Metabolomic Selection in the Progression of Type 2 Diabetes Mellitus: A Genetic Algorithm Approach
Abstract
1. Introduction
2. Materials and Methods
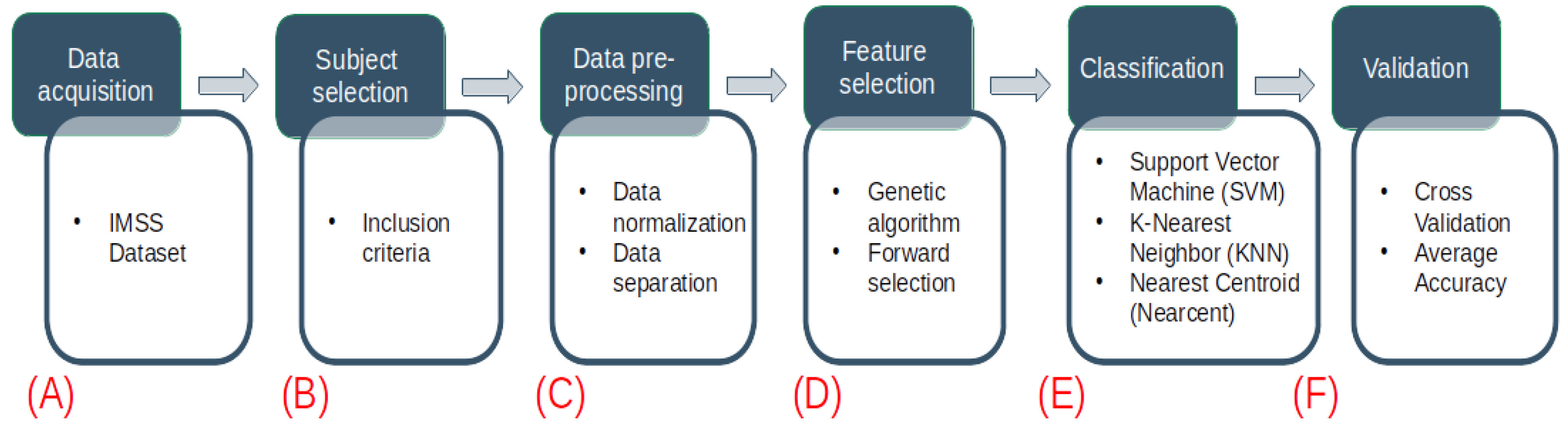
2.1. Sample
2.1.1. Sample Preparation
2.1.2. Quality Control (QC) and Quality Assurance (QA)
2.1.3. Ultra-Performance Liquid Chromatography (UPLC)—Mass Spectrometry Method for Lipid Separation
2.1.4. Data Processing
2.1.5. Metabolite Identification
2.2. IMSS Dataset
2.3. Data Inclusion
2.4. Data Normalization
2.5. Feature Selection
2.6. Model Development
2.7. K-Nearest Neighbors
2.8. Nearest Centroid
2.9. Support Vector Machines
2.10. Implementation
3. Results
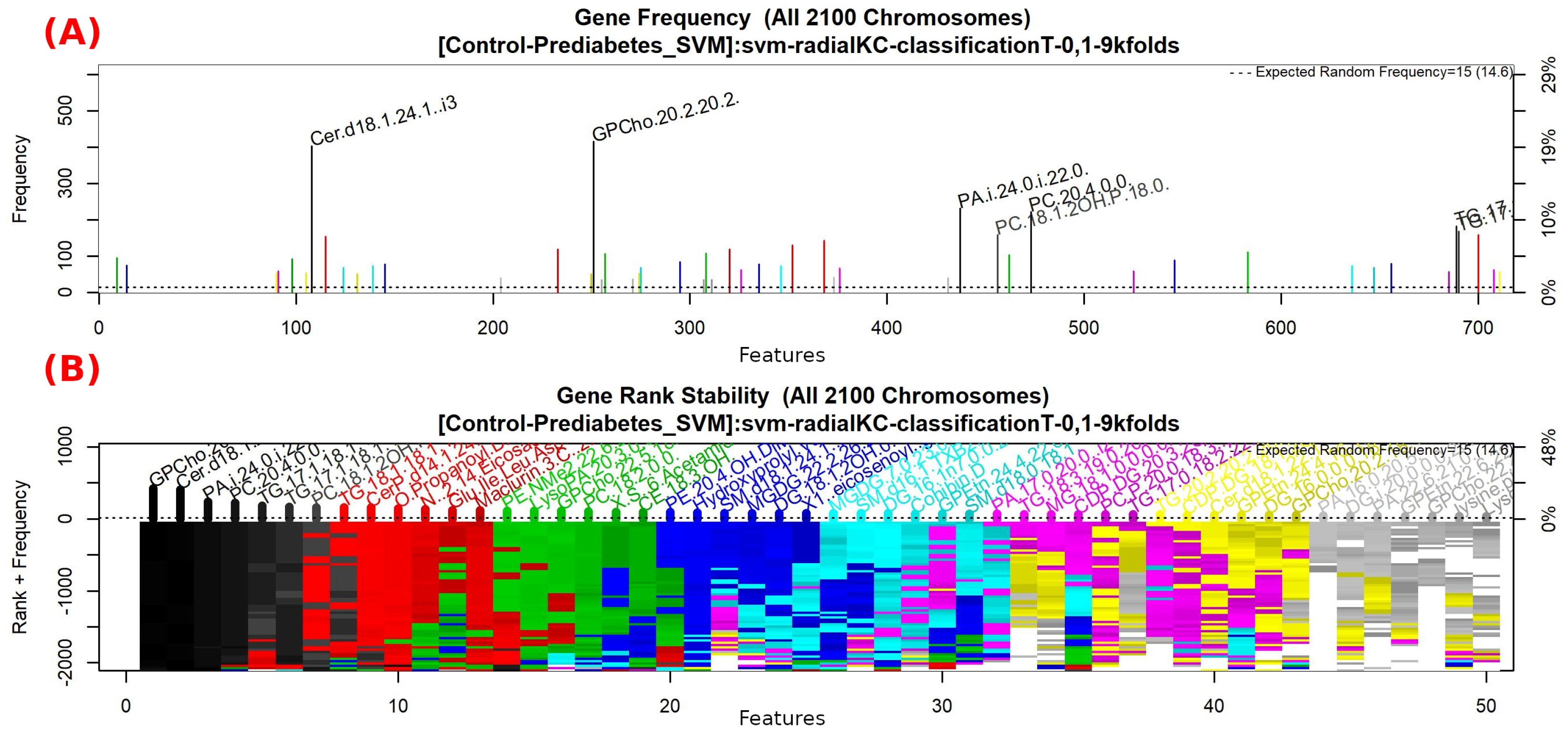
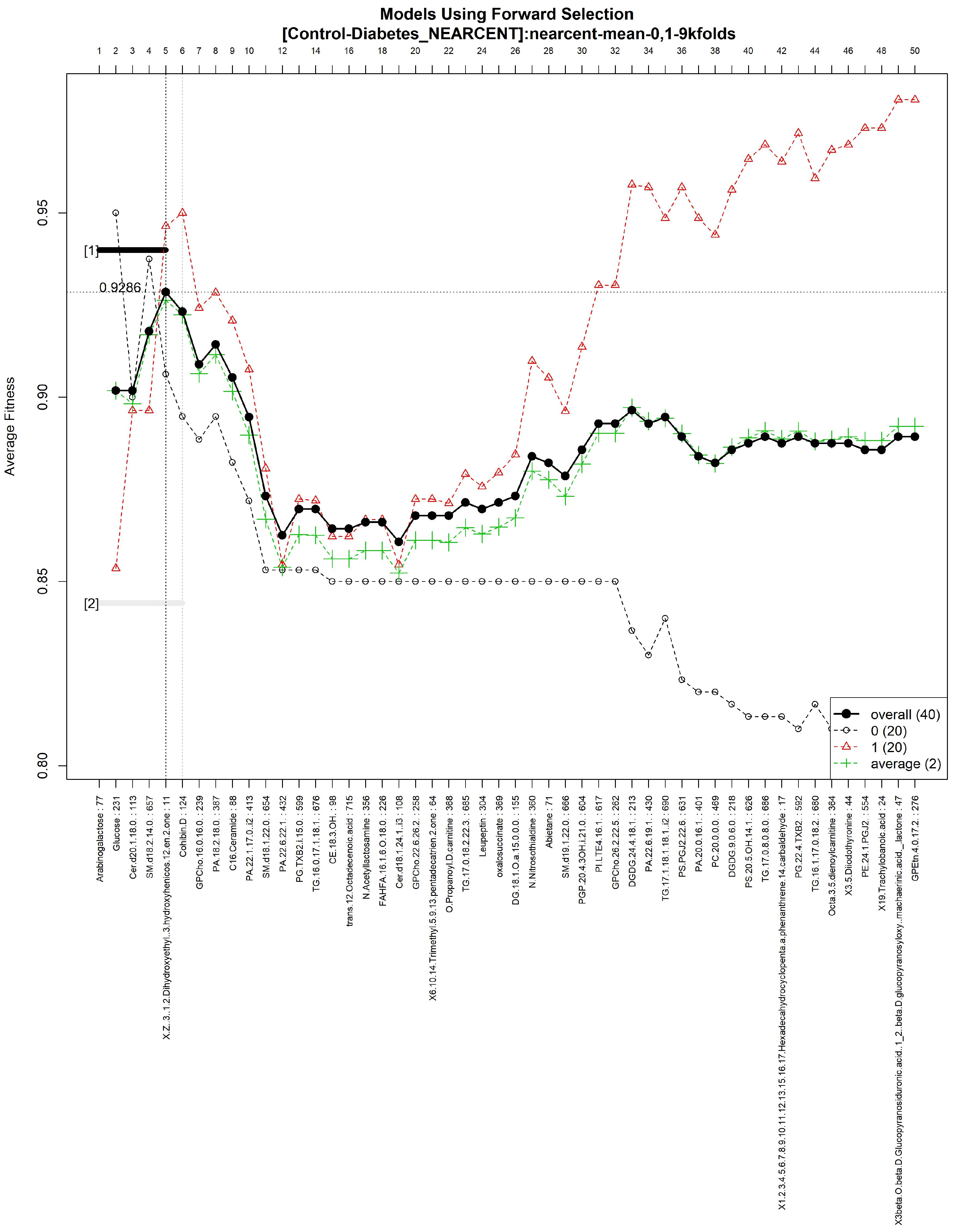
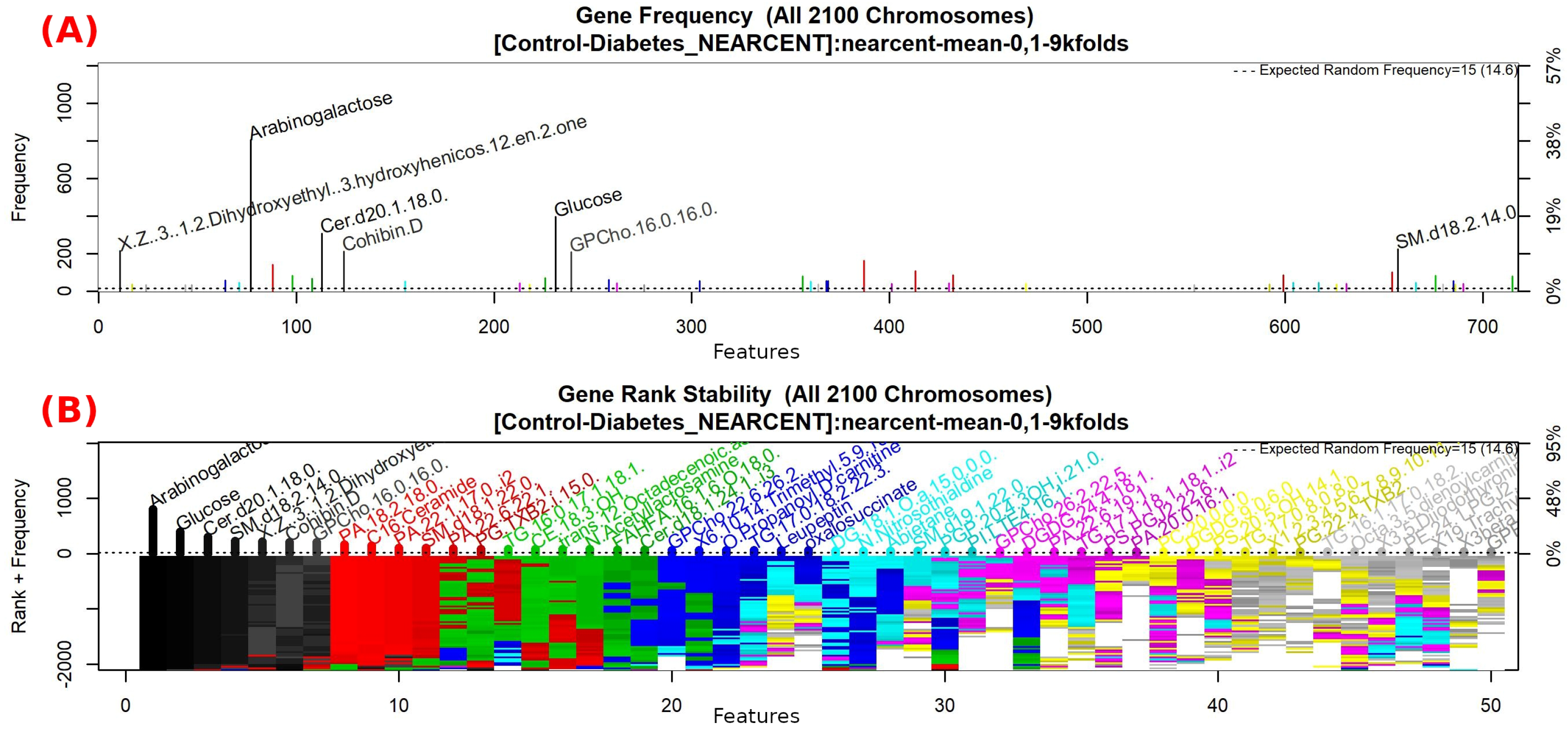
3.1. Galgo Results
3.1.1. GALGO Implementation with the Control-Prediabetes Dataset
3.1.2. GALGO Implementation with the Control-T2DM Dataset
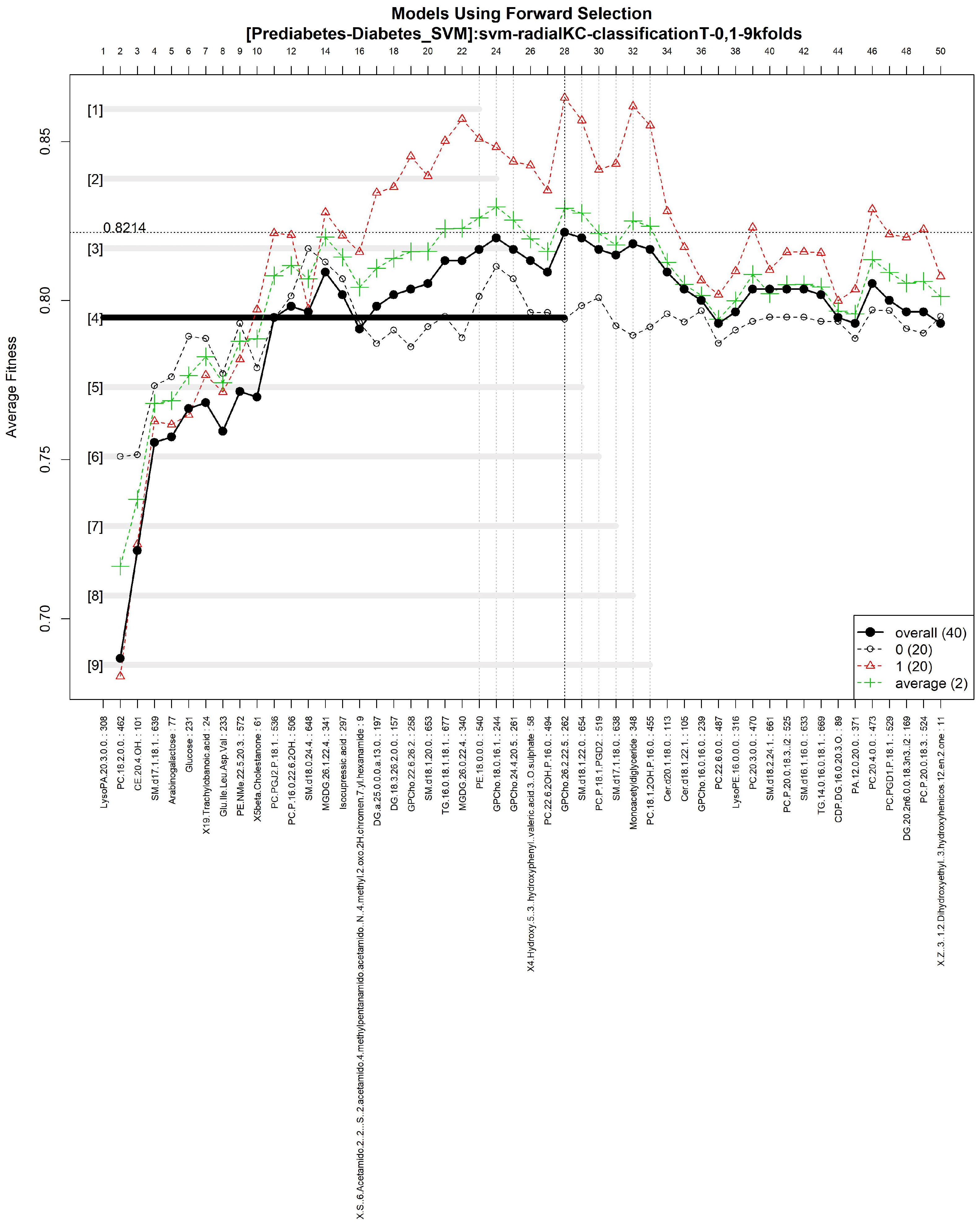
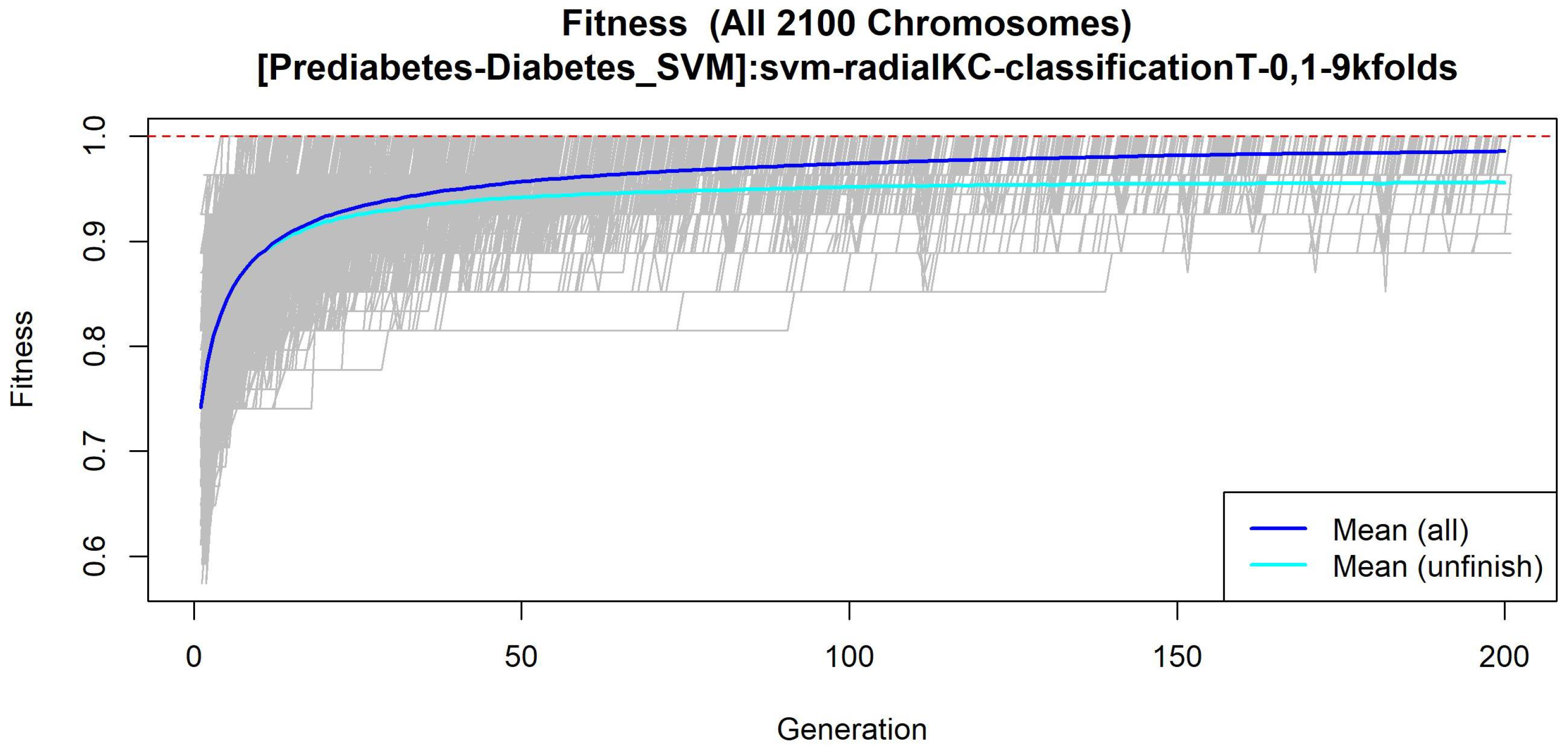
3.1.3. GALGO Implementation with the Prediabetes-T2DM Dataset
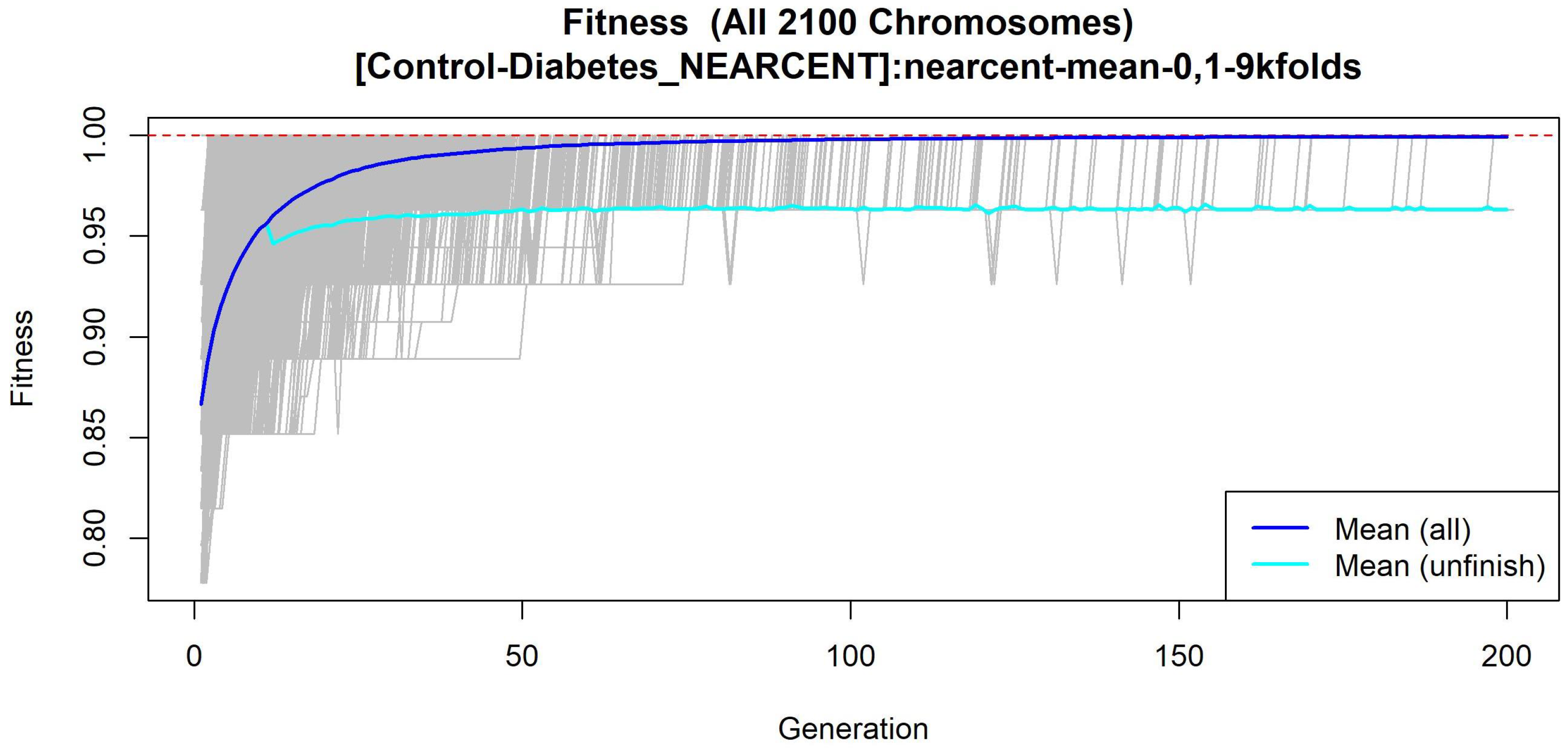
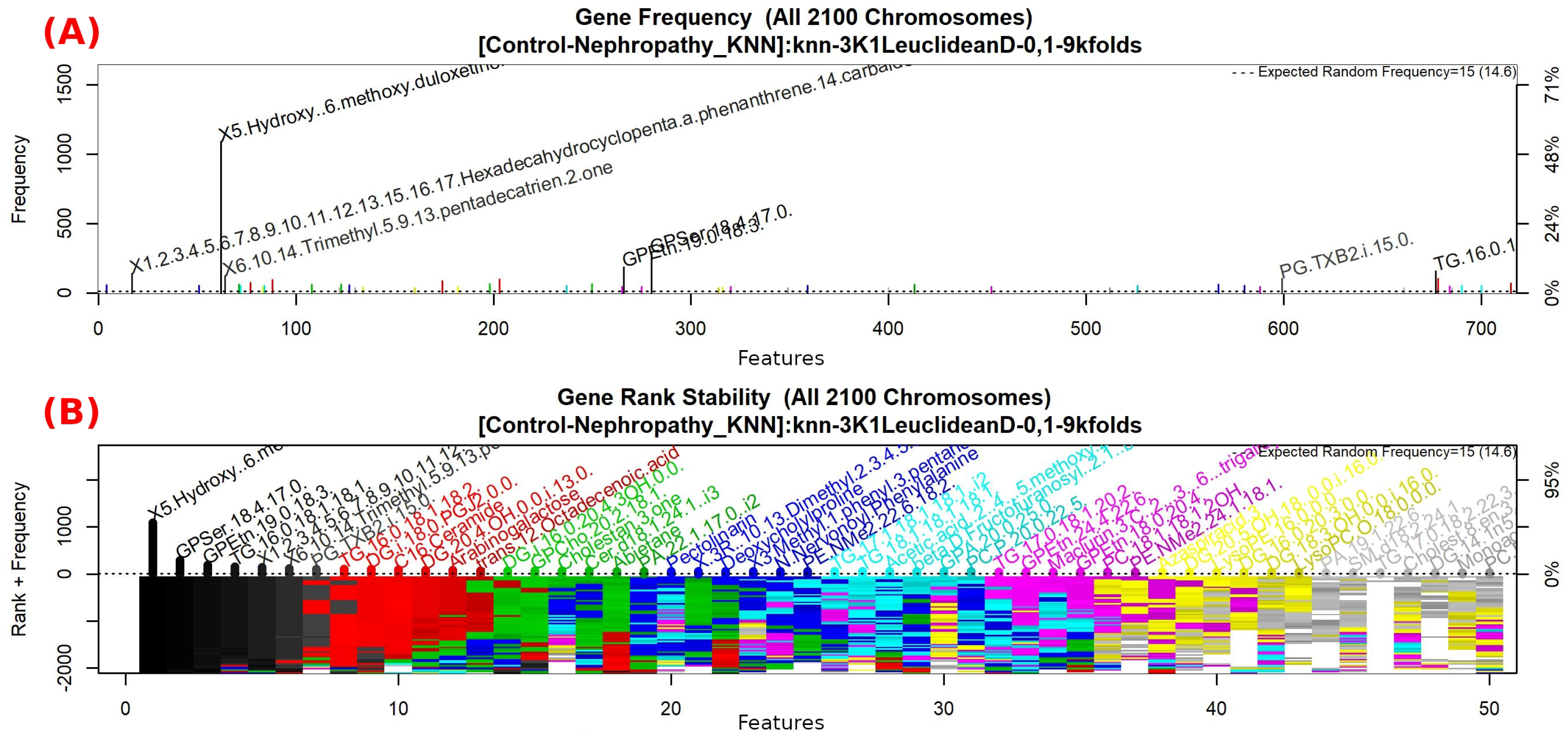
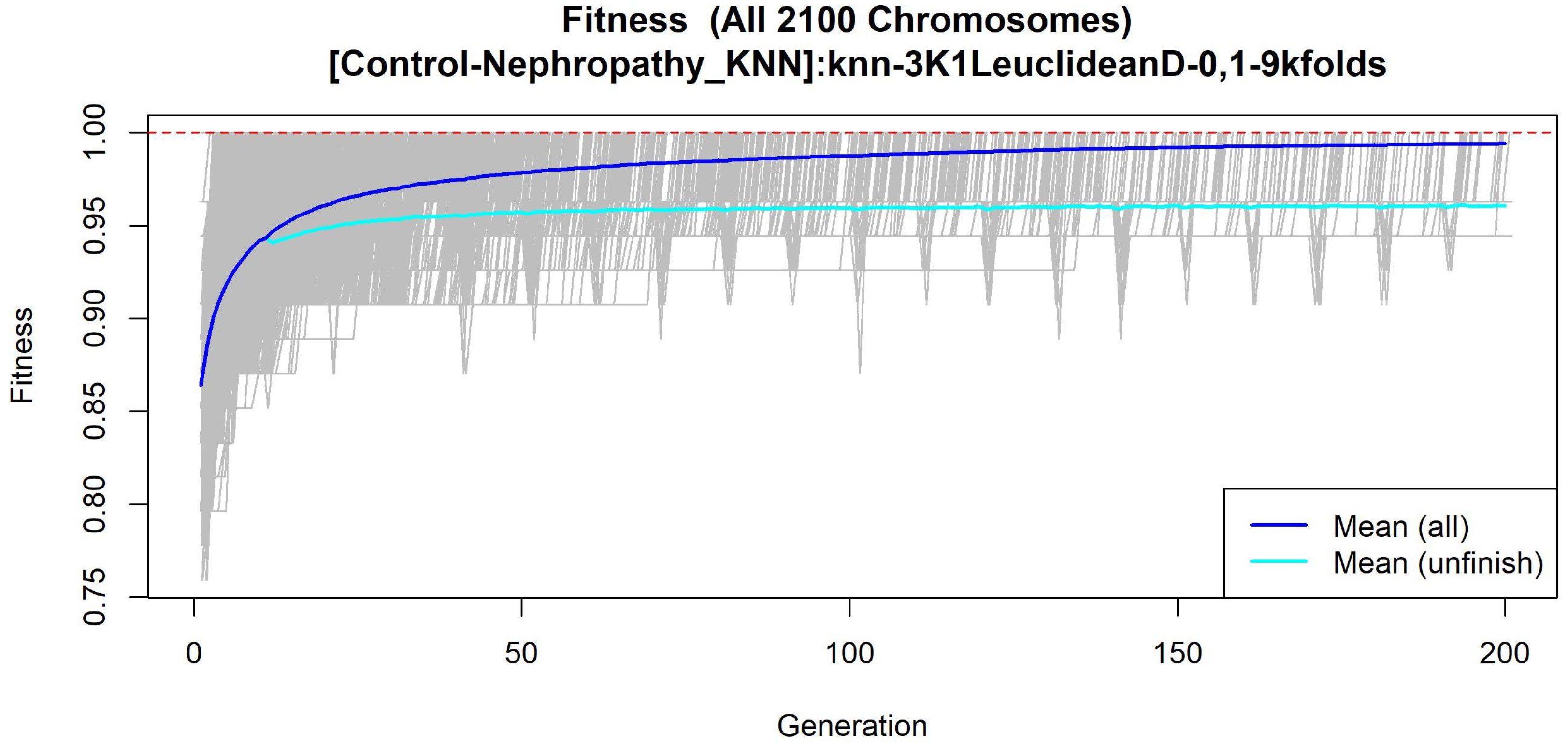
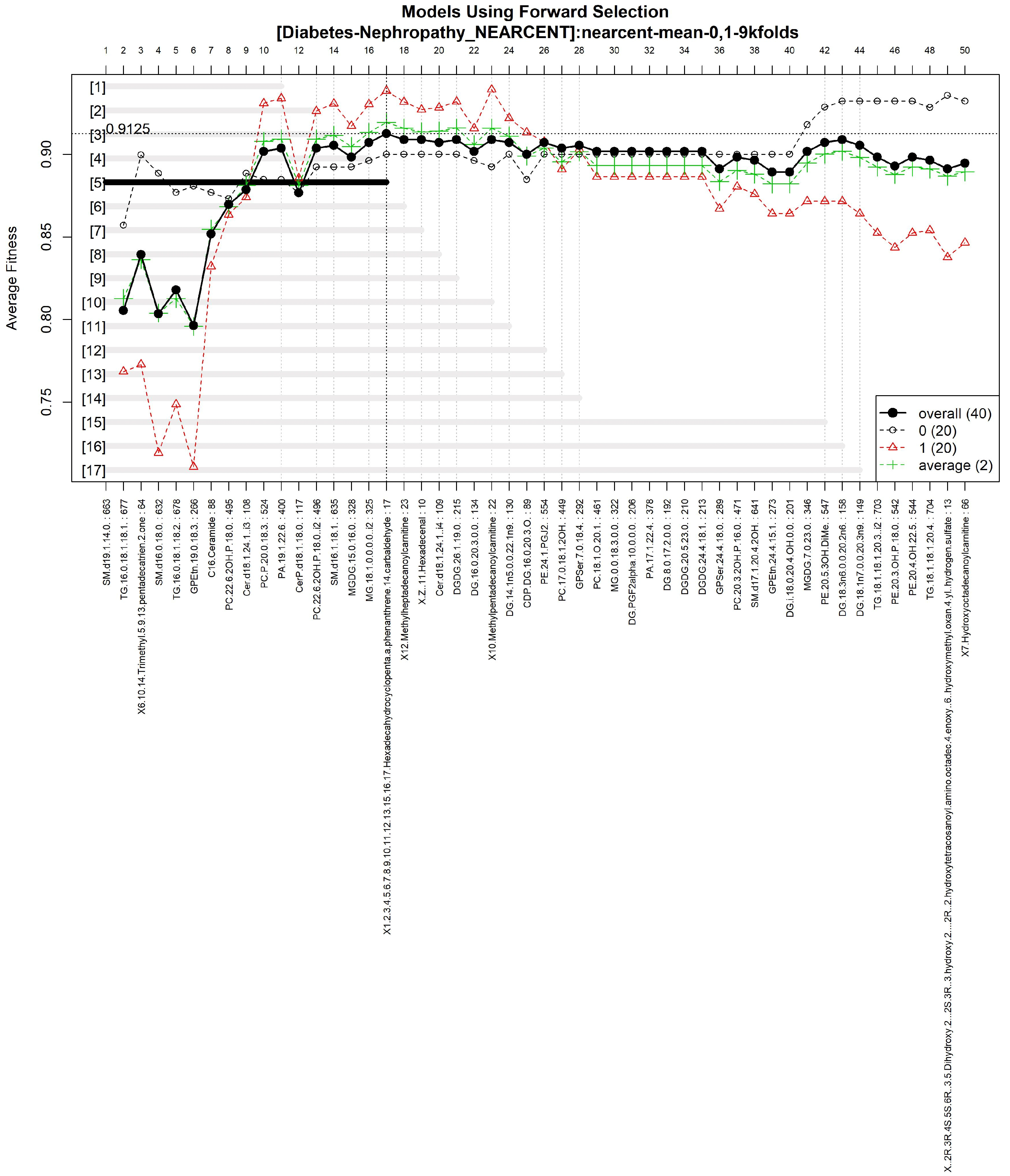
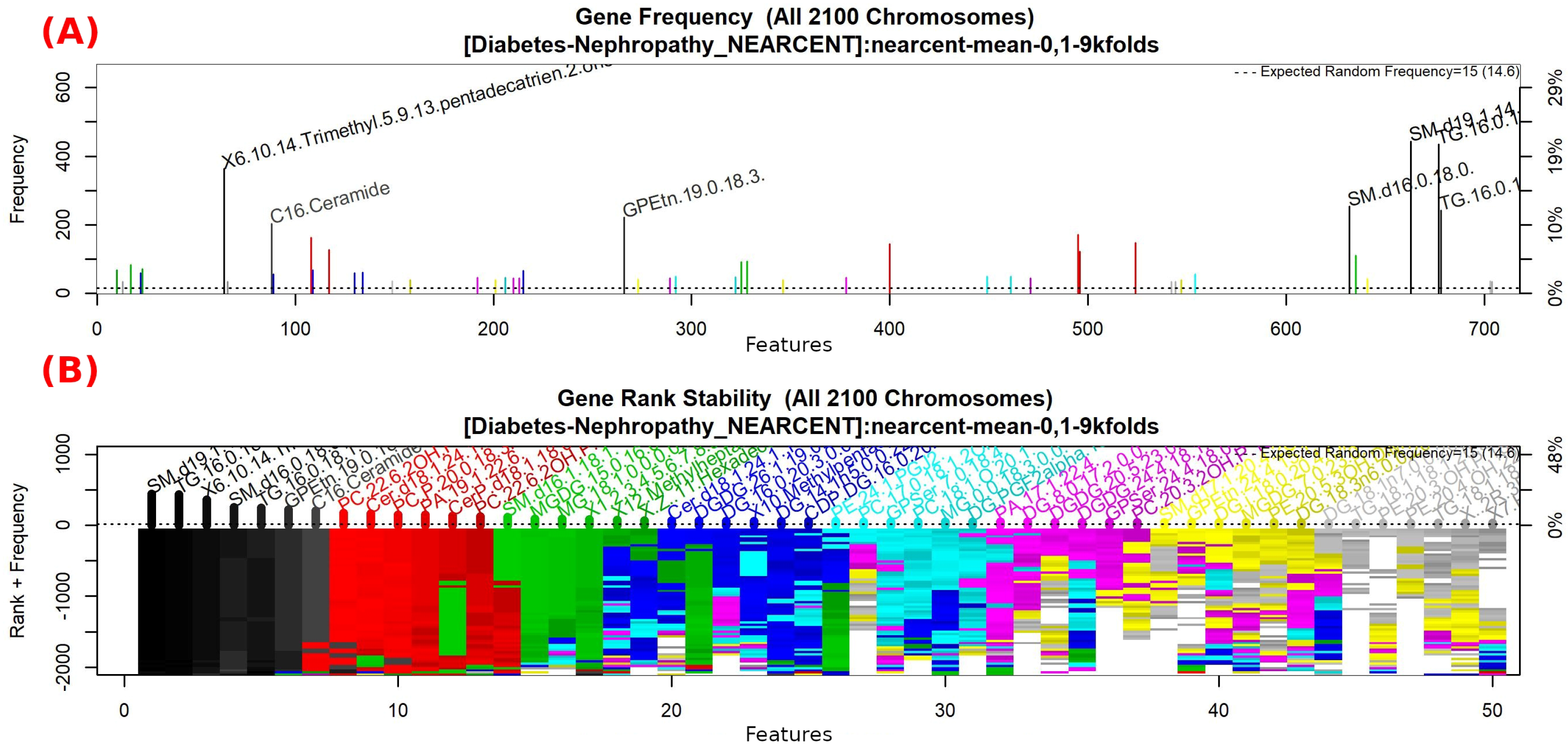
3.1.4. GALGO Implementation with the Control-DN Dataset
3.1.5. GALGO Implementation with the T2DM-DN Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- What Is Diabetes. Available online: https://www.idf.org/aboutdiabetes/what-is-diabetes.html (accessed on 2 October 2022).
- IDF Diabetes Atlas. Tenth Edition. Available online: https://diabetesatlas.org/#:%7E:text=Diabetes%20around%20the%20world%20in%202021%3A&text=Over%203%20in%204%20adults,over%20the%20last%2015%20years (accessed on 2 October 2022).
- Chatterjee, S.; Khunti, K.; Davies, M. Type 2 diabetes. Lancet 2017, 389, 2239–2251. [Google Scholar] [CrossRef]
- International Diabetes Federation. Diabetes Facts & Figures. Available online: https://idf.org/aboutdiabetes/what-is-diabetes/facts-figures.html (accessed on 1 October 2022).
- Schena, F.; Gesualdo, L. Pathogenetic Mechanisms of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2005, 16, S30–S33. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Ma, R. Metabolomics in Diabetes and Diabetic Complications: Insights from Epidemiological Studies. Cells 2021, 10, 2832. [Google Scholar] [CrossRef]
- Drupad, K.; TrivediKatherine, A.; Royston, G. Metabolomics for the masses: The future of metabolomics in a personalized world. New Horiz. Transl. Med. 2017, 3, 294–305. [Google Scholar] [CrossRef]
- Pagnini, C.; Di Paolo, M.C.; Mariani, B.M.; Urgesi, R.; Pallotta, L.; Vitale, M.A.; Villotti, G.; d’Alba, L.; De Cesare, M.A.; Di Giulio, E.; et al. Mayo Endoscopic Score and Ulcerative Colitis Endoscopic Index Are Equally Effective for Endoscopic Activity Evaluation in Ulcerative Colitis Patients in a Real Life Setting. Gastroenterol. Insights 2021, 12, 217–224. [Google Scholar] [CrossRef]
- Li, L.; Krznar, P.; Erban, A.; Agazzi, A.; Martin-Levilain, J.; Supale, S.; Kopka, J.; Zamboni, N.; Maechlerr, P. Metabolomics Identifies a Biomarker Revealing In Vivo Loss of Functional β-Cell Mass Before Diabetes Onset. Diabetes 2019, 68, 2272–2286. [Google Scholar] [CrossRef] [PubMed]
- Diamanti, K.; Cavalli, M.; Pan, G.; Pereira, M.; Kumar, C.; Skrtic, S.; Grabherr, M.; Risérus, U.; Eriksson, J.; Komorowski, J.; et al. Intra- and inter-individual metabolic profiling highlights carnitine and lysophosphatidylcholine pathways as key molecular defects in type 2 diabetes. Sci. Rep. 2019, 9, 9653. [Google Scholar] [CrossRef]
- Park, S.; Sadanala, K.; Kim, E. A Metabolomic Approach to Understanding the Metabolic Link between Obesity and Diabetes. Mol. Cells 2015, 38, 587–596. [Google Scholar] [CrossRef]
- Wishart, D.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef]
- Peddinti, G.; Cobb, J.; Yengo, L.; Froguel, P.; Kravić, J.; Balkau, B.; Tuomi, T.; Aittokallio, T.; Groop, L. Early metabolic markers identify potential targets for the prevention of type 2 diabetes. Diabetologia 2017, 60, 1740–1750. [Google Scholar] [CrossRef]
- Huang, J.; Huth, C.; Covic, M.; Troll, M.; Adam, J.; Zukunft, S.; Prehn, C.; Wang, L.; Nano, J.; Scheerer, M.F.; et al. Machine Learning Approaches Reveal Metabolic Signatures of Incident Chronic Kidney Disease in Individuals With Prediabetes and Type 2 Diabetes. Diabetes 2020, 69, 2756–2765. [Google Scholar] [CrossRef] [PubMed]
- Salihovic, S.; Broeckling, C.D.; Ganna, A.; Prenni, J.E.; Sundström, J.; Berne, C.; Lind, L.; Ingelsson, E.; Fall, T.; Ärnlöv, J.; et al. Non-targeted urine metabolomics and associations with prevalent and incident type 2 diabetes. Sci. Rep. 2020, 10, 16474. [Google Scholar] [CrossRef] [PubMed]
- Roointan, A.; Gheisari, Y.; Hudkins, K.L.; Gholaminejad, A. Non-invasive metabolic biomarkers for early diagnosis of diabetic nephropathy: Meta-analysis of profiling metabolomics studies. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 2253–2272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fuhrer, T.; Ye, H.; Kwan, B.; Montemayor, D.; Tumova, J.; Darshi, M.; Afshinnia, F.; Scialla, J.J.; Anderson, A.; et al. High-Throughput Metabolomics and Diabetic Kidney Disease Progression: Evidence from the Chronic Renal Insufficiency (CRIC) Study. Am. J. Nephrol. 2022, 53, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Dubin, R.F.; Rhee, E.P. Proteomics and Metabolomics in Kidney Disease, including Insights into Etiology, Treatment, and Prevention. Clin. J. Am. Soc. Nephrol. 2019, 15, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.K.; Mageswari, R.U.; Gayathri, A.; Dallfin Bruxella, J.M.; Ishak, M.K.; Mostafa, S.M.; Hamam, H. Barnacles Mating Optimizer with Deep Transfer Learning Enabled Biomedical Malaria Parasite Detection and Classification. Comput. Intell. Neurosci. 2022, 2022, 7776319. [Google Scholar] [CrossRef]
- Kaushik, H.; Singh, D.; Kaur, M.; Alshazly, H.; Zaguia, A.; Hamam, H. Diabetic Retinopathy Diagnosis From Fundus Images Using Stacked Generalization of Deep Models. IEEE Access 2021, 9, 108276–108292. [Google Scholar] [CrossRef]
- Mazhar, M.S.; Saleem, Y.; Almogren, A.; Arshad, J.; Jaffery, M.H.; Rehman, A.U.; Shafiq, M.; Hamam, H. Forensic Analysis on Internet of Things (IoT) Device Using Machine-to-Machine (M2M) Framework. Electronics 2022, 11, 1126. [Google Scholar] [CrossRef]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.K.; Shafi, J.; Choi, J.; Ijaz, M.F. Fine-Tuned DenseNet-169 for Breast Cancer Metastasis Prediction Using FastAI and 1-Cycle Policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Saleh, H.; Sahal, R.; Abuhmed, T.; Islam, S.R.; Ali, F.; Amer, E. Alzheimer’s disease progression detection model based on an early fusion of cost-effective multimodal data. Future Gener. Comput. Syst. 2021, 115, 680–699. [Google Scholar] [CrossRef]
- LIPID MAPS. Available online: https://www.lipidmaps.org/ (accessed on 8 November 2022).
- METLIN. Available online: https://metlin.scripps.edu/ (accessed on 8 November 2022).
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Trevino, V.; Falciani, F. GALGO: An R package for multivariate variable selection using genetic algorithms. Bioinformatics 2006, 22, 1154–1156. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Reyna, A.; Celaya-Padilla, J.; Galván-Tejada, C.; Luna-García, H.; Gamboa-Rosales, H.; Ramirez-Morales, A.; Galván-Tejada, J. Multimodal Early Alzheimer’s Detection, a Genetic Algorithm Approach with Support Vector Machines. Healthcare 2021, 9, 971. [Google Scholar] [CrossRef] [PubMed]
- Celaya-Padilla, J.M.; Villagrana-Bañuelos, K.E.; Oropeza-Valdez, J.J.; Monárrez-Espino, J.; Castañeda-Delgado, J.E.; Oostdam, A.S.H.-V.; Fernández-Ruiz, J.C.; Ochoa-González, F.; Borrego, J.C.; Enciso-Moreno, J.A.; et al. Kynurenine and Hemoglobin as Sex-Specific Variables in COVID-19 Patients: A Machine Learning and Genetic Algorithms Approach. Diagnostics 2021, 11, 2197. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Wu, Q.; Wang, Z.; Chen, G.; Lin, B. Diabetic Retinopathy Prediction by Ensemble Learning Based on Biochemical and Physical Data. Sensors 2021, 21, 3663. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L. K-nearest neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Levner, I. Feature selection and nearest centroid classification for protein mass spectrometry. BMC Bioinform. 2005, 6, 68. [Google Scholar] [CrossRef]
- Karaçalıa, B.; Ramanathb, R.; Snyder, W. A comparative analysis of structural risk minimization by support vector machines and nearest neighbor rule. Pattern Recognit. Lett. 2004, 25, 63–71. [Google Scholar] [CrossRef]
- Zhou, B.; Cheema, A.; Ressom, H. SVM-based spectral matching for metabolite identification. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 756–759. [Google Scholar] [CrossRef]
- Amari, S.; Wu, S. Improving support vector machine classifiers by modifying kernel functions. Neural Netw. 1999, 12, 783–789. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. Artic. 2008, 28, 1–26. [Google Scholar]
- Barberis, E.; Khoso, S.; Sica, A.; Falasca, M.; Gennari, A.; Dondero, F.; Afantitis, A.; Manfredi, M. Precision Medicine Approaches with Metabolomics and Artificial Intelligence. Int. J. Mol. Sci. 2022, 23, 11269. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Yoshioka, K.; Kojima, K.; Yamashita, Y.; Shibahara, T.; Wada, T.; Nangaku, M.; Inagi, R. Potential progression biomarkers of diabetic kidney disease determined using comprehensive machine learning analysis of non-targeted metabolomics. Sci. Rep. 2022, 12, 16287. [Google Scholar] [CrossRef] [PubMed]
- Frohnert, B.; Webb-Robertson, B.; Bramer, L.; Reehl, S.; Waugh, K.; Steck, A.; Norris, J.; Rewers, M. Predictive Modeling of Type 1 Diabetes Stages Using Disparate Data Sources. Diabetes 2019, 69, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Ang, L.; Ho, C.; Soh, S.; Tan, K.; Chan, J.; Godfrey, K.; Chan, S.; Chong, Y.; Eriksson, J.; et al. Machine Learning–Derived Prenatal Predictive Risk Model to Guide Intervention and Prevent the Progression of Gestational Diabetes Mellitus to Type 2 Diabetes: Prediction Model Development Study. JMIR Diabetes 2022, 7, e32366. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Data-Driven Machine-Learning Methods for Diabetes Risk Prediction. Sensors 2022, 22, 5304. [Google Scholar] [CrossRef]
- Li, J.; Guo, C.; Wang, T.; Xu, Y.; Peng, F.; Zhao, S.; Li, H.; Jin, D.; Xia, Z.; Che, M.; et al. Interpretable machine learning-derived nomogram model for early detection of diabetic retinopathy in type 2 diabetes mellitus: A widely targeted metabolomics study. Nutr. Diabetes 2022, 12, 36. [Google Scholar] [CrossRef]
- Wei, H.; Sun, J.; Shan, W.; Xiao, W.; Wang, B.; Ma, X.; Hu, W.; Wang, X.; Xia, Y. Environmental chemical exposure dynamics and machine learning-based prediction of diabetes mellitus. Sci. Total Environ. 2022, 806, 150674. [Google Scholar] [CrossRef]





| Inclusion Criteria |
|---|
| 1. The age of the patients must be over 18 years |
| 2. There will be no distinction in gender, education, ethnicity, race and marital status. |
| 3. The datasets should contain only the metabolomics of each subject. |
| 4. The dataset should distinguish controls from prediabetes, T2DM and DN. |
| 5. The data of each feature in each subject must be complete. |
| Time (min) | %A | %B | Curve |
|---|---|---|---|
| Initial | 60 | 40 | Initial |
| 2 | 57 | 43 | 6 |
| 2.1 | 50 | 50 | 1 |
| 12 | 46 | 54 | 6 |
| 12.1 | 30 | 70 | 1 |
| 18 | 1 | 99 | 6 |
| 18.1 | 60 | 40 | 6 |
| 20 | 60 | 40 | 1 |
| Column temperature 55 °C | |||
| Model | Parameter | Value |
|---|---|---|
| KNN | classification.method chromosomeSize maxSolutions maxGenerations goalFitness | knn 5 2100 200 1 |
| Nearest Centroid | classification.method chromosomeSize maxSolutions maxGenerations goalFitness | nearcent 5 2100 200 1 |
| SVM | classification.method svm.kernel chromosomeSize maxSolutions maxGenerations goalFitness | svm radial 5 2100 200 1 |
| Sub-Dataset | GALGO Model | ML Model |
|---|---|---|
| knn | K-Nearest Neighbours | |
| Control-Prediabetes | nearcent | Nearest Centroid |
| svm | Support Vector Machines | |
| knn | K-Nearest Neighbours | |
| Control-T2DM | nearcent | Nearest Centroid |
| svm | Support Vector Machines | |
| knn | K-Nearest Neighbours | |
| Prediabetes-T2DM | nearcent | Nearest Centroid |
| svm | Support Vector Machines | |
| knn | K-Nearest Neighbours | |
| Control-DN | nearcent | Nearest Centroid |
| svm | Support Vector Machines | |
| knn | K-Nearest Neighbours | |
| T2DM-DN | nearcent | Nearest Centroid |
| svm | Support Vector Machines |
| Sub-Dataset | GALGO Model | ML Model | Average Accuracy |
|---|---|---|---|
| knn | K-Nearest Neighbours | 0.8143 | |
| Control-Prediabetes | nearcent | Nearest Centroid | 0.8321 |
| svm | Support Vector Machines | 0.8464 | |
| knn | K-Nearest Neighbours | 0.9268 | |
| Control-T2DM | nearcent | Nearest Centroid | 0.9286 |
| svm | Support Vector Machines | 0.9036 | |
| knn | K-Nearest Neighbours | 0.7821 | |
| Prediabetes-T2DM | nearcent | Nearest Centroid | 0.7982 |
| svm | Support Vector Machines | 0.8214 | |
| knn | K-Nearest Neighbours | 0.9893 | |
| Control-DN | nearcent | Nearest Centroid | 0.9714 |
| svm | Support Vector Machines | 0.9857 | |
| knn | K-Nearest Neighbours | 0.9054 | |
| T2DM-DN | nearcent | Nearest Centroid | 0.9125 |
| svm | Support Vector Machines | 0.8804 |
| Result Features |
|---|
| “GPCho(20:2/18:1)”, “Cer(d18:1/24:1) i2”, “PA(i-19:0/20:3-2OH)”, “PC(20:3-OH/P-18:1)”, “TG(17:1/17:2/22:5)” |
| Result Features |
|---|
| “Androst-16-ene”, “Ganoderic acid C2”, “Cer(d18:2/20:4-3OH)”, “SM(d18:1/24:1) i2”, “(Z)-11-Hexadecenal” |
| Result Features |
|---|
| “lysine phosphoester”, “PC(18:1-O/20:1)”, “CE(20:4-2OH)”, “SM(d17:1/18:0)”, “Androst-16-ene”, “Ganoderic acid C2”, “12-Methylheptadecanoylcarnitine”, “Glufosinate”, “PE-NMe(22:0/18:1)”, “5alpha-Androsta-16-ene-3-ol”, “PC(PGJ2/DiMe)”, “PC(P-16:0/20:4-3OH)”, “SM(d18:0/18:1)”, “MGDG(26:0/22:4)”, “Isobehenic acid”, “(Melle-4)cyclosporin”, “DG(a-17:0/0:0/8:0) i2”, “DG(18:3/18:1/0:0)”, “GPCho(22:5/18:0)”, “SM(d18:1/18:1)”, “TG(16:0/17:1/18:1)”, “MGDG(24:1/18:1)”, “PE(15:0/18:4)”, “GPCho(16:1/16:1)”, “GPCho(24:1/22:6)”, “4-Hydroperoxycyclophosphamide”, “PC(22:6-2OH/24:0) i2”, “GPCho(24:4/20:5)” |
| Result Features |
|---|
| “5beta-Cholestanone”, “GPSer(18:1/11:0)”, “GPEtn(18:0/20:4)”, “TG(16:0/17:1/18:1)”, “1,1-Dimethylbiguanide”, “5-Pentahydroxy-5-cucurbiten-11-one 3-[glucosyl-(1->6)-glucoside]”, “PG(PGF1alpha/i-16:0)”, “TG(16:0/18:1/18:1)”, “DG(i-18:0/22:6-OH/0:0)”, “Butyl methacrylate”, “DG(20:4/16:0/0:0)”, “Androst-16-ene”, “Threonyltyrosine”, “DG(a-25:0/0:0/a-13:0)”, “GPCho(20:2/18:0)”, “Cholest-8-en-3-ol”, “Cer(d18:1/24:1) i2”, “9-Decenoylcarnitine”, “PA(22:1/17:0)”, “PE(TXB2/DiMe)”, “(2Z,4E,6Z)-Decatrienoylcarnitine”, “delta-24-Cholesterol”, “3-methoxy-4-hydroxy-5-all-trans-hexaprenylbenzoate”, “N-Methylethanolaminium phosphate” |
| Result Features |
|---|
| “SM(d19:0/20:3-OH)”, “TG(16:0/17:1/18:1)”, “5-Pentahydroxy-5-cucurbiten-11-one 3-[glucosyl-(1->6)-glucoside]”, “PS(PGJ2/22:6)”, “TG(16:0/18:1/18:1)”, “GPEtn(18:0/20:4)”, “Butyl methacrylate”, “PC(22:6-2OH/P-16:0)”, “Cer(d18:1/24:1) i2”, “PC(P-20:0/14:1)”, “PA(19:1/16:0)”, “CerP(d15:0/2:0)”, “PC(22:6-2OH/P-18:0)”, “SM(d16:1/18:0)”, “MG(20:4/0:0/0:0)”, “MG(18:1/0:0/0:0)”, “1,1-Dimethylbiguanide” |
| KNN and NEARCENT and SVM | KNN and NEARCENT | KNN and SVM | NEARCENT and SVM |
|---|---|---|---|
| “PC(20:3-OH/P-18:1)” | “TG(17:1/18:1/18:1)” | “GPCho(20:2/18:1)” | |
| “TG(17:1/17:2/22:5)” | “Glufosinate” | “Cer(d18:1/24:1) i2” | |
| “PC(18:1-O/20:1)” | |||
| “DGDG(20:5/14:0)” | |||
| “TG(18:0/18:1/20:2)” | |||
| “lysine phosphoester” | |||
| “TG(17:0/18:1/20:2)” |
| KNN and NEARCENT and SVM | KNN and NEARCENT | KNN and SVM | NEARCENT and SVM |
|---|---|---|---|
| “Androst-16-ene” | “Cholestan-3-one” | “(Z)-11-Hexadecenal” | |
| “Ganoderic acid C2” | “PA(18:1/18:2) i2” | ||
| “Cer(d18:2/20:4-3OH)” | “GPA(26:2/6:0)” | ||
| “SM(d18:1/24:1) i2” | “CE(18:2+=O)” |
| KNN and NEARCENT and SVM | KNN and NEARCENT | KNN and SVM | NEARCENT and SVM |
|---|---|---|---|
| “PC(20:3-OH/P-18:1)” | “Ganoderic acid C2” |
| KNN and NEARCENT and SVM | KNN and NEARCENT | KNN and SVM | NEARCENT and SVM |
|---|---|---|---|
| “5beta-Cholestanone” | “Cer(d18:1/24:1) i2” | “GPSer(18:1/11:0)” | |
| “DG(a-25:0/0:0/a-13:0)” | “PC(18:0/18:1-2OH)” | ||
| “PG(PGF1alpha/i-16:0)” | |||
| “DG(20:4/16:0/0:0)” | |||
| “N-Methylethanolaminium phosphate” | |||
| “GPEtn(18:0/20:4)” | |||
| “TG(16:0/17:1/18:1)” |
| KNN and NEARCENT and SVM | KNN and NEARCENT | KNN and SVM | NEARCENT and SVM |
|---|---|---|---|
| “Butyl.methacrylate” | “SM(d19:0/20:3-OH)” | ||
| “TG(16:0/17:1/18:1)” | “GPEtn(18:0/20:4)” | ||
| “Cer(d18:1/24:1) i2” | “PC(22:6-2OH/P-16:0)” | ||
| “TG(16:0/18:1/18:1)” | “CerP(d15:0/2:0)” | ||
| “1,1-Dimethylbiguanide” | “PC(22:6-2OH/P-18:0)” | ||
| “PS(PGJ2/22:6)” | “SM(d16:1/18:0)” | ||
| “5-Pentahydroxy-5-cucurbiten-11-one 3-[glucosyl-(1->6)-glucoside]” | “PA(19:1/16:0)” |
| Title | Feature Selection Technique | Validation Metric | Result |
|---|---|---|---|
| Machine Learning Approaches Reveal Metabolic Signatures of Incident Chronic Kidney Disease in Individuals With Prediabetes and Type 2 Diabetes [14] | LASSO | AUC | 0.857 |
| Potential progression biomarkers of diabetic kidney disease determined using comprehensive machine learning analysis of non-targeted metabolomics [38] | NON | AUC | 0.775 |
| Predictive Modeling of Type 1 Diabetes Stages Using Disparate Data Sources [39] | Repeated Optimization for Feature Interpretation | AUC | 0.91 |
| Machine Learning–Derived Prenatal Predictive Risk Model to Guide Intervention and Prevent the Progression of Gestational Diabetes Mellitus to Type 2 Diabetes: Prediction Model Development Study [40] | CatBoost tree ensembles | AUC | 0.86 |
| Data-Driven Machine-Learning Methods for Diabetes Risk Prediction [41] | Pearson Correlation, Gain Ratio, Naive Bayes and Random Forest | AUC | 0.942 |
| Interpretable machine learning-derived nomogram model for early detection of diabetic retinopathy in type 2 diabetes mellitus: a widely targeted metabolomics study [42] | Classification and Regression Tree | AUC | 0.95 |
| Environmental chemical exposure dynamics and machine learning-based prediction of diabetes mellitus [43] | Lasso | AUC | 0.78 |
| This Work in Control-Prediabetes | Genetic Algorithm with GALGO-svm | Accuracy | 0.8464 |
| This Work in Control-T2DM | Genetic Algorithm with GALGO-Nearcent | Accuracy | 0.9286 |
| This Work in Prediabetes-T2DM | Genetic Algorithm with GALGO-svm | Accuracy | 0.8214 |
| This Work in Control-DN | Genetic Algorithm with GALGO-knn | Accuracy | 0.9893 |
| This Work in T2DM-DN | Genetic Algorithm with GALGO-nearcent | Accuracy | 0.9125 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgan-Benita, J.; Sánchez-Reyna, A.G.; Espino-Salinas, C.H.; Oropeza-Valdez, J.J.; Luna-García, H.; Galván-Tejada, C.E.; Galván-Tejada, J.I.; Gamboa-Rosales, H.; Enciso-Moreno, J.A.; Celaya-Padilla, J. Metabolomic Selection in the Progression of Type 2 Diabetes Mellitus: A Genetic Algorithm Approach. Diagnostics 2022, 12, 2803. https://doi.org/10.3390/diagnostics12112803
Morgan-Benita J, Sánchez-Reyna AG, Espino-Salinas CH, Oropeza-Valdez JJ, Luna-García H, Galván-Tejada CE, Galván-Tejada JI, Gamboa-Rosales H, Enciso-Moreno JA, Celaya-Padilla J. Metabolomic Selection in the Progression of Type 2 Diabetes Mellitus: A Genetic Algorithm Approach. Diagnostics. 2022; 12(11):2803. https://doi.org/10.3390/diagnostics12112803
Chicago/Turabian StyleMorgan-Benita, Jorge, Ana G. Sánchez-Reyna, Carlos H. Espino-Salinas, Juan José Oropeza-Valdez, Huizilopoztli Luna-García, Carlos E. Galván-Tejada, Jorge I. Galván-Tejada, Hamurabi Gamboa-Rosales, Jose Antonio Enciso-Moreno, and José Celaya-Padilla. 2022. "Metabolomic Selection in the Progression of Type 2 Diabetes Mellitus: A Genetic Algorithm Approach" Diagnostics 12, no. 11: 2803. https://doi.org/10.3390/diagnostics12112803
APA StyleMorgan-Benita, J., Sánchez-Reyna, A. G., Espino-Salinas, C. H., Oropeza-Valdez, J. J., Luna-García, H., Galván-Tejada, C. E., Galván-Tejada, J. I., Gamboa-Rosales, H., Enciso-Moreno, J. A., & Celaya-Padilla, J. (2022). Metabolomic Selection in the Progression of Type 2 Diabetes Mellitus: A Genetic Algorithm Approach. Diagnostics, 12(11), 2803. https://doi.org/10.3390/diagnostics12112803










