Tongue Contour Tracking and Segmentation in Lingual Ultrasound for Speech Recognition: A Review
Abstract
1. Introduction
2. Overview of Ultrasound Imaging in Speech Recognition
3. Evaluation Measures for Tongue Contour Extraction Using Ultrasound
3.1. Mean Sum of Distances (MSD)
3.2. Shape-Based Evaluation
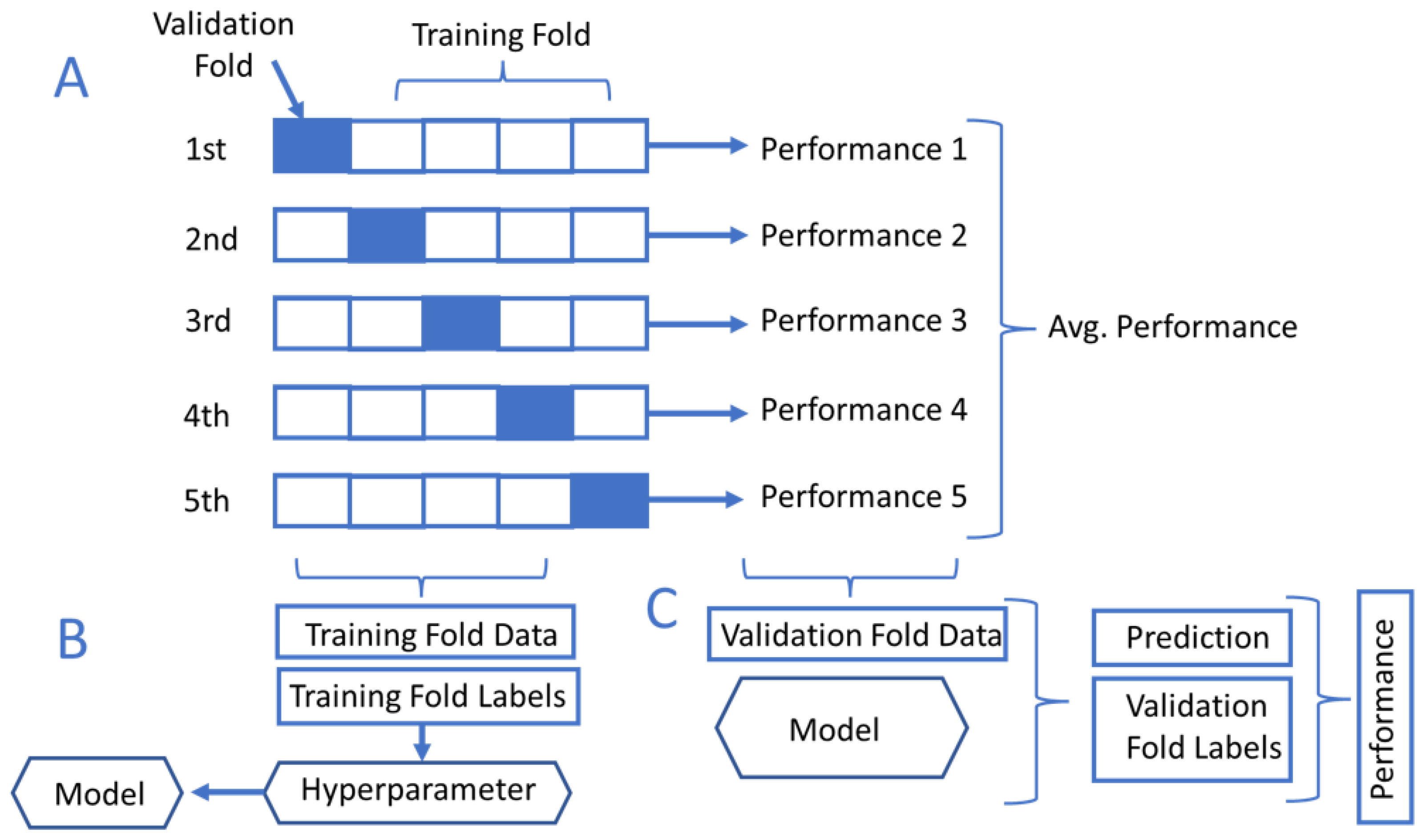
3.3. K-Fold Cross-Validation
3.4. Dice Score Coefficient (DC)
Mean Square Error (MSE)
4. Tongue Contour Tracking Techniques in Ultrasound Images
4.1. Traditional Image Analysis Techniques for Tongue Contour Tracking
4.1.1. Active-Contour-Based Methodologies (Snake Algorithm)
4.1.2. Shape Consistency and Graph-Based Tongue Tracking Methodologies
4.2. Machine-Learning-Based Techniques for Tongue Contour Tracking
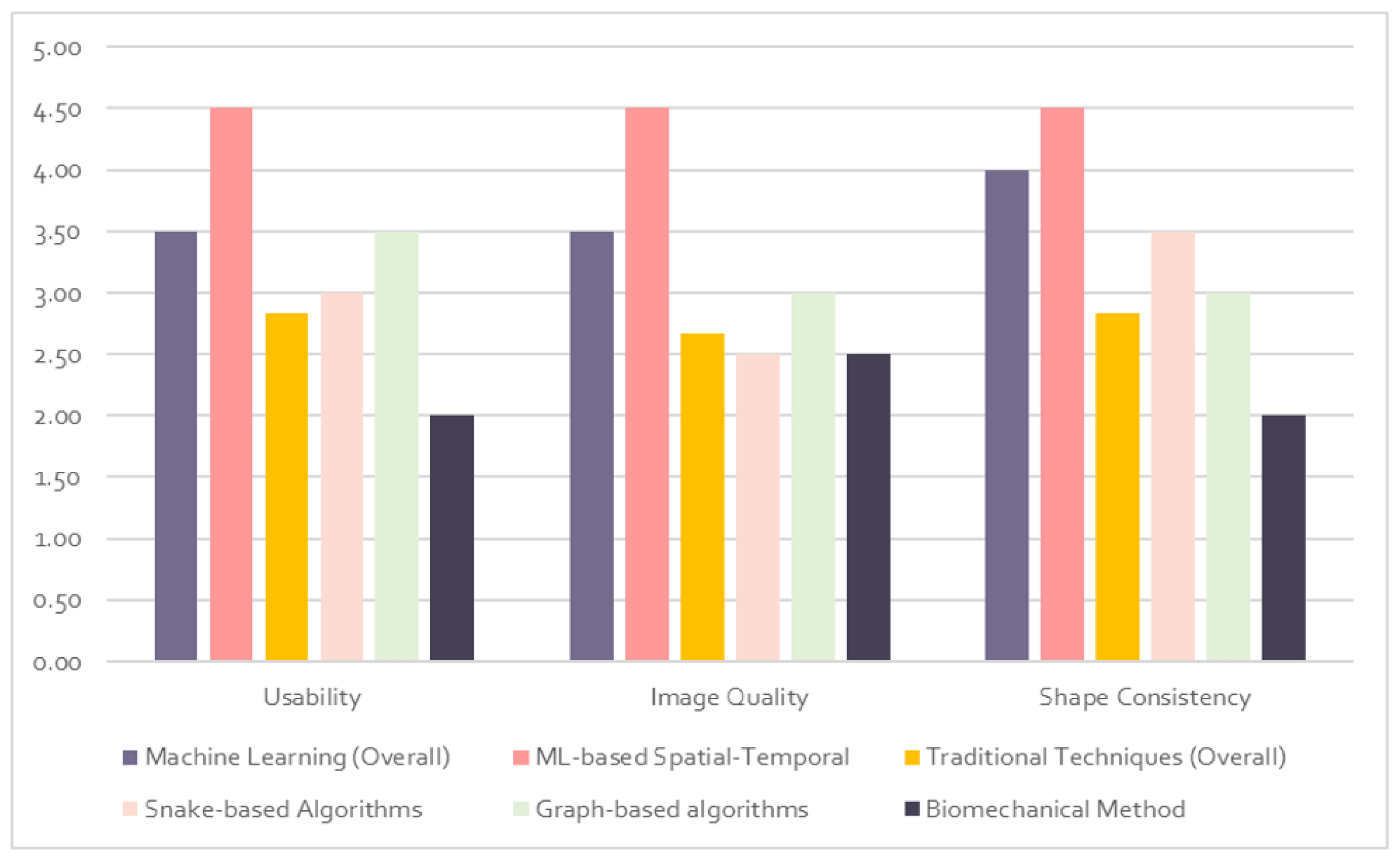
5. Results and Discussion
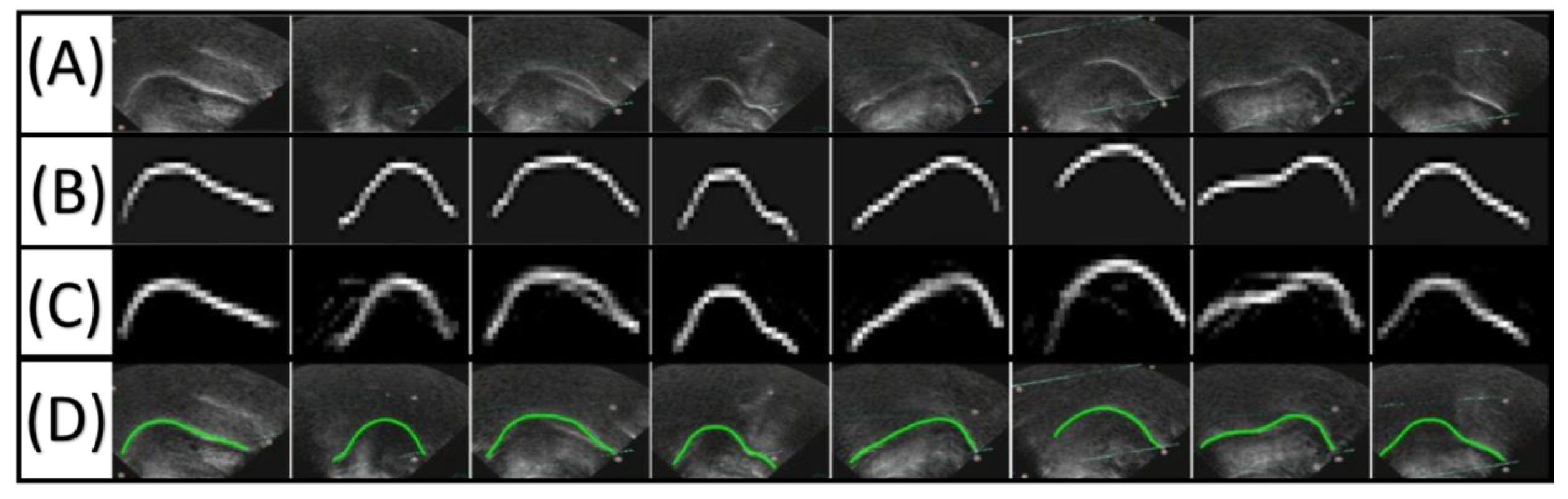
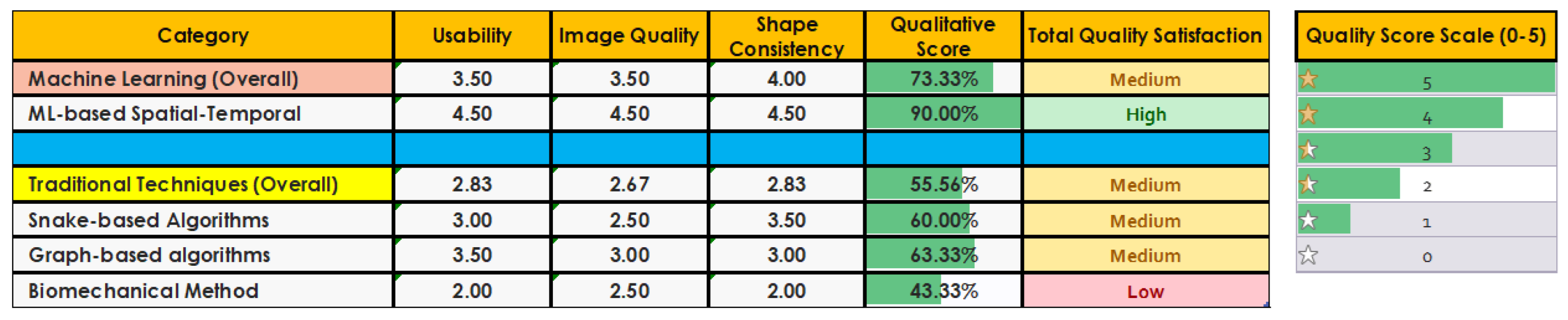
5.1. Qualitative Evaluation
5.2. Quantitative Evaluation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2D | Two dimensions |
| 3D | Three dimensions |
| ASM | Active shape model |
| CNN | Convolutional neural networks |
| CW | Complex wavelet |
| SSIM | Structural similarity index measure |
| CT | Computed tomography |
| DCAE | Denoising convolutional autoencoder |
| EV | Evaluation |
| EPG | Electropalatography |
| EMA | Electromagnetic articulatory |
| ECG | Electrocardiography |
| EEG | Electroencephalography |
| EMG | Electromyography |
| LSTM | Long short-term memory |
| MSD | Mean sum of distances |
| MSE | Mean squared error |
| MRI | Magnetic resonance imaging |
| ML | Machine learning |
| PMA | Permanent magnet articulator |
| PCA | Principal component analysis |
| RMSE | Root-mean-square error |
| SSI | Silent-speech interface |
| US | Ultrasound |
References
- Palmatier, R.W.; Houston, M.B.; Hulland, J. Review articles: Purpose, process, and structure. J. Acad. Mark. Sci. 2018, 46, 1–5. [Google Scholar] [CrossRef]
- Li, M.; Kambhamettu, C.; Stone, M. Automatic contour tracking in ultrasound images. Clin. Linguist. Phon. 2005, 19, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Bressmann, T.; Hamarneh, G. Tongue contour tracking in dynamic ultrasound via higher-order MRFs and efficient fusion moves. Med. Image Anal. 2012, 16, 1503–1520. [Google Scholar] [CrossRef]
- Laporte, C.; Ménard, L. Multi-hypothesis tracking of the tongue surface in ultrasound video recordings of normal and impaired speech. Med. Image Anal. 2018, 44, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Al-hammuri, K. Computer Vision-Based Tracking and Feature Extraction for Lingual Ultrasound. Ph.D. Thesis, University of Victoria, Victoria, BC, Canada, 2019. [Google Scholar]
- Karimi, E.; Ménard, L.; Laporte, C. Fully-automated tongue detection in ultrasound images. Comput. Biol. Med. 2019, 111, 103335. [Google Scholar] [PubMed]
- Cai, J.; Denby, B.; Roussel-Ragot, P.; Dreyfus, G.; Crevier-Buchman, L. Recognition and Real Time Performances of a Lightweight Ultrasound Based Silent Speech Interface Employing a Language Model. In Proceedings of the Interspeech, Florence, Italy, 27–31 August 2011; pp. 1005–1008. [Google Scholar]
- Lee, W.; Seong, J.J.; Ozlu, B.; Shim, B.S.; Marakhimov, A.; Lee, S. Biosignal sensors and deep learning-based speech recognition: A review. Sensors 2021, 21, 1399. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.S.; Eshky, A.; Richmond, K.; Renals, S. Silent versus modal multi-speaker speech recognition from ultrasound and video. arXiv 2021, arXiv:2103.00333. [Google Scholar]
- Stone, M. A guide to analysing tongue motion from ultrasound images. Clin. Linguist. Phon. 2005, 19, 455–501. [Google Scholar] [PubMed]
- Ramanarayanan, V.; Tilsen, S.; Proctor, M.; Töger, J.; Goldstein, L.; Nayak, K.S.; Narayanan, S. Analysis of speech production real-time MRI. Comput. Speech Lang. 2018, 52, 1–22. [Google Scholar]
- Deng, M.; Leotta, D.; Huang, G.; Zhao, Z.; Liu, Z. Craniofacial, tongue, and speech characteristics in anterior open bite patients of East African ethnicity. Res. Rep. Oral Maxillofac. Surg. 2019, 3, 21. [Google Scholar]
- Lingala, S.G.; Toutios, A.; Töger, J.; Lim, Y.; Zhu, Y.; Kim, Y.C.; Vaz, C.; Narayanan, S.S.; Nayak, K.S. State-of-the-Art MRI Protocol for Comprehensive Assessment of Vocal Tract Structure and Function. In Proceedings of the Interspeech, San Francisco, CA, USA, 8–12 September 2016; pp. 475–479. [Google Scholar]
- Köse, Ö.D.; Saraçlar, M. Multimodal representations for synchronized speech and real-time MRI video processing. IEEE/ACM Trans. Audio Speech Lang. Process. 2021, 29, 1912–1924. [Google Scholar] [CrossRef]
- Isaieva, K.; Laprie, Y.; Houssard, A.; Felblinger, J.; Vuissoz, P.A. Tracking the tongue contours in rt-MRI films with an autoencoder DNN approach. In Proceedings of the ISSP 2020—12th International Seminar on Speech Production, Online, 14–18 December 2020. [Google Scholar]
- Zhao, Z.; Lim, Y.; Byrd, D.; Narayanan, S.; Nayak, K.S. Improved 3D real-time MRI of speech production. Magn. Reson. Med. 2021, 85, 3182–3195. [Google Scholar] [CrossRef] [PubMed]
- Xing, F. Three Dimensional Tissue Motion Analysis from Tagged Magnetic Resonance Imaging. Ph.D. Thesis, Johns Hopkins University, Baltimore, MD, USA, 2015. [Google Scholar]
- Höwing, F.; Dooley, L.S.; Wermser, D. Tracking of non-rigid articulatory organs in X-ray image sequences. Comput. Med. Imaging Graph. 1999, 23, 59–67. [Google Scholar] [CrossRef]
- Sock, R.; Hirsch, F.; Laprie, Y.; Perrier, P.; Vaxelaire, B.; Brock, G.; Bouarourou, F.; Fauth, C.; Ferbach-Hecker, V.; Ma, L.; et al. An X-ray database, tools and procedures for the study of speech production. In Proceedings of the ISSP 2011—9th International Seminar on Speech Production, Montreal, QC, Canada, 20–23 June 2011; pp. 41–48. [Google Scholar]
- Yu, J. Speech Synchronized Tongue Animation by Combining Physiology Modeling and X-ray Image Fitting. In Proceedings of the International Conference on Multimedia Modeling, Reykjavik, Iceland, 4–6 January 2017; pp. 726–737. [Google Scholar]
- Guijarro-Martínez, R.; Swennen, G. Cone-beam computerized tomography imaging and analysis of the upper airway: A systematic review of the literature. Int. J. Oral Maxillofac. Surg. 2011, 40, 1227–1237. [Google Scholar] [PubMed]
- Hou, T.N.; Zhou, L.N.; Hu, H.J. Computed tomographic angiography study of the relationship between the lingual artery and lingual markers in patients with obstructive sleep apnoea. Clin. Radiol. 2011, 66, 526–529. [Google Scholar] [PubMed]
- Kim, S.H.; Choi, S.K. Changes in the hyoid bone, tongue, and oropharyngeal airway space after mandibular setback surgery evaluated by cone-beam computed tomography. Maxillofac. Plast. Reconstr. Surg. 2020, 42, 27. [Google Scholar] [CrossRef]
- Sierhej, A.; Verhoeven, J.; Miller, N.R.; Reyes-Aldasoro, C.C. Optimisation strategies for the registration of Computed Tomography images of electropalatography. bioRxiv 2020. [Google Scholar] [CrossRef]
- Guo, X.; Liang, X.; Jin, J.; Chen, J.; Liu, J.; Qiao, Y.; Cheng, J.; Zhao, J. Three-dimensional computed tomography mapping of 136 tongue-type calcaneal fractures from a single centre. Ann. Transl. Med. 2021, 9, 1787. [Google Scholar]
- Yang, M.; Tao, J.; Zhang, D. Extraction of tongue contour in X-ray videos. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 1094–1098. [Google Scholar]
- Luo, C.; Li, R.; Yu, L.; Yu, J.; Wang, Z. Automatic Tongue Tracking in X-ray Images. Chin. J. Electron. 2015, 24, 767–771. [Google Scholar]
- Laprie, Y.; Loosvelt, M.; Maeda, S.; Sock, R.; Hirsch, F. Articulatory copy synthesis from cine X-ray films. In Proceedings of the InterSpeech—14th Annual Conference of the International Speech Communication Association 2013, Lyon, France, 25–29 August 2013. [Google Scholar]
- Berger, M.O.; Erard Mozelle, G.; Laprie, Y. Cooperation of Active Contours and Optical Ow for Tongue Tracking in X-ray Motion Pictures. 1995. Available online: https://members.loria.fr/MOBerger/PublisAvant2004/tongueSCIA95.pdf (accessed on 10 June 2022).
- Thimm, G. Tracking articulators in X-ray movies of the vocal tract. In Proceedings of the International Conference on Computer Analysis of Images and Patterns, Ljubljana, Slovenia, 1–3 September 1999; pp. 126–133. [Google Scholar]
- Koren, A.; Grošelj, L.D.; Fajdiga, I. CT comparison of primary snoring and obstructive sleep apnea syndrome: Role of pharyngeal narrowing ratio and soft palate-tongue contact in awake patient. Eur. Arch. Oto-Rhino 2009, 266, 727–734. [Google Scholar]
- Uysal, T.; Yagci, A.; Ucar, F.I.; Veli, I.; Ozer, T. Cone-beam computed tomography evaluation of relationship between tongue volume and lower incisor irregularity. Eur. J. Orthod. 2013, 35, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, Y.; Ogawa, T.; Ando, E.; Clark, G.T.; Enciso, R. Influence of tongue/mandible volume ratio on oropharyngeal airway in Japanese male patients with obstructive sleep apnea. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2011, 111, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Suzuki, S.; Shiga, M.; Ohbayashi, N.; Kurabayashi, T.; Moriyama, K. Evaluation of tongue volume and oral cavity capacity using cone-beam computed tomography. Odontology 2018, 106, 266–273. [Google Scholar] [PubMed]
- Rana, S.; Kharbanda, O.; Agarwal, B. Influence of tongue volume, oral cavity volume and their ratio on upper airway: A cone beam computed tomography study. J. Oral Biol. Craniofacial Res. 2020, 10, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Eggers, G.; Kress, B.; Rohde, S.; Muhling, J. Intraoperative computed tomography and automated registration for image-guided cranial surgery. Dentomaxillofacial Radiol. 2009, 38, 28–33. [Google Scholar]
- Liu, W.P.; Richmon, J.D.; Sorger, J.M.; Azizian, M.; Taylor, R.H. Augmented reality and cone beam CT guidance for transoral robotic surgery. J. Robot. Surg. 2015, 9, 223–233. [Google Scholar]
- Zhong, Y.W.; Jiang, Y.; Dong, S.; Wu, W.J.; Wang, L.X.; Zhang, J.; Huang, M.W. Tumor radiomics signature for artificial neural network-assisted detection of neck metastasis in patient with tongue cancer. J. Neuroradiol. 2022, 49, 213–218. [Google Scholar]
- Khanal, S.; Johnson, M.T.; Bozorg, N. Articulatory Comparison of L1 and L2 Speech for Mispronunciation Diagnosis. In Proceedings of the 2021 IEEE Spoken Language Technology Workshop (SLT), Shenzhen, China, 19–22 January 2021; pp. 693–697. [Google Scholar]
- Medina, S.; Tome, D.; Stoll, C.; Tiede, M.; Munhall, K.; Hauptmann, A.G.; Matthews, I. Speech Driven Tongue Animation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 20406–20416. [Google Scholar]
- Shaw, J.A.; Oh, S.; Durvasula, K.; Kochetov, A. Articulatory coordination distinguishes complex segments from segment sequences. Phonology 2021, 38, 437–477. [Google Scholar] [CrossRef]
- Hofe, R.; Ell, S.R.; Fagan, M.J.; Gilbert, J.M.; Green, P.D.; Moore, R.K.; Rybchenko, S.I. Small-vocabulary speech recognition using a silent speech interface based on magnetic sensing. Speech Commun. 2013, 55, 22–32. [Google Scholar] [CrossRef]
- Cheah, L.A.; Gilbert, J.M.; Gonzalez, J.A.; Bai, J.; Ell, S.R.; Green, P.D.; Moore, R.K. Towards an Intraoral-Based Silent Speech Restoration System for Post-laryngectomy Voice Replacement. In Proceedings of the International Joint Conference on Biomedical Engineering Systems and Technologies, Rome, Italy, 21–23 February 2016; pp. 22–38. [Google Scholar]
- Gonzalez, J.A.; Green, P.D. A real-time silent speech system for voice restoration after total laryngectomy. Rev. Logop. Foniatría Audiol. 2018, 38, 148–154. [Google Scholar]
- Cheah, L.A.; Gilbert, J.M.; González, J.A.; Green, P.D.; Ell, S.R.; Moore, R.K.; Holdsworth, E. A Wearable Silent Speech Interface based on Magnetic Sensors with Motion-Artefact Removal. In Proceedings of the BIODEVICES, Funchal, Portugal, 19–21 January 2018; pp. 56–62. [Google Scholar]
- Sebkhi, N. A Novel Wireless Tongue Tracking System for Speech Applications. Ph.D. Thesis, Georgia Institute of Technology, Atlanta, GA, USA, 2019. [Google Scholar]
- Lee, A.; Liker, M.; Fujiwara, Y.; Yamamoto, I.; Takei, Y.; Gibbon, F. EPG research and therapy: Further developments. Clin. Linguist. Phon. 2022, 1–21. [Google Scholar] [CrossRef]
- Chen, L.C.; Chen, P.H.; Tsai, R.T.H.; Tsao, Y. EPG2S: Speech Generation and Speech Enhancement based on Electropalatography and Audio Signals using Multimodal Learning. IEEE Signal Process. Lett. 2022. [Google Scholar] [CrossRef]
- Wand, M.; Schultz, T.; Schmidhuber, J. Domain-Adversarial Training for Session Independent EMG-based Speech Recognition. In Proceedings of the Interspeech, Hyderabad, India, 2–6 September 2018; pp. 3167–3171. [Google Scholar]
- Ratnovsky, A.; Malayev, S.; Ratnovsky, S.; Naftali, S.; Rabin, N. EMG-based speech recognition using dimensionality reduction methods. J. Ambient. Intell. Humaniz. Comput. 2021, 1–11. [Google Scholar] [CrossRef]
- Cha, H.S.; Chang, W.D.; Im, C.H. Deep-learning-based real-time silent speech recognition using facial electromyogram recorded around eyes for hands-free interfacing in a virtual reality environment. Virtual Real. 2022, 26, 1047–1057. [Google Scholar] [CrossRef]
- Xiong, D.; Zhang, D.; Zhao, X.; Zhao, Y. Deep learning for EMG-based human-machine interaction: A review. IEEE/CAA J. Autom. Sin. 2021, 8, 512–533. [Google Scholar]
- Hayashi, H.; Tsuji, T. Human–Machine Interfaces Based on Bioelectric Signals: A Narrative Review with a Novel System Proposal. IEEJ Trans. Electr. Electron. Eng. 2022, 17, 1536–1544. [Google Scholar]
- Harada, R.; Hojyo, N.; Fujimoto, K.; Oyama, T. Development of Communication System from EMG of Suprahyoid Muscles Using Deep Learning. In Proceedings of the 2022 IEEE 4th Global Conference on Life Sciences and Technologies (LifeTech), Osaka, Japan, 7–9 March 2022; pp. 5–9. [Google Scholar]
- Zhang, Q.; Jing, J.; Wang, D.; Zhao, R. WearSign: Pushing the Limit of Sign Language Translation Using Inertial and EMG Wearables. Proc. ACM Interact. Mobile Wearable Ubiquitous Technol. 2022, 6, 1–27. [Google Scholar]
- Krishna, G.; Tran, C.; Carnahan, M.; Han, Y.; Tewfik, A.H. Improving eeg based continuous speech recognition. arXiv 2019, arXiv:1911.11610. [Google Scholar]
- Görür, K.; Bozkurt, M.R.; Bascil, M.S.; Temurtas, F. Tongue-operated biosignal over EEG and processing with decision tree and kNN. Acad. Platf.-J. Eng. Sci. 2021, 9, 112–125. [Google Scholar]
- Akshi; Rao, M. Decoding imagined speech using wearable EEG headset for a single subject. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 2622–2627. [Google Scholar]
- Bakhshali, M.A.; Khademi, M.; Ebrahimi-Moghadam, A. Investigating the neural correlates of imagined speech: An EEG-based connectivity analysis. Digit. Signal Process. 2022, 123, 103435. [Google Scholar]
- Koctúrová, M.; Juhár, J. A Novel approach to EEG speech activity detection with visual stimuli and mobile BCI. Appl. Sci. 2021, 11, 674. [Google Scholar] [CrossRef]
- Lovenia, H.; Tanaka, H.; Sakti, S.; Purwarianti, A.; Nakamura, S. Speech artifact removal from EEG recordings of spoken word production with tensor decomposition. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1115–1119. [Google Scholar]
- Krishna, G.; Tran, C.; Yu, J.; Tewfik, A.H. Speech recognition with no speech or with noisy speech. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1090–1094. [Google Scholar]
- Lee, Y.E.; Lee, S.H. Eeg-transformer: Self-attention from transformer architecture for decoding eeg of imagined speech. In Proceedings of the 2022 10th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Korea, 21–23 February 2022; pp. 1–4. [Google Scholar]
- Krishna, G.; Tran, C.; Carnahan, M.; Tewfik, A. Improving EEG based continuous speech recognition using GAN. arXiv 2020, arXiv:2006.01260. [Google Scholar]
- Wilson, I. Using ultrasound for teaching and researching articulation. Acoust. Sci. Technol. 2014, 35, 285–289. [Google Scholar] [CrossRef][Green Version]
- Gick, B.; Bernhardt, B.; Bacsfalvi, P.; Wilson, I.; Zampini, M. Ultrasound imaging applications in second language acquisition. Phonol. Second Lang. Acquis. 2008, 36, 309–322. [Google Scholar]
- Li, S.R.; Dugan, S.; Masterson, J.; Hudepohl, H.; Annand, C.; Spencer, C.; Seward, R.; Riley, M.A.; Boyce, S.; Mast, T.D. Classification of accurate and misarticulated /ar/for ultrasound biofeedback using tongue part displacement trajectories. Clin. Linguist. Phon. 2022, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Eshky, A.; Ribeiro, M.S.; Cleland, J.; Richmond, K.; Roxburgh, Z.; Scobbie, J.; Wrench, A. UltraSuite: A repository of ultrasound and acoustic data from child speech therapy sessions. arXiv 2019, arXiv:1907.00835. [Google Scholar]
- McKeever, L.; Cleland, J.; Delafield-Butt, J. Using ultrasound tongue imaging to analyse maximum performance tasks in children with Autism: A pilot study. Clin. Linguist. Phon. 2022, 36, 127–145. [Google Scholar] [CrossRef]
- Castillo, M.; Rubio, F.; Porras, D.; Contreras-Ortiz, S.H.; Sepúlveda, A. A small vocabulary database of ultrasound image sequences of vocal tract dynamics. In Proceedings of the 2019 XXII Symposium on Image, Signal Processing and Artificial Vision (STSIVA), Bucaramanga, Colombia, 24–26 April 2019; pp. 1–5. [Google Scholar]
- Ohkubo, M.; Scobbie, J.M. Tongue shape dynamics in swallowing using sagittal ultrasound. Dysphagia 2019, 34, 112–118. [Google Scholar] [CrossRef]
- Chen, S.; Zheng, Y.; Wu, C.; Sheng, G.; Roussel, P.; Denby, B. Direct, Near Real Time Animation of a 3D Tongue Model Using Non-Invasive Ultrasound Images. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 4994–4998. [Google Scholar]
- Ji, Y.; Liu, L.; Wang, H.; Liu, Z.; Niu, Z.; Denby, B. Updating the silent speech challenge benchmark with deep learning. Speech Commun. 2018, 98, 42–50. [Google Scholar]
- Denby, B.; Schultz, T.; Honda, K.; Hueber, T.; Gilbert, J.M.; Brumberg, J.S. Silent speech interfaces. Speech Commun. 2010, 52, 270–287. [Google Scholar]
- Gonzalez-Lopez, J.A.; Gomez-Alanis, A.; Doñas, J.M.M.; Pérez-Córdoba, J.L.; Gomez, A.M. Silent speech interfaces for speech restoration: A review. IEEE Access 2020, 8, 177995–178021. [Google Scholar] [CrossRef]
- Noble, J.A.; Boukerroui, D. Ultrasound image segmentation: A survey. IEEE Trans. Med. Imaging 2006, 25, 987–1010. [Google Scholar] [PubMed]
- Huang, H.; Ge, Z.; Wang, H.; Wu, J.; Hu, C.; Li, N.; Wu, X.; Pan, C. Segmentation of Echocardiography Based on Deep Learning Model. Electronics 2022, 11, 1714. [Google Scholar]
- Hu, Y.; Guo, Y.; Wang, Y.; Yu, J.; Li, J.; Zhou, S.; Chang, C. Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med. Phys. 2019, 46, 215–228. [Google Scholar]
- Wang, T.; Lei, Y.; Axente, M.; Yao, J.; Lin, J.; Bradley, J.D.; Liu, T.; Xu, D.; Yang, X. Automatic breast ultrasound tumor segmentation via one-stage hierarchical target activation network. In Proceedings of the Medical Imaging 2022: Ultrasonic Imaging and Tomography, San Diego, CA, USA, 20 February–28 March 2022; Volume 12038, pp. 137–142. [Google Scholar]
- Lei, Y.; He, X.; Yao, J.; Wang, T.; Wang, L.; Li, W.; Curran, W.J.; Liu, T.; Xu, D.; Yang, X. Breast tumor segmentation in 3D automatic breast ultrasound using Mask scoring R-CNN. Med. Phys. 2021, 48, 204–214. [Google Scholar] [PubMed]
- Yang, J.; Tong, L.; Faraji, M.; Basu, A. IVUS-Net: An intravascular ultrasound segmentation network. In Proceedings of the International Conference on Smart Multimedia, Toulon, France, 24–26 August 2018; pp. 367–377. [Google Scholar]
- Du, H.; Ling, L.; Yu, W.; Wu, P.; Yang, Y.; Chu, M.; Yang, J.; Yang, W.; Tu, S. Convolutional networks for the segmentation of intravascular ultrasound images: Evaluation on a multicenter dataset. Comput. Methods Programs Biomed. 2022, 215, 106599. [Google Scholar]
- Allan, M.B.; Jafari, M.H.; Woudenberg, N.V.; Frenkel, O.; Murphy, D.; Wee, T.; D’Ortenzio, R.; Wu, Y.; Roberts, J.; Shatani, N.; et al. Multi-task deep learning for segmentation and landmark detection in obstetric sonography. In Proceedings of the Medical Imaging 2022: Image-Guided Procedures, Robotic Interventions, and Modeling, San Diego, CA, USA, 20–23 February 2022; Volume 12034, pp. 160–165. [Google Scholar]
- Bushra, S.N.; Shobana, G. Obstetrics and gynaecology ultrasound image analysis towards cryptic pregnancy using deep learning-a review. In Proceedings of the 2021 5th International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 6–8 May 2021; pp. 949–953. [Google Scholar]
- Zhang, Z.; Han, Y. Detection of Ovarian Tumors in Obstetric Ultrasound Imaging Using Logistic Regression Classifier With an Advanced Machine Learning Approach. IEEE Access 2020, 8, 44999–45008. [Google Scholar] [CrossRef]
- Gaillard, F. Muscles of the Tongue. Reference Article. Available online: Radiopaedia.org (accessed on 14 August 2022).
- Csapó, T.G.; Xu, K.; Deme, A.; Gráczi, T.E.; Markó, A. Transducer Misalignment in Ultrasound Tongue Imaging. In Proceedings of the 12th International Seminar on Speech Production, Online, 14 December 2020; pp. 166–169. [Google Scholar]
- Ménard, L.; Aubin, J.; Thibeault, M.; Richard, G. Measuring tongue shapes and positions with ultrasound imaging: A validation experiment using an articulatory model. Folia Phoniatr. Logop. 2012, 64, 64–72. [Google Scholar] [CrossRef]
- Raschka, S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv 2018, arXiv:1811.12808. [Google Scholar]
- Stone, M.; Shawker, T.H. An ultrasound examination of tongue movement during swallowing. Dysphagia 1986, 1, 78–83. [Google Scholar]
- Kaburagi, T.; Honda, M. An ultrasonic method for monitoring tongue shape and the position of a fixed point on the tongue surface. J. Acoust. Soc. Am. 1994, 95, 2268–2270. [Google Scholar] [PubMed]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Iskarous, K. Detecting the edge of the tongue: A tutorial. Clin. Linguist. Phon. 2005, 19, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Akgul, Y.S.; Kambhamettu, C.; Stone, M. Extraction and tracking of the tongue surface from ultrasound image sequences. In Proceedings of the 1998 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (Cat. No. 98CB36231), Santa Barbara, CA, USA, 25 June 1998; pp. 298–303. [Google Scholar]
- Akgul, Y.S.; Kambhamettu, C.; Stone, M. Automatic motion analysis of the tongue surface from ultrasound image sequences. In Proceedings of the Workshop on Biomedical Image Analysis (Cat. No. 98EX162), Santa Barbara, CA, USA, 27 June 1998; pp. 126–132. [Google Scholar]
- Akgul, Y.S.; Kambhamettu, C.; Stone, M. Automatic extraction and tracking of the tongue contours. IEEE Trans. Med. Imaging 1999, 18, 1035–1045. [Google Scholar] [CrossRef]
- Qin, C.; Carreira-Perpinán, M.A.; Richmond, K.; Wrench, A.; Renals, S. Predicting Tongue Shapes from a Few Landmark Locations. Available online: http://hdl.handle.net/1842/3819 (accessed on 14 August 2022).
- Xu, K.; Yang, Y.; Stone, M.; Jaumard-Hakoun, A.; Leboullenger, C.; Dreyfus, G.; Roussel, P.; Denby, B. Robust contour tracking in ultrasound tongue image sequences. Clin. Linguist. Phon. 2016, 30, 313–327. [Google Scholar]
- Xu, K.; Gábor Csapó, T.; Roussel, P.; Denby, B. A comparative study on the contour tracking algorithms in ultrasound tongue images with automatic re-initialization. J. Acoust. Soc. Am. 2016, 139, EL154–EL160. [Google Scholar]
- Roussos, A.; Katsamanis, A.; Maragos, P. Tongue tracking in ultrasound images with active appearance models. In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 1733–1736. [Google Scholar]
- Aron, M.; Roussos, A.; Berger, M.O.; Kerrien, E.; Maragos, P. Multimodality acquisition of articulatory data and processing. In Proceedings of the 2008 16th European Signal Processing Conference, Lausanne, Switzerland, 25–29 August 2008; pp. 1–5. [Google Scholar]
- Tang, L.; Hamarneh, G. Graph-based tracking of the tongue contour in ultrasound sequences with adaptive temporal regularization. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition-Workshops, San Francisco, CA, USA, 13–18 June 2010; pp. 154–161. [Google Scholar]
- Loosvelt, M.; Villard, P.F.; Berger, M.O. Using a biomechanical model for tongue tracking in ultrasound images. In Proceedings of the International Symposium on Biomedical Simulation, Strasbourg, France, 16–17 October 2014; pp. 67–75. [Google Scholar]
- Fasel, I.; Berry, J. Deep belief networks for real-time extraction of tongue contours from ultrasound during speech. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 1493–1496. [Google Scholar]
- Jaumard-Hakoun, A.; Xu, K.; Roussel-Ragot, P.; Dreyfus, G.; Denby, B. Tongue contour extraction from ultrasound images based on deep neural network. arXiv 2016, arXiv:1605.05912. [Google Scholar]
- Fabre, D.; Hueber, T.; Bocquelet, F.; Badin, P. Tongue tracking in ultrasound images using eigentongue decomposition and artificial neural networks. In Proceedings of the Interspeech 2015—16th Annual Conference of the International Speech Communication Association, Dresden, Germany, 6–10 September 2015. [Google Scholar]
- Xu, K.; Roussel, P.; Csapó, T.G.; Denby, B. Convolutional neural network-based automatic classification of midsagittal tongue gestural targets using B-mode ultrasound images. J. Acoust. Soc. Am. 2017, 141, EL531–EL537. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Zhu, J.; Styler, W.; Calloway, I.C. Automatic tongue contour extraction in ultrasound images with convolutional neural networks. J. Acoust. Soc. Am. 2018, 143, 1966. [Google Scholar]
- Zhu, J.; Styler, W.; Calloway, I. A CNN-based tool for automatic tongue contour tracking in ultrasound images. arXiv 2019, arXiv:1907.10210. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Mozaffari, M.H.; Lee, W.S. Encoder-decoder CNN models for automatic tracking of tongue contours in real-time ultrasound data. Methods 2020, 179, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Mozaffari, M.H.; Yamane, N.; Lee, W.S. Deep Learning for Automatic Tracking of Tongue Surface in Real-Time Ultrasound Videos, Landmarks instead of Contours. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 2785–2792. [Google Scholar]
- Wen, S. Automatic Tongue Contour Segmentation Using Deep Learning. Ph.D. Thesis, University of Ottawa, Ottawa, ON, Canada, 2018. [Google Scholar]
- Li, B.; Xu, K.; Feng, D.; Mi, H.; Wang, H.; Zhu, J. Denoising convolutional autoencoder based B-mode ultrasound tongue image feature extraction. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 7130–7134. [Google Scholar]
- Zhao, C.; Zhang, P.; Zhu, J.; Wu, C.; Wang, H.; Xu, K. Predicting tongue motion in unlabeled ultrasound videos using convolutional LSTM neural networks. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 5926–5930. [Google Scholar]
- Feng, M.; Wang, Y.; Xu, K.; Wang, H.; Ding, B. Improving ultrasound tongue contour extraction using U-Net and shape consistency-based regularizer. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Virtual, 6–12 June 2021; pp. 6443–6447. [Google Scholar]
- Li, G.; Chen, J.; Liu, Y.; Wei, J. wUnet: A new network used for ultrasonic tongue contour extraction. Speech Commun. 2022, 141, 68–79. [Google Scholar] [CrossRef]
- Kimura, N.; Kono, M.; Rekimoto, J. SottoVoce: An ultrasound imaging-based silent speech interaction using deep neural networks. In Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems, Glasgow, UK, 4–9 May 2019; pp. 1–11. [Google Scholar]
- Povey, D.; Ghoshal, A.; Boulianne, G.; Burget, L.; Glembek, O.; Goel, N.; Hannemann, M.; Motlicek, P.; Qian, Y.; Schwarz, P.; et al. The Kaldi speech recognition toolkit. In Proceedings of the IEEE 2011 Workshop on Automatic Speech Recognition and Understanding, Waikoloa, HI, USA, 11–15 December 2011. [Google Scholar]
- Liu, F.T.; Ting, K.M.; Zhou, Z.H. Isolation forest. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15–19 December 2008; pp. 413–422. [Google Scholar]
- Wang, Z.; Bovik, A.; Sheikh, H.; Simoncelli, E. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Bakurov, I.; Buzzelli, M.; Schettini, R.; Castelli, M.; Vanneschi, L. Structural similarity index (SSIM) revisited: A data-driven approach. Expert Syst. Appl. 2022, 189, 116087. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Bansal, M.; Kumar, M.; Sachdeva, M.; Mittal, A. Transfer learning for image classification using VGG19: Caltech-101 image data set. J. Ambient. Intell. Humaniz. Comput. 2021, 1–12. [Google Scholar] [CrossRef]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Granada, Spain, 16 September 2018; pp. 1–11. [Google Scholar]


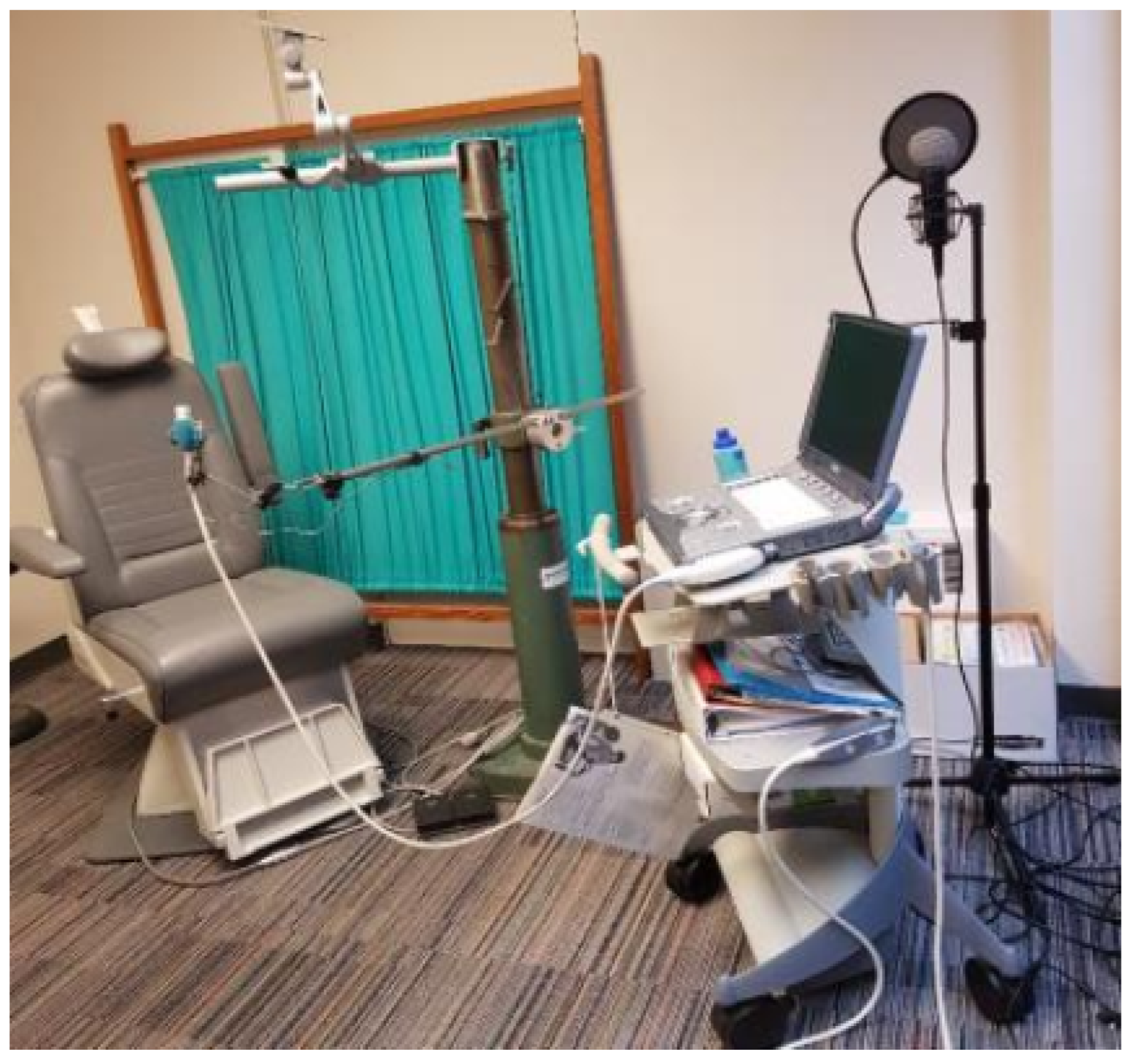

| Method | Category | EV. Measure | EV. Result | Data Type | Core Methodologies | Limitation |
|---|---|---|---|---|---|---|
| EdgeTrack [2] | Traditional | MSD | 0.53–1.0 mm | Tongue US images | Snake algorithm + gradient + local image information and object orientation | Sensitive to noise, computation complexity, can process only 80 US frames in one session |
| TongueTrack [3] | Traditional | MSD | 3 mm | Tongue US | Higher-order Markov random field energy minimization framework | Needs manual reinitialization, sensitive to noise, can process only 500 US frames in one session |
| Tongue shape prediction from landmarks [97] | Traditional | RMSE | 0.2–0.3 mm | Tongue US + EMA or X-ray | Spline interpolation + Landmark mapping using metal pellets | Difficult to use due to the limitation of the data collection |
| Graph-based [102] | Traditional | Mean segmentation error | Dense = 4.49 mm Sparse = 2.23 mm | Tongue US | Image graph-based analysis + adaptive temporal regularization using Markov random field optimization | Computation expensive due to optimizing too many parameters |
| Biomechanical [103] | Traditional | Accuracy | 0.62–0.97 mm | X-ray and US images for tongue and vocal tract | Harris features + optical flow | Sensitive to noise; not practical for ultrasound as it was trained on X-rays |
| Multihypothesis approach [4] | Traditional and machine learning | MSD | 1.69 ± 1.10 mm | Tongue US images | Snake algorithm + particle filter | Computation complexity, needs too many filter parameters to get accurate results |
| Computer vision-based tongue tracking and feature extraction [5] | Traditional | MSD | 0.933 mm | Tongue US images | Image denoising + tongue adaptive localization + feature extraction + data transformation and analysis | Did not use machine learning for feature extraction, which limits the scope of the feature map |
| Fully automate the tongue contour extraction [6] | Traditional | MSD | 1.01–0.63 mm | Tongue US images | Snake algorithm + phase symmetry filter + algorithm resetting | Computation complexity, too many constraints |
| Autotrace [104] | Machine Learning | MSD | 0.73 mm | Tongue US images | Deep learning + translational deep belief network | High computation cost; limited training dataset |
| Enhanced Autotrace [105] | Machine learning | MSD | 1.0 mm | Tongue US images | Deep autoencoder | Autoencoder has limited ability to classify features |
| CNN to automate the tongue segmentation [110] | Machine learning | MSD | U-net = 5.81 mm (Dense U-net) = 5.6 mm | Tongue US images | U-net + Dense U-net | U-net has limited generalization capability (Dense U-net) perform better than U-net but it is slower |
| BowNet and wBowNet [112] | Machine learning | MSD | 1.4mm | Tongue US images | Deep network in landmarks | Lack of global feature extraction of the CNN |
| TongueNet [113] | Machine learning | MSD | 0.31 pixel | Tongue US images | Multiscale contextual information + dilated convolution | Random selection of the annotated landmarks is not efficient in the method |
| DCAE-based B-Mode US [115] | Machine learning | Word error rate | 6.17 % | Tongue US images | Denoising convolutional autoencoder (DCAE) | Autoencoder has a limitation of classifying features in latent space and difficult to be generalized in global context |
| ConvLSTM [116] | Machine learning | MSE and CW-SSIM | MSE = 17.13 CW-SSIM = 0.932 | Tongue US images | CNN + LSTM | Limited memory, predicting up to nine future frames |
| U-NET and shape-consistency-based regularizer [117] | Traditional and machine learning | MSD | (2.243 ± 0.026) mm | Tongue US images | U-net architecture + temporal continuity using shape-consistency-based regularizer | Temporal continuity can be computational expensive for real-time applications |
| wUnet [118] | Machine learning | MSD | 1.18 mm | Tongue US images | U-net architecture + VGG19 block instead of skip connections | VGG19 may add unnecessary features to the network and cause overfitting |
| SottoVoce [119] | Machine learning | Speech recognition success ratio | 65% | Tongue US images + speech audio recording | Deep CNN | Acoustic sensor is not practical for smart systems integration |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-hammuri, K.; Gebali, F.; Thirumarai Chelvan, I.; Kanan, A. Tongue Contour Tracking and Segmentation in Lingual Ultrasound for Speech Recognition: A Review. Diagnostics 2022, 12, 2811. https://doi.org/10.3390/diagnostics12112811
Al-hammuri K, Gebali F, Thirumarai Chelvan I, Kanan A. Tongue Contour Tracking and Segmentation in Lingual Ultrasound for Speech Recognition: A Review. Diagnostics. 2022; 12(11):2811. https://doi.org/10.3390/diagnostics12112811
Chicago/Turabian StyleAl-hammuri, Khalid, Fayez Gebali, Ilamparithi Thirumarai Chelvan, and Awos Kanan. 2022. "Tongue Contour Tracking and Segmentation in Lingual Ultrasound for Speech Recognition: A Review" Diagnostics 12, no. 11: 2811. https://doi.org/10.3390/diagnostics12112811
APA StyleAl-hammuri, K., Gebali, F., Thirumarai Chelvan, I., & Kanan, A. (2022). Tongue Contour Tracking and Segmentation in Lingual Ultrasound for Speech Recognition: A Review. Diagnostics, 12(11), 2811. https://doi.org/10.3390/diagnostics12112811






