Based Knee Osteoarthritis Progression Prediction from Plain Radiographs and Clinical Data. Sci. Rep. 2019, 9, 20038. [Google Scholar] [CrossRef]
- Chang, G.H.; Felson, D.T.; Qiu, S.; Capellini, T.D.; Kolachalama, V.B. Assessment of knee pain from MR imaging using a convolutional Siamese network. Eur. Radiol. 2020, 30, 3538–3548. [Google Scholar] [CrossRef]
- Shamir, L.; Orlov, N.; Eckley, D.M.; Macura, T.; Johnston, J.; Goldberg, I.G. Wndchrm–an Open Source Utility for Biological Image Analysis. Source Code Biol. Med. 2008, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Ashinsky, B.G.; Coletta, C.E.; Bouhrara, M.; Lukas, V.A.; Boyle, J.M.; Reiter, D.A.; Neu, C.P.; Goldberg, I.G.; Spencer, R.G. Machine Learning Classification of OARSI-Scored Human Articular Cartilage Using Magnetic Resonance Imaging. Osteoarthr. Cartil. 2015, 23, 1704–1712. [Google Scholar] [CrossRef] [Green Version]


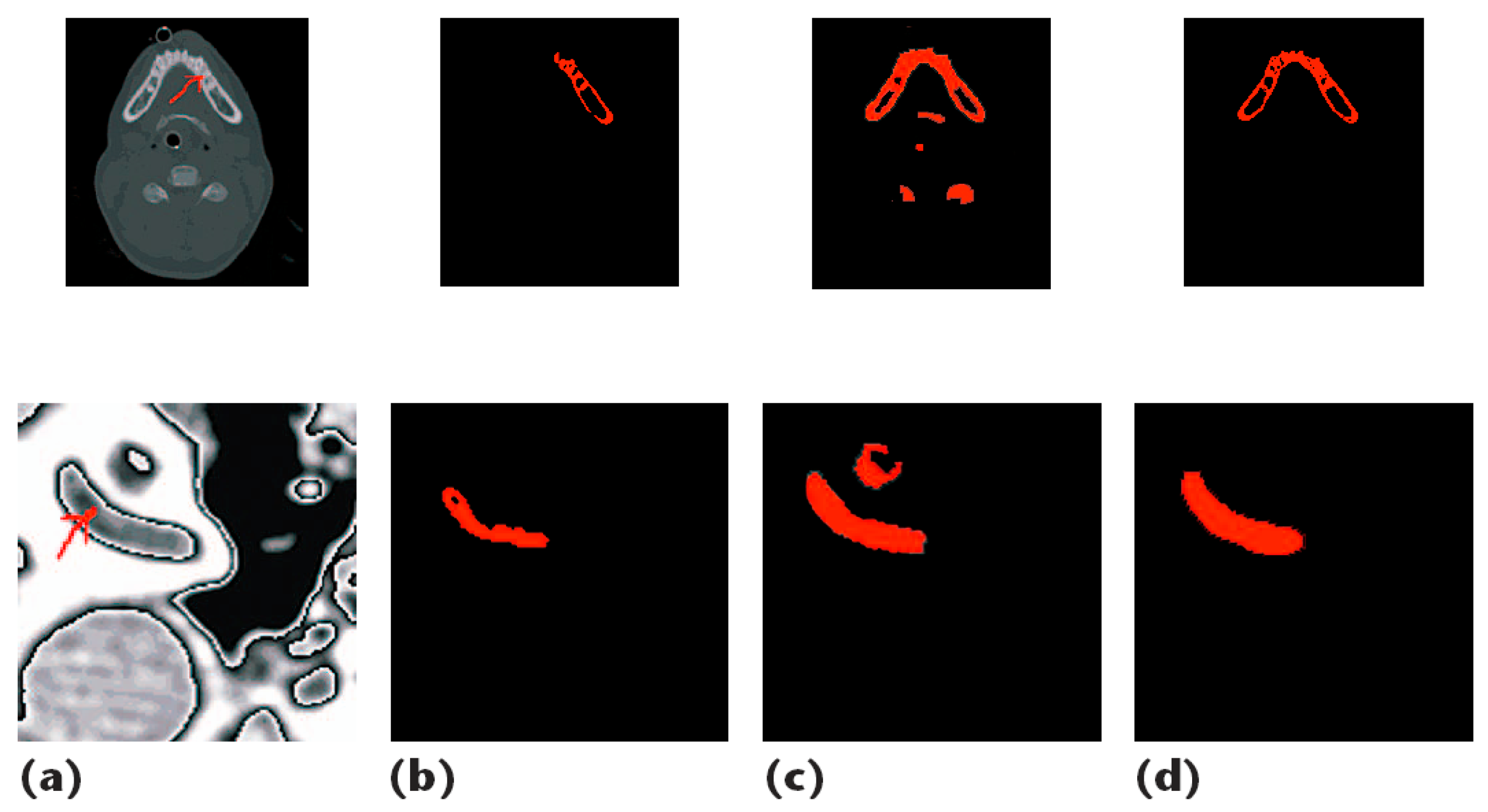

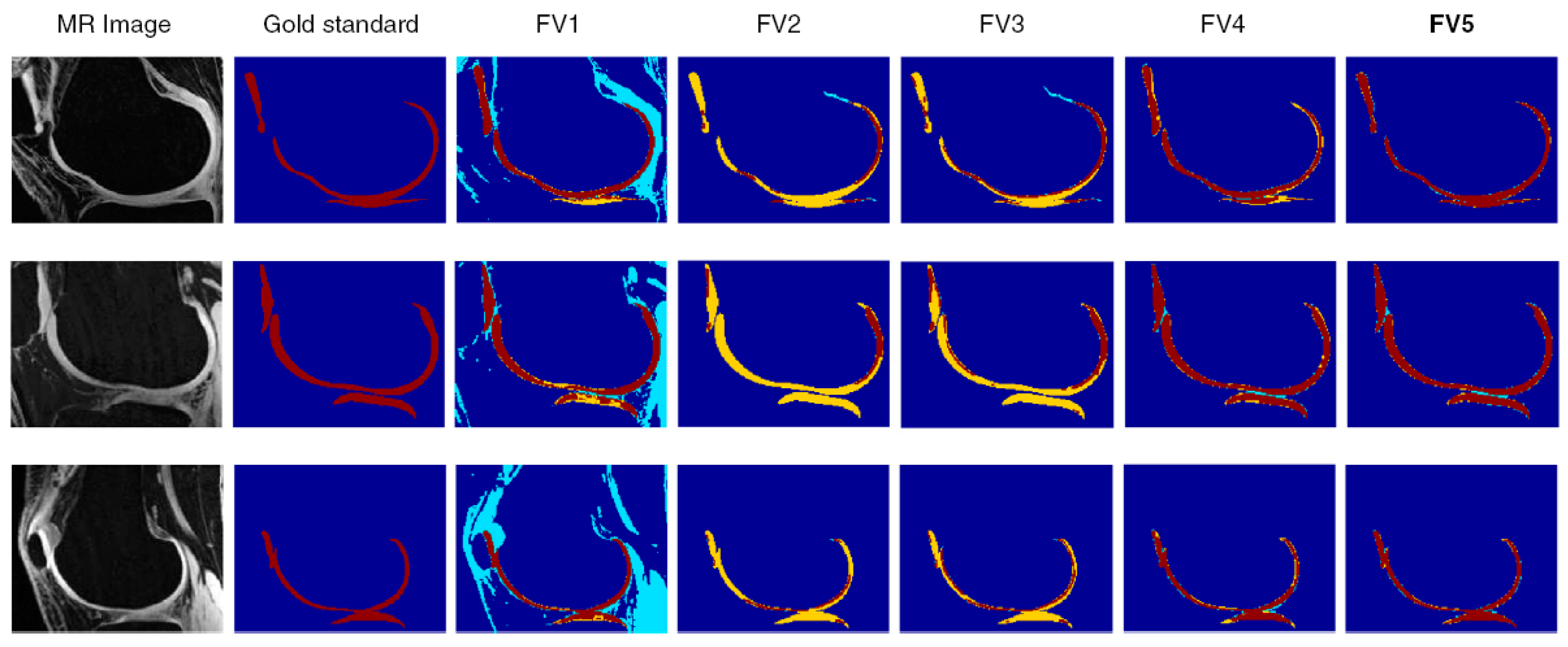


| Ref. | Year | Segmentation Technique | No. of Samples | Sequence Type | Region of Interest | Metric |
|---|---|---|---|---|---|---|
| [32] | 2007 | ASM-SSM | 20 health samples | FS SPGR | Femur/Tibia /Patella | DSC: 0.96(FB); 0.96(TB) and 0.89 (PB) |
| [33] | 2010 | AAM | 80 subjects | DESS | Femur/Tibia /Cartilage | AvgD:0.88 (±0.24) (FB); 0.74 (±0.21) (TB), RMSD: 1.49 (±0.44) (FB); 1.21 (±0.34) (TB) AvgD: 36.3 (±5.3) (FC); 34.6 (±7.9) (TC), RMSD: −25.2 (±10.1) (FC); −9.5 (±18.8) (TC) |
| [34] | 2010 | ASM-AAM | 40 clinical MRI samples | T1 weighted SPGR | Femur/Tibia /Cartilage | AvgD:1.02 (±0.22) (FB); 0.84 (±0.19) (TB), RMSD: 1.54 (±0.30) (FB); 1.24 (±0.28) (TB) AvgD: 34.0 (±12.7) (FC); 29.2 (±8.6) (TC), RMSD: 7.7 (±19.2) (FC); −2.7 (±18.2) (TC) |
| [35] | 2011 | SSM | 40 clinical samples | CTF | Femur/Tibia | For single-object (SSM) DICE: 0.94 (±0.02) (FB); 0.86 (±0.10) (TB) |
| [15] | 2013 | AAM | 178 samples | Sagittal 3-D double-echo | Femur/Tibia /Patella | Odds ratio 12.5 [95% CI 4.0–39.3] for (K/L grade of 0) and [95% CI] 1.8–5.0, p < 0.0001 for OA after 12 months in patients in the lowest tertile grade compared to those in the top tertile grade. |
| [19] | 2010 | LOGISMOS | 69 images | 3D DESS WE | Femur/Tibia/Patella | DSC ± SD: 0.84 ± 0.04(FC);0.80 ± 0.04 (TC); 0.80 ± 0.04 (PC) |
| [45] | 2011 | Graph cuts | 376 images | T2-weighted | Femur/Tibia | DSC: 0.936 (FB); 0.946 (TB); 0.941 (FB + TB) |
| [44] | 2009 | Graph cuts | 8 images | DESS | Femur/Tibia/Patella | DSC: 0.961 (FB); 0.857 (PB); 0.970 (TB); 0.958 |
| [46] | 2010 | Graph cuts | 30 images | T2 sagittal map | Femur/Tibia | Zijdenbos Similarity Index (ZSI) for Avg 95%; Std 0.028; Median 0.96; Min 0.87; Max 0.98. |
| [47] | 2020 | Graph cuts | 65 slices | T1 sequence | Femur/Tibia | Mean Square Error (MSE): 0.19 |
| [50] | 2014 | Multi-atlas | 100 training; 50 test | T1 weighted GRE FS | Femur/Tibia | ASD ± SD: 0.63 ± 0.17 mm (FB); 0.53 ± 0.25 mm (TB) |
| [51] | 2015 | Multi-atlas, KNN | The samples from CCBR OAI and SKI10 were used | T1 weighted Turbo 3D | Tibia | DSC ± SD (training): 0.975 ± 0.010 (TB) |
| [55] | 2011 | Ray casting | 161 samples | GRE FS | Femur/Tibia | DSC ± SD: 0.94 ± 0.05 (FB);0.92 ± 0.07 (TB) |
| [56] | 2007 | Region growing; Level set | 2 samples | T1 weighted | Femur/Tibia/Patella | Sens: 97.05% (FB); 96.95%(TB); 92.69% (PB) Spec: 98.79% (FB); 98.33%(TB); |
| [57] | 2017 | Level set; predefined Threshold | 8 samples | DESS | Femur/Tibia | DSC ± SD: 90.28 ± 2.33% (FB); 91.35 ± 2.22% (TB) |
| [54] | 2005 | FLoG edge detector; Threshold; Wavelet transforms (WT) | 40 samples | GE Signa Horizon LX 1.5 Tesla | Femur/Patella | The results show that the proposed method can segment the femur and patella robustly even under bad imaging conditions. |
| Ref | Year | Data | Dataset | Feature Engineering | Learning Algorithm | Validation | Results |
|---|---|---|---|---|---|---|---|
| [69] | 2019 | X-ray | OAI | ICA | Random forest; Naïve Bayes | Leave-One-Out (LOO) | 87.15% sensitivity; 82.98% accuracy and up to 80.65% for specificity |
| [70] | 2017 | MRI | From hospital | GLCM | SVM with the linear kernel; SVM with RBF kernel; SVM with polynomial kernel | 147 images training 66 images testing | 95.45% accuracy; 95.45% accuracy; 87.8% accuracy |
| [71] | 2018 | MRI | OAI | PCA | SVM Random forest Naïve Bayes ANN | 10-fold cross-validation | For JSL grade prediction the best performance was achieve for random forest AUC = 0.785 and F-measure = 0.743, while for the ANN with AUC = 0.695 and F-measure = 0.796. |
| [72] | 2016 | MRI | OAI | k-means clustering; Neighborhood approximation forests | LOGISMOS Forest Classifier Hierarchical Random | 108 baseline MRIs and 54 patients’ 12-month follow-up scans | 4D cartilage surface positioning errors (in millimeters) |
| [73] | 2018 | Pain scores and X-rays | OAI and MOST | PCA | LASSO regression | 10-fold cross -validation | AUC of 0.86 for Radiographic progression |
| [83] | 2018 | MRI | SKI10 | Not used | CNN | 3D-FSE images and T2 maps | ASD ± SD: 0.56 ± 0.12 (FB); 0.50 ± 0.14 (TB) |
| [84] | 2019 | MRI | SKI10,OAI imorphics, OAI ZIB | Not used | 2D/3D CNN and combination of (SSMs) | 2-fold cross-validation | (i) 74.0 ± 7.7 total score. (ii) DSC: 89.4% (FC). (iii) DIC: 98.6% (FB), 98.5% (TB), 85.6% (TC), 89.9% (FC). |
| [85] | 2020 | MR | National Institutes of Health (NIH), SKI10 | Not used | HNN deep learning | 9-fold cross-validation | DSC ± SD: 0.972 ± 0.054 (FB); 0.947 ± 0.0113 (PB) |
| [86] | 2019 | X-ray | Korea Centers for Disease Control and Prevention (KCDCP) | PCA | Deep Neural Network (DNN) | (66%) train (34%) test, 5F-CV, (50%) train (50%) test | 76.8% AUC |
| [87] | 2020 | X-ray | OAI,MOST | Not used | Ensemble and CNN | 19,704 train 11,743 test | 0.98 Average precision and 0.98 ROC |
| [88] | 2017 | X-ray | OAI,MOST | FCN | CNN | 30% testing 70% training | Accuracy 60.3% for (multi-class Grades 0–4) |
| [89] | 2018 | X-ray | OAI,MOST | FCN | CNN ResNet-34 | 67% train, 11% validation, 22% testing | Accuracy 66.71% (multi-class Grades 0–4) |
| [91] | 2019 | Clinical data, X-ray | OAI,MOST | CNN | Gradient Boosting Machine (GBM) and Logistic Regression (LR) | MOST dataset for testing and OAI dataset for training, 5F-CV | Accuracy 0.79 |
| [90] | 2019 | X-ray | OAI | Cascade | Deep Neural Network (DNN) | 10-fold cross | 82.98% Accuracy 87.15% Sensitivity 80.65% specificity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, S.M.; Mstafa, R.J. A Comprehensive Survey on Bone Segmentation Techniques in Knee Osteoarthritis Research: From Conventional Methods to Deep Learning. Diagnostics 2022, 12, 611. https://doi.org/10.3390/diagnostics12030611
Ahmed SM, Mstafa RJ. A Comprehensive Survey on Bone Segmentation Techniques in Knee Osteoarthritis Research: From Conventional Methods to Deep Learning. Diagnostics. 2022; 12(3):611. https://doi.org/10.3390/diagnostics12030611
Chicago/Turabian StyleAhmed, Sozan Mohammed, and Ramadhan J. Mstafa. 2022. "A Comprehensive Survey on Bone Segmentation Techniques in Knee Osteoarthritis Research: From Conventional Methods to Deep Learning" Diagnostics 12, no. 3: 611. https://doi.org/10.3390/diagnostics12030611
APA StyleAhmed, S. M., & Mstafa, R. J. (2022). A Comprehensive Survey on Bone Segmentation Techniques in Knee Osteoarthritis Research: From Conventional Methods to Deep Learning. Diagnostics, 12(3), 611. https://doi.org/10.3390/diagnostics12030611







